Abstract
The effect of overexpression of the trehalose-6-phosphate (T6P) synthase gene (TPS1) on ethanol fermentation of Saccharomyces cerevisiae has been studied at 30 and 38°C. The activity of T6P synthase and the accumulation of trehalose during ethanol fermentation were significantly improved by overexpression of TPS1, and especially at 38°C. Ethanol produced by transformants with and without TPS1 gene overexpression at 38°C was approx. 60 and 37 g/l, respectively. The fermentation efficiency of transformants with TPS1 gene overexpression at 38°C was similar to that at 30°C. The critical growth temperature was increased from 36 to 42°C by TPS1 gene overexpression. These results indicated that overexpression of the TPS1 gene had a beneficial effect on the fermentation capacity of the title yeast strain at high temperatures.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Saccharomyces cerevisiae is used for industrial ethanol production. However, the temperature must be maintained at about 30°C which necessitates considerable cooling. On the other hand, the simultaneous saccharification and fermentation process, which has been employed by most major ethanol producers, requires higher temperatures to achieve efficient ethanol conversion. Hence, improvement of the thermotolerance of these yeast strains would be beneficial for low-cost bioethanol production. Physical and chemical mutagenesis (Rajoka et al. 2005), adaptation (Balakumar et al. 2001), protoplast fusion (Ezeronye et al. 2001) as well as gene shuffling (Shi et al. 2009) have been employed to increase the thermotolerance of yeast.
The relationship between thermotolerance and trehalose has been studied (Singer and Lindquist 1998), and trehalose is invariably accumulated after heat shock (Estruch 2000; Mahmud et al. 2010). However, studies on the effect of trehalose accumulation and the constitutive synthesis of trehalose on the thermotolerance of yeast during ethanol fermentation remain limited (Soto et al. 1999).
Trehalose-6-phosphate (T6P) synthase converts glucose 6-phosphate and UDP-glucose to trehalose 6-phosphate. Since the trehalose content and the activity of T6P synthase show a positive correlation (Cansado et al. 1998; Hottiger et al. 1987), in this study the influence of T6P synthase gene (TPS1) overexpression on the thermotolerance of the S. cerevisiae strain during ethanol fermentation has been investigated. It was envisaged that this might provide a much simpler means of improving the thermotolerance of industrial yeast for ethanol production. Guo et al. (2010) reported that by overexpressing TPS1 and TPS2 (encoding trehalose-6-phosphate phosphatase), the osmotic stress tolerance, growth rate, and ethanol fermentation ability of the yeast strain were improved; however, the effect on thermotolerance was not studied.
Materials and methods
Yeast strains
The S. cerevisiae strains used in this study are shown in Table 1. Strain 10151 were used as host strains for transformation and strains EP-1 and 14801 were used for preparing chromosomal DNAs.
Construction of pI-RED1-TPS1 plasmid with TPS1 overexpression cassette
Figure 1 shows a flow chart for the construction of pI-RED1-TPS1 plasmid with the TPS1 overexpression cassette. The TPS1 gene was amplified from the chromosomal DNA of strain EP-1 by PCR using the primers TPS1F and TPS1R (Supplementary Table 1). KOD-Plus polymerase (Toyobo, Osaka, Japan) was used according to the manufacturer’s instructions. The constructed plasmid was designated pI-RED1-TPS1 and used for transformations.
Construction of plasmid pI-RED1-TPS1 with TPS1 high-expression cassette. The amplified TPS1 fragment was digested with KpnI and XbaI and inserted into plasmid pAUR123, which had ADH1 promoter and terminator, and had been linearized with KpnI and XbaI. The constructed plasmid was designated pAUR123-TPS1. PADH1-TPS1-TADH1 fragment was obtained by digesting pAUR123-TPS1 with SacI and EcoT22I and inserted into SacI and EcoT22I-digested plasmid pI-RED1 (URA3, YAP1)
Construction of a DNA fragment with the TPS1 overexpression cassette by fusion PCR
Fusion PCR was used to amplify the TPS1 overexpression cassette with URA3 as the marker (Fig. 2). The PURA3-URA3-PADH1-TPS1-TADH1 fragment and the downstream fragment of the URA3 gene were amplified respectively by PCR1 and PCR2. The final fusion fragment (4.6 kb) was amplified by PCR3 and then used for transformations. An approximately 500 bp fragment of PURA3-URA3 containing the 5′-part of the URA3 gene sequence was amplified from strain 14801 for construction of the URA3-recovered strain of 10151.
