Abstract
Mycobacterium tuberculosis PE/PPE family proteins, named after the presence of conserved PE (Pro-Glu) and PPE (Pro-Pro-Glu) domains at N-terminal, are prevalent in M. tuberculosis genome. The function of most PE/PPE family proteins remains elusive. To characterize the function of PE_PGRS18, the encoding gene was heterologously expressed in M. smegmatis, a nonpathogenic mycobacterium. The recombinant PE_PGRS18 is cell wall associated. M. smegmatis PE_PGRS18 recombinant showed differential response to stresses and altered the production of host cytokines IL-6, IL-1β, IL-12p40 and IL-10, as well as enhanced survival within macrophages largely via attenuating the apoptosis of macrophages. In summary, the study firstly unveiled the role of PE_PGRS18 in physiology and pathogenesis of mycobacterium.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Tuberculosis (TB), caused by Mycobacterium tuberculosis (M. tuberculosis) infection, remain as a serious deadly infectious disease and public health concern worldwide. M. tuberculosis can subvert the host immune responses [1] for survival within host. The encoding capacity of M. tuberculosis genome is dedicated to the PE/PPEfamily proteins, including PE, PPE and PE_PGRS subfamilies. Proteins of the PE_PGRS subfamily are largely secreted by ESX-5 secretion system [2]. There are at least two domains in the PE_PGRS protein, PE domain and PGRS domain. The PE domain is required for the subcellular localization and PGRS domain is responsible for the shape and morphology of colony of the mycobacterium [3]. 61 PE_PGRS proteins have been identified. Mostly PE_PGRS proteins contain Calcium-binding motif except Rv0742, Rv0832, Rv0978c, Rv3652, and Rv3812 [4]. Most genes of this family are differentially expressed such as Rv1172c, Rv3652, and Rv0578c, the latter was silent in M. canettii [5]. PE_PGRS62 and PE_PGRS16 showed opposite expression pattern during the infection of M. tuberculosis [6]. Most PE_PGRS subfamily proteins are cell wall or cell membrane associated and involved in virulence of the Mycobacteria [7]. The survival ofMMAR_0242 expressed M. smegmatis within infected cells was enhanced [8]. Two PE-PGRS genes deleted M. marinum mutant can not replicate within macrophages and persist within granuloma [9].
Mycobacterium tuberculosis PE_PGRS18 protein, a typical member of PE_PGRS subfamily with function unknown, was explored in our study. PE_PGRS18 was significantly upregulate within host, namely 20-folds and threefolds increased in macrophages and infected mice respectively [10, 11]. To further reveal the function and mechanism of action of PE_PGRS18 during mycobacteria infection, we constructed the M. smegmatis recombinant expressing PE_PGRS18 and M. smegmatis harboring the vector only. Our study demonstrated that recombinant M. smegmatis expressing PE_PGRS18, altered the production of cytokines, including IL-6, IL- 1β, IL-10 and IL-12p40 in host macrophage. Our study also demonstrated that M. smegmatis recombinant with PE_PGRS18 expression increased the intracellular survival, concomitant with decreased apoptosis of the host macrophages.
Methods and materials
The construction of M. smegmatis recombinant strains
Mycobacterial expression vector pALACE [12] was used for recombinant construction. The PE_PGRS18 gene was amplified from M. tuberculosis genomic DNA using specific primers listed in Table 1 (pALACE-PE_PGRS18-F and pALACE-PE_PGRS18-R). The ClaI-BamHI-digested PCR product was cloned into pALACE to generate pALACE-PE_PGRS18. The plasmids (pALACE and pALACE-PE_PGRS18) were electroporated into M. smegmatis mc2155. The recombinant M. smegmatis strains were selected on MB7H9 solid medium containing 100 μg/ml hygromycin (Hyg). The positive recombinant strains harboring PE_PGRS18 gene were confirmed by PCR amplification. Escherichia coli DH5α used for gene cloning was grown at 37 °C using Luria–Bertani (LB) broth and LB agar with indicated antibiotics supplemented. M. smegmatis mc2 155 was grown in 7H9 medium or supplemented with 0.05% (v/v) Tween 80, 0.2% (w/v) glucose, and 0.5% (v/v) glycerol.
