Abstract
The ectopic expression of cellular retinoic acid binding protein 2 (CRABP2) is associated with various tumorigenesis. However, the effects of CRABP2 on the progression of cervical cancer are still unclear. The current study aimed to investigate the role of CRABP2 in the malignant phenotypes of cervical cancer cells. CRABP2 was artificially regulated in CaSki, SiHa, and C-33A cells. CCK-8 assay and flow cytometry were used to assess the cell proliferation and apoptosis abilities, respectively. Wound healing assay and transwell assay were employed to measure the cell migration and invasion abilities, respectively. The results showed that CRABP2 was highly expressed in cervical carcinoma tissues and cell lines, and its high expression was associated with poor overall survival. Knockdown of CRABP2 promoted the cell apoptosis and inhibited cell proliferation, migration, and invasion in cervical carcinoma cells, whereas CRABP2 overexpression exhibited the opposite results. Mechanically, CRABP2 silencing suppressed the Integrin β1/FAK/ERK signaling via HuR. Treatment with siITGB1 or a FAK inhibitor PF-562271 or an ERK inhibitor FR180204 reversed the promoting effects of CRABP2 on cell proliferation, migration, and invasion. Moreover, the overexpression of CRABP2 reverted the HPV16 E6/E7 knockdown-induced inhibition of cell proliferation, migration, and invasion in cervical cancer cells. These results suggested that HPV16 E6/E7 promoted the malignant phenotypes of cervical cancer by upregulating the expression of CRABP2. In conclusion, CRABP2, upregulated by HPV E6/E7, promoted the progression of cervical cancer through activating the Integrin β1/FAK/ERK signaling pathway via HuR.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cervical cancer is the second largest gynecological tumor preceded by breast cancer (Bedell et al. 2020; Burd 2003). In recent years, the incidence of cervical cancer is rising rapidly (Lei et al. 2020; Moghimi-Dehkordi and Safaee 2012). Especially, the proportion of women suffering from cervical cancer has reached 25% in developing countries (Harro et al. 2001). In the early 1980s, studies have confirmed that the occurrence of cervical cancer is related to the persistent infection of human papillomavirus (HPV)(Burd 2003; Franco 1995). Until 1996, HPV is considered a vital cause of cervical cancer (Burd 2003). The common HPVs related to cervical cancer include HPV 16, HPV 18, HPV 31 and HPV 35(Gecer 2023; Harro et al. 2001). More than 70% of patients with cervical cancer are caused by HPV16 and HPV18(Hariri et al. 2011). HPV16 is the main type that causes cervical cancer infiltration, followed by HPV18(de Sanjosé et al. 2007; Stanley 2010). E6 and E7 are the two key viral genes of HPV16, which can activate host DNA synthesis, stimulate cell cycle progression and replicate the viral genome (Peng et al. 2021; von Knebel Doeberitz et al. 1992). However, the underlying mechanism of HPV E6/E7 in promoting cervical cancer progression is still unclear.
Cellular retinoic acid binding protein 2 (CRABP2), a member of the intracellular lipid-binding protein family, involves in the regulation of cell proliferation, apoptosis, invasion and metastasis (Liu et al. 2022a; Noy 2000, 2010; Sessler and Noy 2005; Tang et al. 2022). Interestingly, CRABP2 is considered to play a different role in the development of various tumors. On one hand, several studies have reported that CRABP2 exerts a promoting function in tumors. For example, CRABP2 promotes the invasion and metastasis of breast cancer cells (Feng et al. 2019). The high expression of CRABP2 is related to the poor prognosis of non-small cell lung cancer and malignant glioma (Kim et al. 2018; Liu et al. 2016). On the other hand, CRABP2 have been found to act as an anti-cancer part in head and neck squamous cell carcinoma (Calmon et al. 2009; Vo and Crowe 1998). These studies suggest that CRABP2 is closely related to the occurrence of human malignant tumors. Notably, a recent study has reported a decreased expression of CRABP2 in HPV16-positive oropharyngeal cancer cells (Martinez et al. 2007). Interestingly, another study has testified the upregulation of CRABP2 in epidermal cells transfected with HPV16 E6/E7(Yang et al. 2019). As a potential downstream factor of HPV16 E6/E7, CRABP2 might have the potential to mediate the pro-cancer role of HPV16 E6/E7 in cervical cancer. The function of CRABP2 in cervical cancer evoked us much interest. However, there were no reports on it.
The aim of this study was to explore the role of CRABP2 in the malignant phenotypes of cervical carcinoma cells. We found that CRABP2 was upregulated in cervical carcinoma tissues by bioinformatics analysis, consistent with the results of cell lines. The expression of CRABP2 was artificially regulated in cervical carcinoma cells. The cell proliferation, apoptosis, migration, and invasion abilities were assessed in the gain or loss of CRABP2. The underlying mechanism was further investigated.
Materials and Methods
Bioinformatics Analysis
The mRNA expression of CRABP2 in various cancer tissues was analyzed according to TCGA dataset by GEDS (http://bioinfo.life.hust.edu.cn/web/GEDS/) and TNMplot (https://tnmplot.com/analysis/) platforms. The results of CRABP2 protein expression by immunohistochemical staining were obtained from The Human Protein Atlas platform (https://www.proteinatlas.org/). The association between CRABP2 and overall survival, disease-special survival, and disease-free interval were analyzed following TCGA dataset by LOGpc platform (https://bioinfo.henu.edu.cn/DatabaseList.jsp).