Preparation of DNA fragment with TPS1 high-expression cassette for homology transformation. PCR1 was carried out using primers URA3Up-F and URA3Down-PADH1-R (Supplementary Table 1) to amplify the PURA3-URA3-PADH1-TPS1-TADH1 fragment (3.6 kb) with the plasmid pI-RED1-TPS1 as template. PCR2 was carried out using primers URA3Down-F and URA3Down-R to amplify the downstream fragment (1 kb) of the URA3 gene with the chromosomal DNA of strain 10151 as the template. The PCR products from PCR1 and PCR2 were purified using Ultraclean PCR clean-up kit (MO BIO Laboratories, Inc., Carlsbad, CA). Primers URA3Up-F and URA3Down-R were used to amplify a 4.6 kb fusion fragment by mixing the PCR1 and PCR2 products as templates
Transformation of plasmid and DNA fragment to the host strain
Yeast transformation was performed by the lithium acetate method (Gietz et al. 2002). Plasmids pI-RED1-TPS1 and pI-RED1 were linearized with Aor51HI (one digestion site in the YAP1 gene) and transformed to the YAP1 gene region by homology transformation in the host strain 10151 to produce TPS1 overexpression strain 10151-pI-RED1-TPS1 and URA3-recovered strain 10151-pI-RED1. Fragments PURA3-URA3-PADH1-TPS1-TADH1-URA3downstream and PURA3-URA3 were transformed to URA3 gene region by homology transformation (Fig. 2) in the host strain 10151 to produce TPS1 overexpression strain 10151-URA3-TPS1 and URA3-recovered strain 10151-URA3. Transformants 10151-pI-RED1 and 10151-URA3 were used to eliminate the possible effect of the URA3 gene on the evaluation of overexpression of the TPS1 gene. The transformants were selected using URA3 as the selection marker on an MM plate (1.7 g YNB/l, 5 g (NH4)2SO4/l, 20 g glucose/l, 20 g agar/l) with the addition of 40 mg adenine/l, 40 mg tryptophan/l and 40 mg histidine/l. Chromosomal DNA of the host strain and all transformants was extracted by using a Gen Toru Kun kit (Takara Shuzo Co., Ltd., Shiga, Japan) and used as a DNA template for PCR check. Primers A123MCS-F and A123MCS-R amplifying fragments between PADH1 and TADH1 were used to check the introduced TPS1 gene in transformant 10151-pI-RED1-TPS1. Primers URA3Up-F and URA3Down-R were used to check the introduced TPS1 gene in transformant 10151-URA3-TPS1.
Ethanol fermentation of host strain and transformants at 30 and 38°C
Ethanol fermentation of host and transformants were carried out at 30 and 38°C using 5% YPD as precultivation medium and 15% YPD as fermenting medium. The fermentation was performed for 48 h and the ethanol concentration, cell number, T6P synthase activity and trehalose concentration were analyzed to evaluate the influence of the overexpression of TPS1 gene.
Growth of transformants at 36–42°C
Growth of strains 10151-URA3 and 10151-URA3-TPS1 was compared at 36, 38, 40, and 42°C under aerobic conditions using a TVS062CA biophotorecorder (Advantec, Tokyo, Japan) with 2% YPD medium. Growth was started with an initial cell number of 1 × 106 cells/ml, which was approximately 1/10 of that used in the ethanol fermentation experiments.
Analytical methods
Ethanol in the supernatant of centrifuged broth (2,500×g at 4°C for 10 min) was assayed by GC with 2-propanol as internal standard (Tang et al. 2006). The total number of cells and the number of viable cells were calculated by the Methylene Blue staining method using a hemocytometer. The activity of T6P synthase and the concentration of trehalose were determined as described previously (Hottiger et al. 1987). One unit of T6P synthase was determined as that responsible for the synthesis of 1 μmol trehalose 6-phosphate in 1 min at 35°C. HPLC equipped with a Shodex Sugar SC1821 column and an RI detector was used for trehalose determination.