Detection the expression of PE_PGRS18 in M. smegmatis
The M. smegmatis recombinant strains harboring His-tagged PE_PGRS18 (Ms_PE_PGRS18) and vector pALACE (Ms_Vec) were cultured in MB7H9 liquid medium supplemented with 100 μg/ml Hyg. At the OD600 = 0.6, the recombinant strains were subjected to 28 mM acetamide (Aladdin, China) for protein expression. In summary, the recombinants with Ms_PE_PGRS18 and Ms_Vec were collected after 16 h acetamide induction using centrifugation at the speed of 3000×g for 10 min, 4 °C. The harvested cells were washed and then sonicated in cold PBS. After sonication, the whole cell lysate preparation was centrifuged at the speed of 20,000×g to isolate the insoluble pellets in the bottom. The separated fractions were separated by SDS-PAGE and further detected by Western blot analysis with using specific anti-His monoclonal antibody (TIANGEN, China). The blots were formed when incubated with IgG-HRP, an anti-mouse IgG monoclonal antibody labeled with horse radish peroxidase (TIANGEN, China).
Recombinant M. smegmatis infected macrophages
The THP-1 cells was cultivated in RPMI1640 medium (Invitrogen) added with 2 mM 10% (v/v) FBS, 100 U/ml penicillin, l-glutamine and 100 μg/ml streptomycin (Invitrogen) and cultured in humidified incubator supplied with 5% CO2 at 37 °C. Macrophages were seeded at 1 × 106 cells per well in 12-well tissue culture plates or at 1 × 105 cells per well in 24-well tissue culture plates. The suspension THP-1 cells were transformed into anchorage-dependent macrophage after 48 h treatment with 100 μg/ml phorbol 12-myristate 13-acetate (PMA, Sigma). Cells were infected with Ms_Vec or Ms_PE_PGRS18 at MOI of 25. Gentamicin was supplied into culture at the concentration of 200 ug/ml after 4 h infection to change new culture medium. After 6, 24, 48, and72 h infection, the culture supernatants were gathered for NO measurement. The production of NO were quantified by commercially NO kit (Nanjing Institute of biological engineering) according to standard procedure. For the detection of bacterial survival within macrophage, the infected macrophages were washed by PBS for three times and dissolved by 0.05% SDS. The cell lysates were tenfold series diluted and then spotted on 7H9 agar.
Recombinant bacteria survival under diverse in vitro stresses
Recombinant M. smegmatis strains were cultivated into optimal concentration in MB7H9 medium containing 100 μg/ml Hyg. After 16 h induction by acetamide, Ms_Vec and Ms_PE_PGRS18 were exposed to stresses. Ms_Vec and Ms_PE_PGRS18 were exposed to 0.05% SDS for 1, 2, 3, and 4 h. In addition, by adding HCl into 7H9 medium, pH gradient was prepared and sterilized in High Temperature Sterilizing Oven. After the treatment, the recombinant strains were tenfold dilutions and plotted onto MB7H9 agar containing Hyg. CFU were counted after 3 days.
Scanning electron microscopy
Ms_PE_PGRS18 and Ms-Vec cells were grown for about 24 h until the OD600 = 1.0 in MB7H9 medium. Cell were harvested by centrifugation and the harvested pellets were washed three times using 1× PBS then resuspended in 2.5% glutaraldehyde solution. The samples were dehydrated by a graded series of ethanol. After critical point drying, samples were sputtered with platinum and observed by a scanning electron microscopy (SEM; FEI Quanta 200).