Cell Culture
SiHa (HPV16+), CaSki (HPV16+), C-33 A (HPV−), HeLa (HPV18+) and HaCaT cells were purchased from Procell (Wuhan, China). Short tandem repeat profiling and PCR Mycoplasma Test Kit (Wanleibio, Shenyang, China) were used to last confirm the cell lines without other cell lines and mycoplasma contamination on June, 2022. C-33 A, HaCaT, HeLa, and SiHa cells were cultured in MEM medium (Solarbio, Beijing, China) supplemented with 10% fetal bovine serum (FBS). CaSki cells were cultured in RPMI-1640 medium (Hyclone, Logan, UT, USA) within 10% FBS. All cells were cultured in an incubator with 5% CO2 at 37 °C. We wonder whether CRABP2 could exert an effect on the malignant phenotypes of both HPV16+ and HPV16− cells. Thus, CaSki, SiHa, and C-33 A cells were chosen for the further investigation.
Cell Transfection
For the overexpression of CRABP2, the CDS of CRABP2 was subcloned and inserted into pcDNA3.1 vector. Blank vectors were used as the negative control. C-33 A cells were transfected with the CRABP2-overexpressed plasmid. siRNAs sequences targeting CRABP2 or ELAV like RNA binding protein 1 (HuR) were synthetized by Genescript (Nanjing, China). SiHa and CaSki cells were transfected with CRABP2 siRNA. To investigate the role of integrin β1 signaling in the CRABP2-mediated effects, C-33 A cells were co-transfected with ITGB1 siRNA and CRABP2-overexpressed plasmid. To explore the role of HuR in the relationship between CRABP2 and integrin β1, C-33 A cells were co-transfected with CRABP2-overexpressed plasmid and HuR siRNA1/2. Then, to explore whether HPV16 E6/E7 participated in the regulation of CRABP2, SiHa cells were co-transfected with HPV16 E6/E7 siRNA and CRABP2-overexpressed plasmid. Lipofectamine 3000 reagent (Invitrogen, Carlsbad, California, USA) was used for cell transfection. The sequences of siRNA were listed in Table 1.
RNA Extraction and qRT-PCR
TRI pure (BioTeke, Beijing, China) was used to isolate total RNA from cell lines according to the manufacturer’s protocol. Then, SuperScript M-MLV reverse transcriptase (BioTeke) was used for the reverse transcription of total RNA into cDNA. The amplification was performed using SYBR Green PCR reagents (Solarbio). β-actin was served as an internal control. The relative expression levels were analyzed by 2−∆∆CT method. The primer sequences were listed in Table 2.
Western Blot and Co-immunoprecipitation
RIPA buffer (Beyotime, Shanghai, China) was used to extract proteins of cell lines, and the BCA kit (Beyotime) was used to determine the protein concentration. Then the protein was separated by SDS-PAGE electrophoresis and transferred to polyvinylidene fluoride membranes (Millipore, Billerica, MA, USA). After blocked with 5% skim milk at room temperature for 1 h, the membranes were probed with primary antibodies at 4 °C overnight. The antibodies were as follows: anti-CRABP2 (1: 500, ABclonal, Wuhan, China), anti-HuR (1;1000; proteintech, Wuhan, China), anti-Integrin β1 (1: 500, Wanleibio, Shenyang, China), anti-p-FAK (1: 1000, ABclonal), anti-FAK (1: 500, Wanleibio), anti-p-ERK (1: 1000, Wanleibio), anti-ERK (1: 1000, Wanleibio), and anti-β-actin (1: 2000, proteintech). Subsequently, the membrane was incubated with secondary antibodies HRP-conjugated goat anti-rabbit IgG (1: 10,000; proteintech) or HRP-conjugated goat anti-mouse IgG (1: 10,000; proteintech) for 40 min at 37 °C. Bands were visualized by gel imaging system (LIU YI Biotech, Beijing, China).
Immunoprecipitation assay was performed to detect the endogenous protein interaction. Briefly, the CRABP2 antibodies were crosslinked to protein G-agarose beads (Millipore). Protein lysates were incubated with resin-antibody complex at 4 °C. Then, the complex was rinsed with buffer, and the protein was separated on the SDS-PAGE electrophoresis.
Cell Counting Kit-8 (CCK-8) Assay
Cells were seeded in a 96-well plate at a density of 3 × 103 cells per well. After transfection, cells were treated with/without PF-562,271 (10 µM, Beyotime) or FR180204 (40 µM, Beyotime). Then, Cell viability was determined by CCK-8 kit (Sigma, St. Louis, MO, USA) according to the manufacturer’s instructions. Subsequently, the optical density (OD) values at the wavelength of 450 nm.
Wound Healing Assay
Cells were seeded into a 6-well plate. When the cell density reached about 90%, cells were cultured in a serum-free medium, followed by 1 µg/mL of mitomycin C (Sigma) treatment for 1 h. Subsequently, a 200 µL pipette tip was used to scratch a wound across the center of the cell well. Then, cells were visualized at 0 and 24 h under a microscope and the wound healing ratio of each group was calculated.