Results and discussion
Plasmid and DNA fragment construction and transformation
Plasmid pI-RED1-TPS1 was constructed according to Fig. 1 and checked by restriction endonuclease digestion (data not shown). PURA3-URA3-PADH1-TPS1-TADH1-URA3 downstream fragment (4.6 kb) was obtained by fusion PCR as described in Fig. 2. Transformants 10151-pI-RED1-TPS1, 10151-pI-RED1, 10151-URA3-TPS1, and 10151-URA3 were successfully obtained by transforming plasmid pI-RED1-TPS1, plasmid pI-RED1, DNA fragment PURA3-URA3-PADH1-TPS1-TADH1-URA3 downstream, and DNA fragment PURA3-URA3, respectively, with the host strain 10151. A PCR check was performed to confirm the successful introduction (data not shown).
Comparison of the ethanol fermentation capacities of the host strain and transformants
The host strain 10151 and four transformants each with three strains were grown at 30 and 38°C to evaluate the effect of high expression of the TPS1 gene on thermotolerance during ethanol fermentation. Since three strains of each type of transformant had the same performance, only the data of one strain of each transformant are shown in Fig. 3.
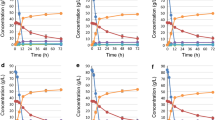
Comparison of the activity of trehalose-6-phosphate synthase (a), the concentration of trehalose (b), the concentration of ethanol (c), and the total cell number and the ratio of active cells (d) of host strain 10151 (circles), URA3-recovered strains 10151-pI-RED1 (squares) and 10151-URA3 (stars), and TPS1 overexpression strains 10151-pI-RED1-TPS1 (triangles) and 10151-URA3-TPS1 (diamonds), during ethanol fermentation at 30 and 38°C using 15% YPD medium. Cells grown on a 2% YPD plate were transferred to 100 ml of a pre-cultivation medium (5% YPD) in a 500 ml flask. Pre-cultivation was performed aerobically at 30°C for 16 h with shaking at 160 rpm using a rotary shaker. A 10 ml inoculum of pre-cultivation broth was added to 90 ml of fermentation medium (15% YPD) in a 200 ml flask. Flasks were immersed in water baths thermostatted at 30 or 38°C and the cultivation media were stirred using magnetic stirrers. Aliquots of approximately 10 ml of the broth were sampled for analysis. All parameters were determined in duplicate. The absolute difference between duplicate measurements of ethanol concentration was within 1% and the absolute differences between duplicate measurements of cell number, trehalose-6-phosphate synthase activity, and trehalose concentration were within 3%. Data are means of two replications
As shown in Fig. 3a and b, at both 30 and 38°C, high activity of trehalose-6P-synthase resulted in a high concentration of trehalose, which is consistent with the results of previous studies (Cansado et al. 1998; Hottiger et al. 1987). No obvious difference was found in the activity of T6P synthase and the trehalose concentration for strains 10151, 10151-URA3, and 10151-pI-RED1, indicating that the expression of URA3 and YAP1 genes did not affect the synthesis of trehalose. The similar results for 10151-pI-RED1-TPS1 and 10151-URA3-TPS1 also suggested that high expression of the YAP1 gene did not make a significant contribution to trehalose synthesis. The activity of T6P synthase and trehalose concentration at 38°C were higher than those at 30°C for all strains. Higher temperature induced higher expression of the TPS1 gene, as reported previously (Mahmud et al. 2010; Bell et al. 1992). However, at both 30 and 38°C, the TPS1 overexpression strains, 10151-pI-RED1-TPS1 and 10151-URA3-TPS1, had higher activity of T6P synthase and accumulation of trehalose after 18 and 24 h than the other three strains without TPS1 overexpression. The constitutive expression of the TPS1 gene contributed to the difference. The differences between the TPS1 overexpression strains and other strains were especially notable at 38°C. The concentration of trehalose in strains 10151-pI-RED1-TPS1 and 10151-URA3-TPS1 was approx. 350 μg/108 cells after 18 h and 230 μg/108 cells at 24 h, compared to 160 μg/108 cells after 18 h and 72 μg/108 cells after 24 h for the three strains without TPS1 overexpression.