Subcellular fractionation of recombinant M. smegmatis
Recombinant Ms_Vec and Ms_PE_PGRS18 constructs were grown and subjected to cell fractionation separation as described in [13], with minor modifications. Usually, the acetamide-induced recombinant Ms_PE_PGRS18 and Ms_Vec were subjected to sonication. The whole cell lysates were centrifuged at the speed of 3000×g for 5 min at 4 °C to discard intact cells and cell fragments. The supernatants were ultra-centrifuged at the speed of 27,000×g for 30 min, at 4 °C. After ultra-centrifugation, the pellets (cell wall fraction) and the supernatants (cell membrane and cytosol fractions) can be easily separated. The pellets were suspended by using PBS. Equivalent proteins from pellets and supernatants fraction were subjected to Western blot to detect the expression of Ms_PE_PGRS18. Mycobacteria GroEL2 protein served as cytosol marker.
Cytokines measurement
Culture supernatants were harvested after infection of macrophages with Ms_PE_PGRS18 or Ms_Vec for 6, 24, 48, and 72 h. The RNA was extracted from the infected cells using RNA extraction kit (TIANGEN) and mRNA level of cytokines was measured by RT-PCR using PCR primers listed in Table 1.
Apoptosis analysis
Annexin V/propidium iodide (PI) staining kit was used to assess Apoptotic cells according to the manufacturer’s instructions (BD Pharmingen Inc., San Diego, CA, USA). The cells were stained with FITC-conjugated with Annexin V and PI. Using aFACSCanto II (Becton–Dickinson, San Jose, CA, USA) with FACSDiva to analyze the stained cells and the results were analyzed using the FlowJo software (BD Biosciences, Mountain View, CA, USA).
Statistical analysis
GraphpadPrism6 software was used for the analysis of differences between experimental and control group. Statistical significance (P-value)was decided using Student’s t test. P-values less than 0.05 were statistically significant. *P < 0.05, **P < 0.01, and ***P < 0.001.
Results
Expression of PE_PGRS18 in M. smegmatis
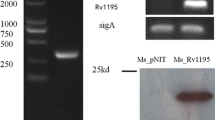
Mycobacterium tuberculosis PE_PGRS subfamily member PE_PGRS18 (42 kDa) encoding gene is 1374 bp. To explore the function of PE_PGRS18, Ms_PE_PGRS18 recombinant and Ms_Vec strains were constructed. Ms_PE_PGRS18 expressed a His-tagged PE_PGRS18 protein harboring pALACE-PE_PGRS18, while Ms_Vec only harbored the vector with Hyg resistant marker gene [12]. Both Ms_Vec and Ms-PE_PGRS18 were cultivated in MB7H9 medium complemented with Hyg. Presence of PE_PGRS18 gene in Ms_PE_PGRS18 strain was confirmed by the PCR amplification (Fig. 1a) and presence of ∼43 kDa PE_PGRS18-His protein in the Ms_PE_PGRS18 strain, while absent in Ms_Vec strain conformed by the Western blot (Fig. 1b).
Expression of PE_PGRS18 in recombinant M. smegmatis. a Ms_PE_PGRS18 and Ms_Vec were grown at 37 °C in MB7H9 liquid media to OD600 = 0.8, then were subjected to PCR amplification to detect the presence of PE_PGRS18 gene. b Lysates were prepared from acetamide-induced bacterial cells and subjected to Western blot analysis to detect His-tagged PE_PGRS18 protein using mouse anti-His antibody
PE_PGRS18 is associated with the mycobacterial cell envelope
At least 29 PE/PPE proteins were predicted to be involved in the “cell wall and cell processes” [14], including the PE_PGRS18. To experimentally test this, scanning electron microscopy was used to compare the morphology difference between Ms_PE_PGRS18 and Ms_Vec. The Ms_PE_PGRS18 was more rough as compare to Ms_Vec (Fig. 2b, d), consistent with the colony morphology of Ms-PE_PGRS18 and control (Fig. 2a, c). We assumed that PE_PGRS18 might be attached with the cell wall of M. smegmatis. In order to further identify this hypothesis, we constructed the M. smegmatis expression His-tagged PE_PGRS18 strain. Recombinant M. smegmatis strains including Ms_PE_PGRS18 and Ms_Vec were subjected to cell fractionation experiment, finally we used Western blot to determine their subcellular localization experiment. We found the PE_PGRS18 protein was localizated in the cell wall and nothing can be found in cytoplasm fraction (Fig. 2e). As expected, cytoplasmic heat-shock protein GroEL2 was detected only in the cytoplasm of M. smegmatis .