Transwell Assay
The transwell invasion assay was performed as shown in the manufacturer’s protocol. Briefly, the inserts (Corning, NY, USA) coated with the Matrigel matrix (Corning) were placed in a 24-well plate. Then, the upper chamber of the insert was added with 200 µL of cell suspension (3 × 104 cells per well). The lower chamber was added with 800 µL of medium containing 10% FBS. Then, cells were fixed in 4% paraformaldehyde for 25 min and then stained with 0.4% crystal violet for 5 min. Subsequently, cells were photographed and counted.
Flow Cytometry Assay
The cell apoptosis assay was performed by using the Annexin V-FITC Apoptosis Detection Kit (Beyotime) according to the user’s instructions. Briefly, the cells were collected and stained with 5 µL Annexin V-FITC and 10 µL propidium iodide (PI) for 15 min in the dark. Cell apoptosis assay was performed using a NovoCyte flow cytometer (ACEA Biosciences, San Diego, California, USA).
Statistical Analysis
Comparison between two groups was assessed using the unpaired t-test. One-way or two-way ANOVA was applied to compare the differences among multiple groups. P < 0.05 was considered statistically significant. All the experiments were repeated for triplicate. Data were shown as mean ± SD. All statistical analyses were performed using GraphPad Prism 8.0 (GraphPad, San Diego, CA, USA).
Results
CRABP2 was Upregulated in Cervical Cancer Tissues and Cell Lines
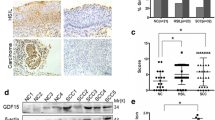
As shown in Fig. 1a, c, and d, bioinformatics analysis demonstrated that both the mRNA and protein expression of CRABP2 was up-regulated in cervical cancer according to TCGA dataset. There was a trend that high expression of CRABP2 was linked with poor overall survival, disease-special survival, and disease-free interval (Fig. 1b). Consistent with the results of high expression of CRABP2 in cervical cancer tissues, cervical cancer cells (CaSki, SiHa, HeLa, and C-33 A) exhibited higher level of CPABP2 mRNA and protein expression than that of human keratinocyte lines (HaCaT) (Fig. 1e). Then, we artificially regulated the expression of CRABP2 in CaSki, SiHa, and C-33 A cells. Decreased expression of CRABP2 mRNA and protein levels were observed in CRABP2-silenced CaSki and SiHa cells, and CRABP2-overexpressed C-33 A cells exhibited the opposite results (Fig. 1f-i). Two fragments with the highest interference efficiency were selected for subsequent experiments.
CRABP2 was upregulated in cervical carcinoma tissues and cell lines. (a) The expression of CRABP2 in various cancer tissues, and the data was obatined from TCGA datasets on GEDS platform. (b) High expression of CRABP2 was associated with poor overall survival, disease-special survival, and disease-free interval, and the data was collected from TCGA dataset on LOGpc platform. (c) CRABP2 mRNA was upregulated in cervical carcinoma tissues, and the data was obatined from TCGA dataset on TNMplot platform. (d) Protein expression of CRABP2 was increased in cervical carcinoma tissues, which was detected by immunohistochemical staining. The data was obatined from The Human Protein Atlas platform. Scale bar: 200 μm. (e) mRNA and protein expression of CRABP2 in HaCaT, HeLa, CaSki, SiHa, and C-33 A cells. (f) The mRNA levels of CRABP2 in CRABP2-silenced CaSki and SiHa cells. (g-i) The mRNA and protein levels of CRABP2 in CRABP2-silenced CaSki and SiHa cells as well as CRABP2-upregulated C-33 A cells. N = 3. **p < 0.01 vs. NC siRNA or Vector; ##p < 0.01 vs. HaCaT
CRABP2 Silence Inhibited Cell Proliferation and Induced Cell Apoptosis of Cervical Cancer Cells
We further explored the function of CRABP2 on cervical cancer cell proliferation and apoptosis abilities. The results of CCK-8 assay revealed that loss of CRABP2 exerted an inhibitory effect on the proliferation of CaSki and SiHa cells, and CRABP2 overexpression led to the contrary results in C-33 A cells (Fig. 2a-c). Then, cell apoptotic ability was promoted by CRABP2-silencing in CaSki and SiHa cells (Fig. 2d-g). Collectively, these data indicated that CRABP2 knockdown repressed cell proliferation and induced cell apoptosis of cervical cancer cell lines.
CRABP2 silence inhibited cell proliferation and induced cell apoptosis of cervical carcinoma cells. (a-c) CCK-8 assay was used to examine the cell viability of CRABP2-silenced CaSki cells and SiHa cells, as well as CRABP2-overexpressed C-33 A cells. (d-g) Flow Cytometry was performed to detect cell apoptosis of CaSki and SiHa cells. N = 3. *p < 0.05, **p < 0.01 vs. NC siRNA or Vector
CRABP2 Promoted Cell Migration and Invasion of Cervical Cancer Cells
To evaluate the effect of CRABP2 on cervical cancer cell migration and invasion, wound healing assay and transwell invasion assay were performed, respectively. A lower migration rate was observed in CRABP2-silenced CaSki and SiHa cells, indicating that loss of CRABP2 repressed cervical cancer cell migration. The result in C-33 A cells with CRABP2 overexpression was opposite (Fig. 3a-d). Meanwhile, cervical cancer cell invasion ability was suppressed by CRABP2 deficiency in CaSki and SiHa cells and was promoted by CRABP2 overexpression in C-33 A cells (Fig. 3e-h). Taken together, the results revealed that CRABP2 acted as a promoted part in cervical cancer cell migration and invasion.