At 30°C, both TPS1 overexpression strains, 10151-pI-RED1-TPS1 and 10151-URA3-TPS1, completed their fermentation after 18 h and the ethanol with both strains reached 60 g/l (Fig. 3c). However, the fermentation rates for the other three strains without TPS1 overexpression were slower and the fermentation continued up to 48 h, whereupon ethanol was approx. 57 g/l. These results suggest that the ethanol fermentation rate was improved by overexpression of the TPS1 gene. Compared to the results at 30°C, the differences among the strains were more significant at 38°C. The fermentation with both TPS1 overexpression strains, 10151-pI-RED1-TPS1 and 10151-URA3-TPS1, was complete after 24 h and ethanol was 60 g/l, which was the same as that at 30°C. However, the fermentation rates and fermentation efficiencies of the other strains without TPS1 overexpression were decreased compared to those at 30°C. Strain 10151 ceased fermentation after 24 h and ethanol was only 27 g/l. The other two strains with URA3 recovered continued their fermentation up to 48 h giving ethanol at approx. 38 g/l. The results indicated that TPS1 overexpression significantly improved the ethanol fermentation rate and ethanol fermentation efficiency of strain 10151, and essentially similar results were obtained at 30 and 38°C. URA3 recovery also contributed to the improvement to some extent; however, it was almost negligible compared to the effect of TPS1 overexpression.
Differences in total cell numbers were not apparent for all of the strains at 30°C, though the ratios of viable cells for strains 10151-pI-RED1-TPS1 and 10151-URA3-TPS1 were a little higher than those for the other three strains after 18 and 24 h, whereupon the fermentation was in fact complete (Fig. 3d). However, the differences were notable at 38°C. The total cell number and the ratio of viable cells for both TPS1 overexpression strains were higher than those for the other three strains during the whole fermentation period. The ratio of viable cells for both TPS1 overexpression strains remained over 85% up to 24 h, which therefore led to much higher fermentation rates and efficiencies, as indicated by the production of ethanol.
Based on the above results, TPS1 overexpression increased the activity of T6P synthase and the synthesis of trehalose, which led to a higher cell concentration with a higher viable cell ratio. The ethanol fermentation rate and the fermentation efficiency at 38°C were hence significantly improved.
Comparison of growth of transformants with and without TPS1 overexpression under high-temperature conditions
As shown in Fig. 4, strain 10151-URA3 did not grow at 38, 40 or 42°C. However, strain 10151-URA3-TPS1 grew even at 42°C though the growth rate decreased incrementally with increasing temperature. Overexpression of the TPS1 gene increased the temperature tolerance of strain 10151 from 36 to at least 42°C. This suggested that the improvement of growth contributed remarkably to the improvement of ethanol fermentation at high temperatures.
Growth of transformants 10151-URA3 (a) and 10151-URA3-TPS1 (b) in 2% YPD medium supplemented with adenine at 36°C (circles), 38°C (squares), 40°C (triangles) and 42°C (stars). 2% YPD supplemented with 40 mg adenine/l was used as growth medium. Cells pre-cultivated at 30°C using 2% YPD supplemented with uracil and adenine were used for inoculation. 100 μl of diluted pre-cultured broth containing 5 × 106 cells/ml was inoculated into 5 ml aliquots of growth medium in L-type tubes. The cultivation was performed at 50 rpm at different temperatures for 24 h
In conclusion, strains with overexpression of the TPS1 gene have been constructed by introducing a PADH1-TPS1-TADH1 cassette into the chromosome of the host strain 10151. Under ethanol fermentation conditions, the activity of T6P synthase and the accumulation of trehalose were significantly improved by overexpression of the TPS1 gene. The ethanol fermentation performance of transformants with overexpression of the TPS1 gene at 38°C was similar to that at 30°C, indicating that TPS1 gene overexpression had a remarkable effect in improving the fermentation capacity of the title yeast strain at high temperatures. Growth improvement is considered to be the crucial factor for the improvement of ethanol fermentation, as the critical temperature of growth was increased from 36 to 42°C by TPS1 gene overexpression. However, further study is needed to investigate whether this improvement is reproducible on industrial yeast strains.