PE_PGRS18 is a cell envelope-associated protein and can change the surface of M. smegmatis. a and c Overexpression PE_PGRS18 could impact the colony morphology of M. smegmatis compared with control. b and d Scanning electron micrographs of Ms_Vec and Ms_PE_PGRS18. Experiments were performed three times and similar results were obtained. e Subcellular fractionation of M. smegmatis induced to express PE_PGRS18-His showed localization to the cell wall. Subcellular fractions were separated by SDS-PAGE and proteins were detected with an anti-His antibody. The cytoplasm marker of Mycobacterium GroEL2 served as a control
PE_PGRS18 enhances the intracellular survival of recombinants
Many PE/PPE proteins are involved in the virulence M. tuberculosis and promote its survival within macrophages [15–17]. Our interest with PE_PGRS18 and its impact on the survival of M. smegmatis in host as other PE/PPE proteins. The survival rate of recombinant Ms_PE_PGRS18 and Ms_Vec was compared to confirm if PE_PGRS18 increase the survival of M. smegmatis within macrophages. PMA-differentiated THP-1 macrophages were infected with Ms_Vec or Ms_PE_PGRS18 at an MOI of 25. There was a significant difference in intracellular survival of bacteria in macrophages between Ms_PE_PGRS18 and Ms_Vec at 24, 48 h, after infection (Fig. 3a), implying that PE_PGRS18 can increase the survival of M. smegmatis within macrophages during the early stage of infection.
The survival of recombinant M. smegmatis within macrophages. a Differentiated macrophages were infected with Ms_Vecor Ms_PE_PGRS18 as described in the "Methods" section, respectively. The macrophages were washed and lysed using 0.025% SDS at indicated times. Lysates were plated on MB7H9 medium containing 100 μg/ml Hyg to determine the bacterial number. CFUs were enumerated after the incubation for 3–4 days at 37 °C. b In vitro growth of recombinant Ms_Vec and Ms_PE_PGRS18 after treatment with different pH gradient. The bacteria were collected by centrifugation and resuspended to an OD600 of 0.5 into 5 ml MB 7H9 (pH 5 or 3). All cultures were again incubated at 37 °C, and then tenfold serial dilutions of Ms_Vecand Ms_PE_PGRS18 were spotted on MB 7H9 containing Hyg. c Survival of Ms_Vec and Ms_PE_PGRS18 after exposure to 0.05% SDS. Re-suspended 5 ml recombinant Ms_Vec and Ms_PE_PGRS18(OD600 = 0.5) were exposed to 0.05% SDS for 1, 2, 3, and 4 h. The cultures were serially diluted and plated onto MB 7H10 plates and the colonies counted after 3–4 days of incubation at 37 °C. The data shown are means ± SD of triplicate wells. Similar results were obtained in three independent experiments. *P < 0.05
To gain further insight on the impact of PE_PGRS18 on the survival of M. smegmatis upon exposure to antimicrobial factors, we determined the CFU of recombinant Ms_PE_PGRS18 and Ms_Vec under different surface stress and acid conditions. As shown in Fig. 3b, no significant difference between Ms_Vec and Ms_PE_PGRS18 under in vitro acid stress (0, 3, 6, 9 h after treatment) was spotted. As Fig. 3c shown, bacterial number was significantly decreased at 0.05% SDS and Ms_PE_PGRS18 was more sensitive to 0.05% SDS as compare to Ms_Vec.
Ms_PE_PGRS18 decreased the death of macrophage
Mycobacterium tuberculosis could mediate host cell death. Alternatively, infection can cause macrophages apoptosis to maintain an intact cell membrane, to decrease pathogen viability and strengthen host immunity. To determine whether PE_PGRS18 attenuated the macrophage apoptosis, PMA-differentiated macrophages were infected with Ms_Vec or Ms_PE_PGRS18 for 24 h, and the infected macrophages were subjected to fluorescence microscopy for cell apoptosis analysis (Fig. 4a). Results showed that PE_PGRS18 overexpression markedly decreased the M. smegmatis infection-induced apoptosis of THP-1 cells compared with Ms_Vec infected THP-1 cells. Flow cytometry validated this result (Fig. 4b). Data suggested that PE_PGRS18 promoted the intracellular survival of M. smegmatis by suppressing apoptosis in macrophages.