CRABP2 promoted cell migration and invasion of cervical carcinoma cells. (a-d) Wound healing assay was carried out to measure the cell migration of CRABP2-silenced CaSki and SiHa cells, as well as CRABP2-overexpressed C-33 A cells (× 100). Scale bar: 200 μm. (e-h) Transwell assay was used to assess the cell invasion of CaSki, SiHa, and C-33 A cells (× 200). Scale bar: 100 μm. N = 3. **p < 0.01 vs. NC siRNA; ##p < 0.01 vs. Vector
CRABP2 Deficiency Inhibited the Progression of Cervical Cancer by Suppressing the Integrin β1/FAK/ERK Signaling Pathway via HuR
We further investigated the underlying mechanism of the pro-cancer effects of CRABP2. Our results showed that CRABP2 deficiency inhibited the expression of Integrin β1, p-FAK and p-ERK in CaSki and SiHa cells, indicating that CRABP2 deficiency inhibited the Integrin β1/FAK/ERK signaling pathway (Fig. 4a). Furthermore, the mRNA level of ITGB1 was decreased in ITGB1-silenced C-33 A cells (Fig. 4b). The results of CCK-8 assay showed that the CRABP2 overexpression-induced promotion of cell viability was suppressed by ITGB1 silencing, FAK antagonist PF-562,271 or ERK antagonist FR180204 (Fig. 4c). Then, the facilitated cell migration and invasion abilities in C-33 A cells with CRABP2 upregulation were inhibited by ITGB1 knockdown, PF-562,271 or FR180204 (Fig. 4d-g). Therefore, these data demonstrated that the loss of CRABP2 inhibited the proliferation, migration, and invasion of cervical cancer cells via suppressing the Integrin β1/FAK/ERK signaling pathway.
CRABP2 deficiency inhibited the progression of cervical cancer by suppressing the Integrin β1/FAK/ERK signaling pathway. (a) Western blot bands of Integrin β1, p-FAK, FAK, p-ERK and ERK in CRABP2-silenced CaSki and SiHa cells. (b) The expression of ITGB1 mRNA in C-33 A cells transfected with siITGB1 (c) CCK-8 assay was used to examine the cell viability of CRABP2-overexpressed C-33 A cells transfected with siITGB1 or treated with PF-562,271 or RF-180,204. (d, e) Wound healing assay was carried out to measure the cell migration of C-33 A cells (× 100). Scale bar: 200 μm. (f, g) Transwell assay was used to assess the cell invasion of C-33 A cells (× 200). Scale bar: 100 μm. N = 3. ##p < 0.01 vs. Vector; *<0.05, **p < 0.01 vs. NC siRNA or CRABP2-OE
Next, the relationship between CRABP2 and Integrin β1 evoked us much attention. We explored the interaction of the two proteins. Co-immunoprecipitation demonstrated that there were no physical interactions between them in C-33 A cells (Fig. 5a), suggesting that the regulation of Integrin β1 by CRABP2 might be indirect. Interestingly, we found that CRABP2 bound with HuR (Fig. 5b), which was an upstream factor of Integrin β1. Then, CRABP2 was upregulated in C-33 A cells transfected with CRABP2-overexpressed vector (Fig. 5c). HuR was downregulated in C-33 A cells transfected with HuR siRNA 1/2 (Fig. 5d). C-33 A cells were further co-transfected with CRABP2-overexpressed vectors and HuR siRNA1/2. The knockdown of HuR reversed the CRABP2 overexpression-induced upregulation of HuR, Integrin β1, p-FAK, and p-ERK (Fig. 5e), implying that HuR is needed by CRABP2 to promote Integrin β1/FAK/ERK signaling.
CRABP2 enhances the Integrin β1/FAK/ERK signaling by the upregulation of HuR. (a-b) The interaction of CRABP2 and Integrin β1 or HuR in C-33 A cells was detected by co-immunoprecipitation, respectively. (c) mRNA and protein expression of CRABP2 in C-33 A cells transfected with CRABP2 overexpressed vectors (CRABP2OE). (d) mRNA and protein expression of HuR in C-33 A cells transfected with HuR siRNA1/2 (HuRsiRNA1/2). (e) Western blot bands of Integrin β1, p-FAK, FAK, p-ERK and ERK in C-33 A cells co-transfected with CRABP2OE and HuRsiRNA1/2. N = 3. **p < 0.01 vs. Vector or NC siRNA
HPV16 E6/E7 Deficiency Inhibited the Malignant Phenotypes of Cervical Cancer Cells via Downregulating CRABP2
To explore the upstream regulator of CRABP2, SiHa cells were co-transfected HPV16 E6/E7 siRNA and CRABP2-overexpressed plasmid. We first detected the expression of HPV16 E6/E7 and CRABP2 in HPV16 E6/E7-silenced CaSki and SiHa cells. Our results showed a decreased expression level of HPV16 E6/E7 and CRABP2 (Fig. 6a-f). The results of CCK-8 assay showed a decreased cell viability curve in SiHa cells with HPV16 E6/E7 deficiency, which was increased by CRABP2 overexpression (Fig. 6g). The wound healing assay and transwell invasion assay demonstrated that HPV16 E6/E7 deficiency repressed the cervical cancer migration and proliferation capacity, which was reversed by CRABP2 overexpression (Fig. 6h-k). Collectively, our results indicated that HPV16 E6/E7 deficiency inhibited cervical cancer cell proliferation, migration, and invasion by downregulating the expression of CRABP2.
