Abstract
A FAD-dependent glucose dehydrogenase (FADGDH) mutant with narrow substrate specificity was constructed by site-directed mutagenesis. Several characteristics of FADGDH, such as high catalytic activity and high electron transfer ability, make this enzyme suitable for application to glucose sensors. However, for further applications, improvement of the broad substrate specificity is needed. In this paper, we mutated two residues, Asn475 and Ala472, which are located near the putative active site of the catalytic subunit of FADGDH and have been predicted from the alignment with the active site of glucose oxidase. Of the 38 mutants constructed, Ala472Phe and Asn475Asp were purified and their activities were analyzed. Both mutants showed a higher specificity toward glucose compared to the wild type enzyme.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The self-monitoring of blood glucose (SMBG) is an important component of the modern therapy for diabetes mellitus. The currently available glucose monitoring devices are based on enzymatic reactions, combined with either electrochemical detection or color-developing reactions. The current progress in electronics and sensor fabrication technology resulted in drastic decrease in both time and the blood sample volume required for the measurement. Together with the introduction of alternate-site testing, diabetics are being relieved of the pain and troublesome procedure of SMBG.
However, there are still a limited number of enzymes suitable for enzymatic glucose sensing systems; two such enzymes are glucose oxidase (GOD) and pyrroloquinoline quinone glucose dehydrogenase (PQQGDH). The present authors have been engaged in the screening and engineering of enzymes suitable for glucose sensor development and we have previously reported on the screening of thermostable cofactor-binding glucose dehydrogenase from a soil bacterium, Burkhordelia cepacia (Sode et al. 1996). This GDH possesses FAD in its catalytic subunit. The catalytic subunit alone, FADGDH, shows dye-mediated GDH activity with pronounced thermal stability. But this enzyme is originally an oligomeric enzyme, a FADGDH complex is composed of a catalytic subunit harboring FAD in its redox center (the α subunit), an electron transfer subunit, a covalently attached multi-heme harboring cytochrome c (the β subunit), and a small subunit with unknown function (the γ subunit). This enzyme shows high thermostability and electron transfer ability. Recently, we have also reported on the cloning of these genes and the functional expression of FADGDH in Escherichia coli (Inose et al. 2003; Tsuya et al. 2006).
We recently reported on the development of a screen-printed, carbon electrode-based, disposable glucose sensor strip, using both FADGDH catalytic subunit and FADGDH complex (Yamaoka and Sode 2007a, b). Thanks to the high catalytic efficiency and electron transfer ability, whole-blood glucose can be measured within 1 s using this enzyme sensor and a 150 nl whole-blood sample. Moreover, the sensor reading was stable for more than 60 days even at 70°C.
However, the substrate specificity of the FADGDH is broad and this enzyme catalyzes the oxidaton of monosaccarides and dissacarides such as maltose into corresponding lactones. Therefore, narrowing of the substrate specificity of FADGDH toward glucose is one of the properties to be improved for further application for glucose sensor development.
In this paper, we present the construction of an engineered FADGDH with narrower substrate specificity toward glucose compared to the wild type enzyme. Although the 3D structure of this enzyme is not yet available, we predicted the region where the active site of this enzyme localized based on the primary structure alignment with fungi derived glucose oxidases (GOXs) (Wohlfahrt et al. 1999). By focusing on two residues where we observe the differences with GOXs active sites, we have carried out maximum mutagenesis and characterized the variants. We have succeeded in the construction of engineered FADGDH catalytic subunit showing very high specificity toward glucose.
Materials and methods
Site-directed mutagenesis
The expression vector for FADGDH pTrcγαβ and pBBJMccm vector which expresses the Escherichia coli ccmABCDEFGH genes essential for the maturation of cytochrome c, was previously constructed (Tsuya et al. 2006). Site-directed mutagenesis was accomplished using the Quick-Change mutagenesis kit (Stratagene) according to the manufacture’s instructions with the oligonucleotides and each complementary oligonucleotides. The mutations were confirmed by sequencing with the ABI Prism BigDye Terminator cycle sequencing kit v3.0 on an ABI Prism 310 Genetic Analyzer (Applied Biosytems). The primer sequences used in this study is shown in Fig. 1.