The MS_PE_PGRS18 attenuated the apoptosis of macrophages. a An analysis of Annexin V-positive cells by FACS flow cytometry after Ms_Vec and MS_PE_PGRS18 stimulation in macrophages. b An apoptosis assay in Ms_Vec and Ms_PE_PGRS18 stimulated macrophages was performed with FLICA staining (green) according to the manufacturer’s instructions. Statistically significant differences are indicated; *p = 0.05, **p = 0.01 and ***p = 0.001
PE_PGRS18 selectively regulates the expression of IL-6, IL- 1β, IL-10 and IL-12p40
To determine whether PE_PGRS18 play a role in subverting the early immune responses, PMA-differentiated THP-1macrophages were infected with Ms_Vec and Ms_PE_PGRS18 for 6, 24, 48, and 72 h. The infected macrophages were harvested to quantify the expression of cytokines by RT-PCR. Total RNA was extracted from the infected macrophages at different intervals (6, 24, 48, and 72 h). Macrophages infected with Ms_PE_PGRS18 expressed significant lower amounts of the cytokines IL-6, IL- 1β and IL-10 than Ms_Vec after infection 24 h. In addition, IL12P40 was increased after infection 24, 48 h (Fig. 5a–d).
PE_PGRS18 increased the expression of IL-6, IL-1β, IL-10 and IL12P40 in infected macrophages. We collected cells after 6, 24, 48 h of infection, then RT-PCR analysis of IL-6 (a), IL-1β (b) and IL-10 (c) and IL12P40 (d) mRNA level was performed. Similar results were obtained from three independent experiments.*P < 0.05, **P < 0.01, and ***P < 0.001
Enhanced intracellular survival by PE_PGRS18 is not associated with the increased production of NO
NO as an important determinant of bacillary burden in hosts [18, 19]. To determine whether enhanced intracellular survival by PE_PGRS18 is associated with the increased production of NO, we measured the production of NO in Ms_Vec and Ms_PE_PGRS18 infected THP-1 macrophages after 6, 24, 48 and 72 h infection by Griess reagent method (Fig. 6a). There is no significant difference for the NO production between the PE_PGRS18 overexpression M. smegmatis and control.
Amounts of NO produced by macrophages infected with recombinant M. Smegmatis. Culture supernatants were collected from THP-1 infected at a MOI of 25:1 with MS_Vec or MS_PE_PGRS18 and the release of NO was assessed at 6, 24, 48, and 72 h. Data are shown as means ± SD of triplicate wells. Similar results were obtained in three independent experiments
Discussion
Abundant PE/PPE family genes is a salient feature of M. tuberculosis genome [20]. In our study, we successfully constructed the Ms_PE_PGRS18 recombinant M. smegmatis strain and identified PE_PGRS18 as a cell wall associated protein. Macrophage is the main residence of infected M. tuberculosis, macrophage can phagocytose the invaded pathogens via multiple mechanisms [21]. M. tuberculosis can escape the attack of host macrophages and persist within hostile environment, including NO, iron-deprived conditions and low pH conditions [22]. Ms_PE_PGRS18 is more sensitive to SDS compared with Ms_Vec. PE_PGRS18 increased the intracellular survival of M. smegmatis. This enhanced intracellular survival seemed irrelevant to the elevated SDS resistance. PE_PGRS18 can inhibit cell apoptosis with unknown mechanism.