HPV16 E6/E7 deficiency inhibited cervical carcinoma cell proliferation, migration, and invasion by downregulating CRABP2. (a, b) The expression of HPV16 E6/E7 mRNA in HPV16 E6/E7-silenced CaSki and SiHa cells. (c, d) The expression of CRABP2 mRNA in CaSki cells and SiHa cells. (e, f) Western blot bands of CRABP2. (g) CCK-8 assay was used to examine the cell viability of SiHa cells co-transfected with HPV16 E6/E7 siRNA and CRABP2-OE. (h, i) Wound healing assay was carried out to measure the cell migration of SiHa cells (× 100). Scale bar: 200 μm, (j, k) Transwell assay was used to assess the cell invasion of SiHa cells (× 200). Scale bar: 100 μm. N = 3. **p < 0.01 vs. NC siRNA; ##p < 0.01 vs. HPV16 E6/E7 siRNA + Vector
Discussion
In the current study, we found that CRABP2 was highly expression in cervical cancer tissues according to TCGA dataset, which was consistent with the results in cervical cancer cell lines. The loss of CRABP2 repressed the cell proliferation, migration, and invasion, as well as induced apoptosis abilities by downregulating the Integrin β1/FAK/ERK signaling via HuR. CRABP2 overexpression led to the opposite results. Then, we found that the pro-cancer effects of HPV16 E6/E7 were at least partly mediated by the upregulation of CRABP2. The finding implied that CRABP2 might be a novel therapeutic target in the treatment of cervical cancer.
CRABP2 has been reported to upregulated in many cancers including Wilms tumor, pancreatic ductal adenocarcinoma, bladder cancer, bladder cancer, lung cancer, and serous sarcoma (Gupta et al. 2006, 2008; Han et al. 2014; Jin et al. 2013; Toyama et al. 2012; Xiao et al. 2014). In glioblastoma, increased expression of CRABP2 is associated with poor prognosis (Liu et al. 2016). The similar results were obtained in the current study. Herein, we found that the mRNA and protein levels of CRABP2 were upregulated in cervical cancer tissues according to TCGA dataset by LOGpc and The Human Protein Atlas platforms. It is better to testify the expression changes of CRABP2 in clinical samples of cervical cancer, which is a limitation of the current study. Moreover, it has been confirmed that CRABP2 depletion promotes cell apoptosis ability in hepatocellular carcinoma (Chen et al. 2020). Xie et al. have found that CRABP2 contributes to the progression of ovarian cancer by accelerating the epithelial mesenchymal transition (Xie et al. 2022). These studies suggest the pro-tumor potential of CRABP2. Similarly, herein, we found that the knockdown of CRABP2 repressed the malignant phenotypes of cervical cancer cells, whereas CRABP2 overexpression obtained the contrary effects.
Altered expression levels of Integrin β1 is frequently found in cancers, where Integrin β1 acts as a vital role in supporting the malignant properties and regulating the switch of tumor cells from a formant state to active metastasis (Ganguly et al. 2013; Guo and Giancotti 2004; Teng et al. 2013; Wu et al. 2019). Integrin β1 could be activated to recruited and auto-phosphoryate FAK, and further regulate various signaling including the FAK/ERK pathway (Wang et al. 2020) Wu et al. have reported that CRABP2 facilitates the progression of lung cancer by suppressing the Integrin β1/FAK/ERK signaling (Wu et al. 2019). Consistent with the results, our findings showed that ITGB1 siRNA, PF-562,271 (a FAK inhibitor), FR180204 (an ERK inhibitor) reversed the CRABP2-induced proliferation, migration, invasion of cervical cancer cells, indicating that the tumorigenic effects of CRABP2 were at least partly mediated by the regulation of Integrin β1/FAK/ERK signaling pathway. The underlying mechanism of the relationship between CRABP2 and Integrin β1 remained unclear. We found that there were no physical interactions between them in C-33 A cells, suggesting that the upregulation of Integrin β1 by CRABP2 might be indirect. Interestingly, CRABP2 was bound to HuR, which is a known upstream factor of Integrin β1(Mukherjee et al. 2009) (Wu et al. 2019). Wu et al. have reported that the pro-cancer role of CRABP2 in lung cancer is mediated by the promotion of Integrin β1 via HuR (Wu et al. 2019). The similar results were obtained in the current study. The depletion of HuR reversed the CRABP2 overexpression-induced promotion of Integrin β1/FAK/ERK signaling, implying that HuR is needed by CRABP2 to elevate Integrin β1 expression. HuR could stabilize the transcripts of targeted genes by interacting with specific sequences within their 3’UTR (López de Silanes et al. 2004). Interaction with CRABP2 enhances the affinity of HuR toward targeted mRNA (including the HuR-encoded gene Elavl1) and facilitates the transcript stabilization in breast cancer cells (Vreeland et al. 2014). We preferred that similar mechanism might be existed in the regulation of CRABP2 on HuR and downstream Integrin β1 in cervical cancer cells. This part of research evoked us much attention and it will be explored in our further investigation.