References
Balakumar S, Arasaratnam V, Balasubramaniam K (2001) Isolation and improvement of a thermotolerant Saccharomyces cerevisiae strain. World J Microbiol Biotechnol 17:739–746
Bell W, Klaassen P, Ohnacker M, Boller T, Herweijer M, Schoppink P, Van der Zee P, Wiemken A (1992) Characterization of the 56-kDa subunit of yeast trehalose-6-phosphate synthase and cloning of its gene reveal its identity with the product of CIF1, a regulator of carbon catabolite inactivation. Eur J Biochem 209:951–959
Cansado J, Vicente-Soler J, Soto T, Fernandez J, Gacto M (1998) Trehalose-6P synthase is essential for trehalase activation triggered by glucose, nitrogen source or heat shock, but not by osmostress, in Schizosaccharomyces pombe. Biochim Biophys Acta 1381:271–278
Estruch F (2000) Stress-controlled transcription factors, stress-induced genes and stress tolerance in budding yeast. FEMS Microbiol Rev 24:469–486
Ezeronye OU, Okerentugba PO (2001) Optimum conditions for yeast protoplast release and regeneration in Saccharomyces cerevisiae and Candida tropicalis using gut enzymes of the giant African snail Achatina achatina. Lett Appl Microbiol 32:190–193
Gietz RD, Woods RA (2002) Transformation of yeast by lithium acetate single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol 350:87–96
Guo ZP, Zhang L, Ding ZY, Shi GY (2010) Minimization of glycerol synthesis in industrial ethanol yeast without influencing its fermentation performance. Metab Eng. doi:10.1016/j.ymben.2010.11.003
Hottiger T, Schmutz P, Wiemken A (1987) Heat-induced accumulation and futile cycling of trehalose in Saccharomyces cerevisiae. J Bacteriol 169:5518–5522
Mahmud SA, Hirasawa T, Shimizu H (2010) Differential importance of trehalose in Saccharomyces cerevisiae in response to various environmental stresses. J Biosci Bioeng 109:262–266
Rajoka MI, Ferhan M, Khalid AM (2005) Kinetics and thermodynamics of ethanol production by a thermotolerant mutant of Saccharomyces cerevisiae in a microprocessor- controlled bioreactor. Lett Appl Microbiol 40:316–321
Shi DJ, Wang CL, Wang KM (2009) Genome shuffling to improve thermotolerance, ethanol tolerance and ethanol productivity of Saccharomyces cerevisiae. J Ind Microbiol Biotechnol 36:139–147
Singer MA, Lindquist S (1998) Thermotolerance in Saccharomyces cerevisiae: the Yin and Yang of trehalose. Trends Biotechnol 16:460–468
Soto T, Fernández J, Vicente-soler J, Cansado J, Gacto M (1999) Accumulation of trehalose by overexpression of tps1, coding for trehalose-6-phosphate synthase, causes increased resistance to multiple stress in the fission yeast Schizosaccharomyces pombe. Appl Environ Microbiol 65:2020–2024
Tang YQ, An MZ, Liu K, Nagai S, Shigematsu T, Morimura S, Kida K (2006) Ethanol production from acid hydrolysate of wood biomass using the flocculating yeast Saccharomyces cerevisiae strain KF-7. Proc Biochem 41:909–914
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
An, MZ., Tang, YQ., Mitsumasu, K. et al. Enhanced thermotolerance for ethanol fermentation of Saccharomyces cerevisiae strain by overexpression of the gene coding for trehalose-6-phosphate synthase. Biotechnol Lett 33, 1367–1374 (2011). https://doi.org/10.1007/s10529-011-0576-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-011-0576-x