PE vaccine and the PGRS domain have been shown to influence the immune recognition of the PE antigen [23]. It has been reported that PE_PGRS33-induced apoptotic signaling in T cells regulated by the Smac-dependent activation of caspases [24, 25], and also mediate macrophage apoptosis by TLR2-dependent release of TNF-ɑ [26]. PE_PGRS33DNA induced a strong CD8+ cytotoxic lymphocyte and Th1-type response, resulting in increased IFN-γ and reduced interleukin-4 production [27]. PE_PGRS33 and PE_PGRS26 increased the IL-10 level, while IL-12 and NO was down-regulated. PE_PGRS33 and Rv1759c induced the production of IFN-γ, could be a good candidate as a new subunit vaccine [28]. Conversely, PE_PGRS16 can upregulate the IL-12 and NO level both in macrophage cultures and in vivo [29]. PE_PGRS30 also decreased the production of IL-12, TNF-ɑ and IL-6 [30]. Immunoregulatory molecules, IL-6 [31], IL-1β [32] and IL-12 [33] are important components of the host immune defense against mycobacterial infection [34], M. tuberculosis effectors can suppress the expression of pro-inflammatory cytokines to increase its survival in macrophages [35–37]. In this study, we found that Ms_PE_PGRS18 might significantly decrease the production of IL-6, IL-1β production after infection 24 h in comparison with Ms_Vec. M. smegmatis expressing PE_PGRS18 could remarkably up-regulated the production of IL-12P40 and inhibit the apoptosis of macrophage, lead to enhanced intracellular survival of nonpathogen M. smegmatis in macrophages. In summary, these results showed a potential role of PE_PGRS18 in M. tuberculosis virulence.
References
Lin PL, Flynn JL (2010) Understanding latent tuberculosis: a moving target. J Immunol 185(1):15–22
Abdallah AM, Verboom T, Weerdenburg EM et al (2009) PPE and PE_PGRS proteins of Mycobacterium marinum are transported via the type VII secretion system ESX-5. Mol Microbiol 73(3):329–340
Delogu G, Pusceddu C, Bua A et al (2004) Rv1818c-encoded PE_PGRS protein of Mycobacterium tuberculosis is surface exposed and influences bacterial cell structure. Mol Microbiol 52(3):725–733
Bachhawat N, Singh B (2007) Mycobacterial PE PGRS proteins contain calcium-binding motifs with parallel β-roll folds. Genom Proteom Bioinform 5(3–4):236–241
Talarico S, Zhang L, Marrs CF et al (2008) Mycobacterium tuberculosis PE_PGRS16 and PE_PGRS26 genetic polymorphism among clinical isolates. Tuberculosis (Edinb) 88(4):283–294
Dheenadhayalan V, Delogu G, Sanguinetti M et al (2006) Variable expression patterns of Mycobacterium tuberculosis PE_PGRS genes: evidence that PE_PGRS16 and PE_PGRS26 are inversely regulated in vivo. J Bacteriol 188(10):3721–3725
Banu S, Honoré N, Saint-Joanis B et al (2002) Are the PE-PGRS proteins of Mycobacterium tuberculosis variable surface antigens? Mol Microbiol 44(1):9–19
Singh VK, Berry L, Bernut A et al (2016) A unique PE_PGRS protein inhibiting host cell cytosolic defenses and sustaining full virulence of Mycobacterium marinum in multiple hosts. Cell Microbiol 18(11):1489–1507
Ramakrishnan L, Federspiel NA, Falkow S (2000) Granuloma-specific expression of Mycobacterium virulence proteins from the glycine-rich PE-PGRS family. Science 288(5470):1436–1439
Srivastava V, Jain A, Srivastava BS et al (2008) Selection of genes of Mycobacterium tuberculosis upregulated during residence in lungs of infected mice. Tuberculosis 88(3):171–177
Meena, LS (2014) An overview to understand the role of PE_PGRS family proteins in Mycobacterium tuberculosis H 37 Rv and their potential as new drug targets. Biotechnol Appl BioChem 62(2):145–153
Lakshminarayan H, Narayanan S, Bach H et al (2008) Molecular cloning and biochemical characterization of a serine threonine protein kinase, PknL, from Mycobacterium tuberculosis. Protein Expr Purif 58(2):309–317