As the most important factors in HPV infection, the expression levels of oncoproteins E6 and E7 can change the HPV-mediated cervical lesion types (Ma et al. 2015; Scheurer et al. 2005). Briefly, E6 protein can promote the ubiquitin-dependent degradation of p53 protein and other pro-apoptotic proteins whereas E7 protein can destroy the normal cell cycle and allow tumor growth via binding to retinoblastoma protein (pRB)(de Freitas et al. 2014; Dyson et al. 1989; Scheffner et al. 1990). Furthermore, Yang et al. have found that CRABP2 is upregulated in human keratinocytes transfected with HPV16(Yang et al. 2019). The results indicate the potential of HPV16 in the regulation of CRABP2, which evokes us much attention. We wonder whether the tumorigenic role of HPV16 E6/E7 protein in cervical cancer was associated with CRABP2. Herein, we revealed that HPV16 E6/E7 deficiency exerted an anti-tumor effect in cervical cancer via downregulating the expression of CRABP2, which might offer a novel target for cervical cancer treatment in the future. Furthermore, HPV16 E6/E7 has the properties to regulate the expression of oncogenes on the transcription or post-translation levels. Yin et al. have found that reduced promoter methylation status of downstream gene was detected in cervical cancer cells with ectopic expression of HPV16 E6/E7, which might lead to its aberrant expression (Yin et al. 2017). Liu et al. have reported that HPV16 E6/E7 stabilizes downstream protein expression by decreasing its poly-ubiquitination in cervical cancer cells (S Liu et al. 2022b). These studies indicate that HPV16 E6/E7 upregulates the expression of downstream factors by the induction of transcription activation or the promotion of protein stability. Given the observation of the changes of CRABP2 mRNA and protein, it is possible that HPV16 E6/E7 upregulates CRABP2 by enhancing its transcription or both transcription and post-translation levels. We attach much importance to the underlying mechanism of the regulation of HPV16 E6/E7 on CRABP2, which will be explored in our further investigation.
Conclusion
CRABP2 was upregulated in cervical cancer tissues and cell lines. Knockdown of CRABP2 repressed the cell proliferation, migration, and invasion of cervical cancer cells. The opposite results were obtained by CRABP2 overexpression. Depletion of CRABP2 induced the apoptosis of cervical cancer cells. The pro-cancer effects of CRABP2 were mediated by promoting the Integrin β1/FAK/ERK signaling pathway via the upregulation of HuR. Moreover, CRABP2 was the downstream factor of HPV E6/E7 to facilitate the malignant phenotypes of cervical cancer cells.
Data Availability
The data of this work is available on request to the corresponding author.
References
Bedell SL, Goldstein LS, Goldstein AR, Goldstein AT (2020) Cervical Cancer screening: past, Present, and Future. Sex Med Reviews 8:28–37
Burd EM (2003) Human papillomavirus and Cervical cancer. Clin Microbiol Rev 16:1–17
Calmon MF, Rodrigues RV, Kaneto CM, Moura RP, Silva SD, Mota LD, Pinheiro DG, Torres C, de Carvalho AF, Cury PM, Nunes FD, Nishimoto IN, Soares FA, da Silva AM, Kowalski LP, Brentani H, Zanelli CF, Silva WA Jr., Rahal P, Tajara EH, Carraro DM, Camargo AA, Valentini SR (2009) Epigenetic silencing of CRABP2 and MX1 in head and neck tumors. Neoplasia 11:1329–1339
Chen Q, Tan L, Jin Z, Liu Y, Zhang Z (2020) Downregulation of CRABP2 Inhibit the Tumorigenesis of Hepatocellular Carcinoma In Vivo and In Vitro. BioMed research international 2020: 3098327
de Freitas AC, Coimbra EC, Leitão Mda C (2014) Molecular targets of HPV oncoproteins: potential biomarkers for cervical carcinogenesis. Biochim Biophys Acta 1845:91–103
de Sanjosé S, Diaz M, Castellsagué X, Clifford G, Bruni L, Muñoz N, Bosch FX (2007) Worldwide prevalence and genotype distribution of cervical human papillomavirus DNA in women with normal cytology: a meta-analysis. Lancet Infect Dis 7:453–459
Dyson N, Howley PM, Münger K, Harlow E (1989) The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 243:934–937
Feng X, Zhang M, Wang B, Zhou C, Mu Y, Li J, Liu X, Wang Y, Song Z, Liu P (2019) CRABP2 regulates invasion and Metastasis of Breast cancer through hippo pathway dependent on ER status. J Experimental Clin cancer Research: CR 38:361