Preparation of cell extracts and purification of FADGDH wild type and mutants
E. coli DH5α was co-transformed with the mutated pTrcγαβ expression vector and pBBJMccm (Tsuya et al. 2006), which encodes the E. coli ccmABCDEFGH genes essential for the maturation of cytochrome c. The co-transformed E. coli DH5α was grown aerobically at 37°C in LB medium containing 50 μg ampicillin/ml and kanamycin. After reaching an OD660 value of 0.6, cells were induced with 0.5 mM IPTG and the incubation was continued at 30°C for 10 h. Cells were harvested by centrifugation and resuspended in 10 mM PPB (pH 7.0) containing 0.1% Triton X-100, PPB and lysed by sonication. The lysate was centrifuged at 10,000g at 4°C for 10 min and the supernatant was recentrifuged at 40,000g at 4°C for 90 min. The crude extract was obtained by resuspending the pellet in 20 mM Tris/HCl, pH 7.5, containing 1% Triton X-100 and dialyzing the sample overnight at 4°C against 20 mM Tris/HCl buffer, pH 7.5, containing 0.1% Triton X-100. For purification, the dialyzed sample was applied to a DEAE-5PW column (Tosoh, Japan) equilibrated with 20 mM Tris/HCl, pH 7.5, containing 0.1% Triton X-100. The absorbed protein was eluted with a 0–0.5 M linear NaCl gradient. The active fractions were pooled and dialyzed overnight at 4°C against 20 mM Tris/HCl buffer, pH 7.5, containing 0.1% Triton X-100 and applied to a Superdex-200 10/300GL (Amersham Biosciences) equilibrated with 20 mM Tris/HCl, pH 7.5, containing 0.2 M NaCl and 0.1% Triton X-100. Pooled fractions were dialyzed at 4°C against 10 mM PPB, pH 7.0, containing 0.1% Triton X-100.
Enzyme assay
FADGDH activity was assayed in 100 mM PPB, pH 7.0, containing 0.1% Tritron X-100, 0.6 mM 2,6-dichrolophenolindophenol and 6 mM phenazine methosulfate at 37°C in the presence of various concentrations of substrates. The activity was calculated by monitoring the decrease in absorbance of DCIP at 600 nm. One unit of enzyme was defined as the oxidation of 1 μmol glucose per min under standard assay conditions.
Results and discussion
Alignment analyses of FADGDH catalytic site
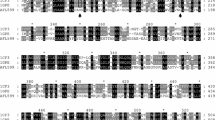
The FADGDH catalytic subunit, the α subunit, is categorized in the GMC oxidoreducatase family, based on its primary structure homology and conserved FAD binding site. Among enzymes in the GMC oxidoreducatase family of which 3D structures are available, glucose oxidases (GOXs) from fungi shows 13.5% identity and 24.4% similarity with α subunit (Wohlfahrt et al. 1999). However, these GOXs show significant high homology with α subunit around their substrate binding sites. Figure 2 shows the primary structure alignment of the substrate binding sites of GOXs with α subunit. The residues recognizing the first hydroxyl group of glucose, which is the catalytic site of GOXs, the residues recognizing the third hydroxyl group of glucose and also the other two residues around this site are conserved in the α subunit. This comparison strongly suggested that this region in α subunit serves its putative active site and substrate binding sites. Significant divergences were observed at the α subunit Ala472 and at Asn475. In GOXs, the corresponding residues are Arg (Arg516 in Aspergillus niger GOX and Arg510 in Penicillium amagasakiense GOD) for Ala472 and Tyr (Tyr513 in A. niger GOX) or Trp (Trp519 in P. amagasakiense GOD), respectively. Considering that GOXs catalyze the oxidation of glucose with very high specificity, whereas FADGDH can catalyze the oxidation of maltose, the differences in the substrate specificity among these enzymes may depend on the differences in the residues in this region. Therefore, we focused on these two residues, Ala472 and Asn475 in FADGDH α subunit as the residue may affect the substrate specificity.
Maximum mutagenesis studies on Ala472 and Asn475
In order to investigate the effect of amino acid substitutions into the substrate specificity of FADGDH α subunit, we introduced maximum mutagenesis into Ala472 and Asn475. We have constructed 38 mutants (19 mutants for Ala472 position and 19 mutants for Asn475 position, respectively), and the activity toward glucose and maltose, as well as the ratio of enzymatic activity toward maltose versus glucose were investigated using crude enzyme preparation (Table 1). Among 19 mutants of Ala472 position, 16 mutants showed higher specificity toward glucose compared with wild type. Ala472 residue in FADGDH α subunit corresponds to Arg516 in A. niger GOX and Arg510 in P. amagasakiense GOD. Ala472Arg also showed specificity toward glucose. However, among these Ala472 mutants, Ala472Phe showed the highest specificity toward glucose. Ala472Phe showed only 8% of relative activity for maltose compared with for glucose, whereas wild type enzyme shows about 50% of the relative activity. Besides, majority of the Asn475 variants showed significant loss of their catalytic activity toward glucose. Asn475 residue in FADGDH α subunit corresponds to Tyr513 in A. niger GOX and Trp519 in P. amagasakiense GOD. Both Asn475Tyr and Asn475Trp showed similar or even broader substrate specificity compared with those of wild type. This may be consistent with the fact that 513 and 519 position in GOX is not conserved. Considering that Asn475 is very close to the putative catalytic center of this enzyme, His476, the alteration of Asn475 residue may significantly affect its catalytic site structure. However, Asn475Asp showed 15% of the relative activity for maltose compared with for glucose, without significant loss of the activity toward glucose. Therefore, we chose Ala472Phe and Asn475Asp for further detailed characterization.