Franco EL (1995) Cancer causes revisited: human papillomavirus and cervical neoplasia. J Natl Cancer Inst 87:779–780
Ganguly KK, Pal S, Moulik S, Chatterjee A (2013) Integrins and Metastasis. Cell Adh Migr 7:251–261
Gecer M (2023) High-risk human papillomavirus (hrHPV) prevalence and genotype distribution among Turkish women. J Cytol 40:42–48
Guo W, Giancotti FG (2004) Integrin signalling during tumour progression. Nat Rev Mol Cell Biol 5:816–826
Gupta A, Williams BR, Hanash SM, Rawwas J (2006) Cellular retinoic acid-binding protein II is a direct transcriptional target of MycN in neuroblastoma. Cancer Res 66:8100–8108
Gupta A, Kessler P, Rawwas J, Williams BR (2008) Regulation of CRABP-II expression by MycN in Wilms Tumor. Exp Cell Res 314:3663–3668
Han SS, Kim WJ, Hong Y, Hong SH, Lee SJ, Ryu DR, Lee W, Cho YH, Lee S, Ryu YJ, Won JY, Rhee H, Park JH, Jang SJ, Lee JS, Choi CM, Lee JC, Lee SD, Oh YM (2014) RNA sequencing identifies novel markers of non-small cell Lung cancer. Lung cancer (Amsterdam Netherlands) 84:229–235
Hariri S, Unger ER, Sternberg M, Dunne EF, Swan D, Patel S, Markowitz LE (2011) Prevalence of genital human papillomavirus among females in the United States, the National Health and Nutrition Examination Survey, 2003–2006. J Infect Dis 204:566–573
Harro CD, Pang YY, Roden RB, Hildesheim A, Wang Z, Reynolds MJ, Mast TC, Robinson R, Murphy BR, Karron RA, Dillner J, Schiller JT, Lowy DR (2001) Safety and immunogenicity trial in adult volunteers of a human papillomavirus 16 L1 virus-like particle vaccine. J Natl Cancer Inst 93:284–292
Jin BY, Fu GH, Jiang X, Pan H, Zhou DK, Wei XY, Zhou L, Chung L, Zheng SS (2013) CRABP2 and FABP5 identified by 2D DIGE profiling are upregulated in human Bladder cancer. Chin Med J 126:3787–3789
Kim DJ, Kim WJ, Lim M, Hong Y, Lee SJ, Hong SH, Heo J, Lee HY, Han SS (2018) Plasma CRABP2 as a Novel Biomarker in patients with Non-small Cell Lung Cancer. J Korean Med Sci 33:e178
Lei J, Ploner A, Elfström KM, Wang J, Roth A, Fang F, Sundström K, Dillner J, Sparén P (2020) HPV Vaccination and the risk of Invasive Cervical Cancer. N Engl J Med 383:1340–1348
Liu RZ, Li S, Garcia E, Glubrecht DD, Poon HY, Easaw JC, Godbout R (2016) Association between cytoplasmic CRABP2, altered retinoic acid signaling, and poor prognosis in glioblastoma. Glia 64:963–976
Liu CL, Hsu YC, Kuo CY, Jhuang JY, Li YS, Cheng SP (2022a) CRABP2 is Associated with thyroid Cancer recurrence and promotes Invasion via the Integrin/FAK/AKT pathway. Endocrinology 163.
Liu S, Song L, Yao H, Zhang L (2022b) HPV16 E6/E7 stabilize PGK1 protein by reducing its poly-ubiquitination in Cervical cancer. Cell Biol Int 46:370–380
López de Silanes I, Zhan M, Lal A, Yang X, Gorospe M (2004) Identification of a target RNA motif for RNA-binding protein HuR. Proc Natl Acad Sci U S A 101:2987–2992
Ma C, Zeng C, Jin L, Yang Y, Li P, Chen L, Wang J (2015) GSK3β mediates the carcinogenic effect of HPV16 in Cervical cancer. Sci Rep 5:16555
Martinez I, Wang J, Hobson KF, Ferris RL, Khan SA (2007) Identification of differentially expressed genes in HPV-positive and HPV-negative oropharyngeal squamous cell carcinomas. Eur J Cancer 43:415–432
Moghimi-Dehkordi B, Safaee A (2012) An overview of Colorectal cancer survival rates and prognosis in Asia. World J Gastrointest Oncol 4:71–75
Mukherjee N, Lager PJ, Friedersdorf MB, Thompson MA, Keene JD (2009) Coordinated posttranscriptional mRNA population dynamics during T-cell activation. Mol Syst Biol 5:288
Noy N (2000) Retinoid-binding proteins: mediators of retinoid action. Biochem J 348 Pt 3:481–495
Noy N (2010) Between death and survival: retinoic acid in regulation of apoptosis. Annu Rev Nutr 30:201–217
Peng S, Ferrall L, Gaillard S, Wang C, Chi WY, Huang CH, Roden RBS, Wu TC, Chang YN, Hung CF (2021) Development of DNA Vaccine Targeting E6 and E7 Proteins of Human Papillomavirus 16 (HPV16) and HPV18 for Immunotherapy in Combination with Recombinant Vaccinia Boost and PD-1 Antibody. mBio 12
Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM (1990) The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63:1129–1136
Scheurer ME, Tortolero-Luna G, Adler-Storthz K (2005) Human papillomavirus Infection: biology, epidemiology, and prevention. Int J Gynecol Cancer 15:727–746