Characterization of Ala472Phe and Asn475Asp
We purified Ala472Phe and Asn475Asp and each was subjected to substrate specificity analyses. Figure 3 shows the correlations between the glucose or maltose concentration and the enzymatic activities for Ala472Phe, Asn475Asp and wild type enzyme. The kinetic parameters of these mutants toward glucose are K m = 17 mM, V max = 590 U/mg for Ala472Phe, and K m = 28 mM, V max = 770 U/mg for Asn475Asp, respectively. Both purified variants showed better substrate specificity than wild type enzyme, as were expected from the results obtained using crude enzyme sample. Ala472Phe and Asn475Asp showed almost no detectable activity toward maltose at the concentration lower than 2 mM. Due to the low activity toward maltose, the kinetic parameters of these mutants toward maltose were not able to be determined. Considering that the activity toward maltose above 5 mM is proportional to maltose concentration and did not show typical saturation curve within the range we have investigated (lower than 10 mM), the K m values of these mutants toward maltose should be very high. Recent issue on maltose interference in the blood glucose monitoring suggest that potent presence of up to 5 mM maltose in a blood of the dialysis patients due to the degradation of the glucose polymer icodextrin, which has become widely used in continuous ambulatory peritoneal dialysis (Janssen et al. 1998). Therefore, we compared the enzymatic activity toward glucose and maltose at 5 mM. The results are summarized in Table 2. The wild type enzyme showed about 26% of the relative activity for 5 mM maltose compared with for glucose, Ala472Phe showed less than 4% of the relative activity for 5 mM maltose. Although it is not perfect, this single mutation resulted in the significant improvement of the substrate specificity of FADGDH catalytic subunit. Asn475Asp showed about 18% of the relative activity for maltose. Although these mutants showed higher glucose specificity versus wild type enzyme, their activity toward 5 mM glucose are about 22% of those for wild type enzyme, the amino acid substitution at these sites simultaneously resulted in the decrease of their catalytic efficiencies.
These results suggested that by introducing mutations into Ala472 and Asp475, the construction of enzymes with high specificity toward glucose is possible. The fact that these mutations significantly affected the substrate specificity of this enzyme strongly supports that our prediction of the putative substrate binding site of FADGDH catalytic subunit is correct. The prediction of 3D structure of the FADGDH catalytic subunit based on the putative substrate binding sites information, together with the combination of further strategic mutations on the basis of our finding, the glucose specific FADGDH with higher activity will be constructed.
References
Inose K, Fujikawa M, Yaniazaki T, Kojima K, Sode K (2003) Cloning and expression of the gene encoding catalytic subunit of thermostable glucose dehydrogenase from Burkholderia Cepacia in Escherichia coli. Biochim Biophys Acta (Proteins and Proteomics) 1645:133–138
Janssen W, Harff G, Caers M, Schellekens A (1998) Positive interference of icodextrin metabolites in some enzymatic glucose methods. Clin Chem 44:2379–2380
Sode K, Tsugawa W, Yamazaki T, Watanabe M, Ogasawara N, Tanaka M (1996) A novel thermostable glucose dehydrogenase varying temperature properties by altering its quaternary structures. Enzyme Microbial Technol 19:82–85
Tsuya T, Ferri S, Fujikawa M, Yamaoka H, Sode K (2006) Cloning and functional expression of glucose dehydrogenase complex of Burkholderia Cepacia in Escherichia coli. J Biotechnol 123:127–136
Wohlfahrt G, Witt S, Hendle J, Schomburg D, Kalisz HM, Hecht HJ (1999) 1.8 and 1.9 a resolution structures of the Penicillium amagasakiense and Aspergillus niger glucose oxidases as a basis for modelling substrate complexes. Acta Cryst D55:969–977
Yamaoka H, Sode K (2007a) SPCE based glucose sensor employing novel thermostable glucose dehydrogenase, FADGDH: blood glucose measurement with 150 nL sample in one-second. J Diabetes Sci Technol 1:28–35
Yamaoka H, Sode K (2007b) A disposable electrochemical glucose sensor using catalytic subunit of novel thermostable glucose dehydrogenase. Open Biotechnol J 1:26–30
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yamaoka, H., Yamashita, Y., Ferri, S. et al. Site directed mutagenesis studies of FAD-dependent glucose dehydrogenase catalytic subunit of Burkholderia cepacia . Biotechnol Lett 30, 1967–1972 (2008). https://doi.org/10.1007/s10529-008-9777-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-008-9777-3