Sessler RJ, Noy N (2005) A ligand-activated nuclear localization signal in cellular retinoic acid binding protein-II. Mol Cell 18:343–353
Stanley M (2010) Pathology and epidemiology of HPV Infection in females. Gynecol Oncol 117:S5–10
Tang X, Liang Y, Sun G, He Q, Hou Z, Jiang X, Gao P, Qu H (2022) Upregulation of CRABP2 by TET1-mediated DNA hydroxymethylation attenuates mitochondrial apoptosis and promotes oxaliplatin resistance in gastric cancer. Cell Death Dis 13:848
Teng YC, Lee CF, Li YS, Chen YR, Hsiao PW, Chan MY, Lin FM, Huang HD, Chen YT, Jeng YM, Hsu CH, Yan Q, Tsai MD, Juan LJ (2013) Histone demethylase RBP2 promotes lung tumorigenesis and cancer Metastasis. Cancer Res 73:4711–4721
Toyama A, Suzuki A, Shimada T, Aoki C, Aoki Y, Umino Y, Nakamura Y, Aoki D, Sato TA (2012) Proteomic characterization of ovarian cancers identifying annexin-A4, phosphoserine aminotransferase, cellular retinoic acid-binding protein 2, and serpin B5 as histology-specific biomarkers. Cancer Sci 103:747–755
Vo HP, Crowe DL (1998) Transcriptional regulation of retinoic acid responsive genes by cellular retinoic acid binding protein-II modulates RA mediated Tumor cell proliferation and invasion. Anticancer Res 18:217–224
von Knebel Doeberitz M, Rittmüller C, zur Hausen H, Dürst M (1992) Inhibition of tumorigenicity of Cervical cancer cells in nude mice by HPV E6-E7 anti-sense RNA. Int J Cancer 51:831–834
Vreeland AC, Yu S, Levi L, de Barros Rossetto D, Noy N (2014) Transcript stabilization by the RNA-binding protein HuR is regulated by cellular retinoic acid-binding protein 2. Mol Cell Biol 34:2135–2146
Wang C, Zhang S, Liu J, Tian Y, Ma B, Xu S, Fu Y, Luo Y (2020) Secreted pyruvate kinase M2 promotes Lung Cancer Metastasis through activating the integrin Beta1/FAK signaling pathway. Cell Rep 30:1780–1797e1786
Wu JI, Lin YP, Tseng CW, Chen HJ, Wang LH (2019) Crabp2 promotes Metastasis of Lung Cancer cells via HuR and integrin β1/FAK/ERK signaling. Sci Rep 9:845
Xiao W, Hong H, Awadallah A, Yu S, Zhou L, Xin W (2014) CRABP-II is a highly sensitive and specific diagnostic molecular marker for pancreatic ductal adenocarcinoma in distinguishing from benign pancreatic conditions. Hum Pathol 45:1177–1183
Xie T, Tan M, Gao Y, Yang H (2022) CRABP2 accelerates epithelial mesenchymal transition in serous Ovarian cancer cells by promoting TRIM16 methylation via upregulating EZH2 expression. Environ Toxicol 37:1957–1967
Yang R, Klimentová J, Göckel-Krzikalla E, Ly R, Gmelin N, Hotz-Wagenblatt A, Řehulková H, Stulík J, Rösl F, Niebler M (2019) Combined transcriptome and Proteome Analysis of Immortalized Human keratinocytes Expressing Human Papillomavirus 16 (HPV16) oncogenes reveals novel key factors and networks in HPV-Induced Carcinogenesis. mSphere 4.
Yin F, Wang N, Wang S, Yu F, Sun X, Yu X, Luo B, Zhao C, Wang Y (2017) HPV16 oncogenes E6 or/and E7 may influence the methylation status of RASSFIA gene promoter region in Cervical cancer cell line HT-3. Oncol Rep 37:2324–2334
Acknowledgements
The study was sponsored by Natural Science Foundation of China (82102822), Shenzhen Longgang Fund for Science and Technology Project (LGKCYLWS2020101), Shenzhen Science and Technology R&D Fund (JCYJ20210324131607019), College-level Scientific Research Program of Taizhou Polytechnic College (TZYKYZD-23-1), Social Development Project on Science and Technology Support Program of Taizhou (TS202320), the National Science Fund for Distinguished Young Scholars (82102822).
Author information
Authors and Affiliations
Contributions
Jiaxin Liu, conceptualization, methodology, writing - original draft preparation; Lu Tang and Wenzhu Chu, conceptualization, formal analysis and investigation; Lanlan Wei, conceptualization, writing - review and editing, and supervision. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethic Approval
Not applicable.
Consent to Participate
Not applicable.
Consent to Publish
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, J., Tang, L., Chu, W. et al. Cellular Retinoic Acid Binding Protein 2 (CRABP2), Up-regulated by HPV E6/E7, Leads to Aberrant Activation of the Integrin β1/FAK/ERK Signaling Pathway and Aggravates the Malignant Phenotypes of Cervical Cancer. Biochem Genet 62, 2686–2701 (2024). https://doi.org/10.1007/s10528-023-10568-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-023-10568-6