Deng W, Li W, Zeng J et al (2014) Mycobacterium tuberculosis PPE family protein Rv1808 manipulates cytokines profile via co-activation of MAPK and NF-kappaB signaling pathways. Cell Physiol Biochem 33(2):273–288
Mazandu GK, Mulder NJ (2012) Function prediction and analysis of Mycobacterium tuberculosis hypothetical proteins. Int J Mol Sci 13(6):7283–7302
Bottai D, Di Luca M, Majlessi L et al (2012) Disruption of the ESX-5 system of Mycobacterium tuberculosis causes loss of PPE protein secretion, reduction of cell wall integrity and strong attenuation. Mol Microbiol 83(6):1195–1209
Tiwari BM, Kannan N, Vemu L et al (2012) The Mycobacterium tuberculosis PE proteins Rv0285 and Rv1386 modulate innate immunity and mediate bacillary survival in macrophages. Plos ONE 7(12):e51686–e51686
Singh SK, Tripathi DK, Singh PK et al (2012) Protective and survival efficacies of Rv0160c protein in murine model of Mycobacterium tuberculosis. Appl Microbiol Biotechnol 97(13):5825–5837
Gupta D, Sharma S, Singhal J et al (2010) Suppression of TLR2-induced IL-12, reactive oxygen species, and inducible nitric oxide synthase expression by Mycobacterium tuberculosis antigens expressed inside macrophages during the course of infection. J Immunol 184(10):5444–5455
Voskuil MI, Schnappinger D, Visconti KC et al (2003) Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J Exp Med 198(5):705–713
Fishbein S, Wyk N, Warren RM et al (2015) Phylogeny to function: PE/PPE protein evolution and impact on Mycobacterium tuberculosis pathogenicity. Mol Microbiol 96(5):901–916
Leemans JC, Thepen T, Weijer S et al (2005) Macrophages play a dual role during pulmonary tuberculosis in mice. J Infect Dis 191(1):65–74
Flynn JL, Chan J (2003) Immune evasion by Mycobacterium tuberculosis: living with the enemy. Curr Opin Immunol 15(4):450–455
Delogu G, Brennan MJ (2001) Comparative immune response to PE and PE_PGRS antigens of Mycobacterium tuberculosis. Infect Immun 69(9):5606–5611
Dheenadhayalan V, Delogu G, Brennan MJ (2006) Expression of the PE_PGRS 33 protein in Mycobacterium smegmatis triggers necrosis in macrophages and enhanced mycobacterial survival. Microbes Infect 8(1):262–272
Cadieux N, Parra M, Cohen H et al (2011) Induction of cell death after localization to the host cell mitochondria by the Mycobacterium tuberculosis PE_PGRS33 protein. Microbiology 157(Pt 3):793–804
Chaitra MG, Shaila MS, Nayak R (2007) Evaluation of T-cell responses to peptides with MHC class I-binding motifs derived from PE_PGRS 33 protein of Mycobacterium tuberculosis. J Med Microbiol 56(Pt 4):466–474
Singh PP, Parra M, Cadieux N et al (2008) A comparative study of host response to three Mycobacterium tuberculosis PE_PGRS proteins. Microbiology 154(Pt 11):3469–3479
Chatrath S, Gupta VK, Dixit A et al (2016) PE_PGRS30 of Mycobacterium tuberculosis mediates suppression of proinflammatory immune response in macrophages through its PGRS and PE domains. Microbes Infect 18(9):536–542
Basu S, Pathak SK, Banerjee A et al (2007) Execution of macrophage apoptosis by PE_PGRS33 of Mycobacterium tuberculosis is mediated by Toll-like receptor 2-dependent release of tumor necrosis factor-alpha. J Biol Chem 282(2):1039–1050
Campuzano J, Aguilar D, Arriaga K et al (2007) The PGRS domain of Mycobacterium tuberculosis PE_PGRS Rv1759c antigen is an efficient subunit vaccine to prevent reactivation in a murine model of chronic tuberculosis. Vaccine 25(18):3722–3729
Jang S, Uematsu S, Akira S et al (2004) IL-6 and IL-10 induction from dendritic cells in response to Mycobacterium tuberculosis is predominantly dependent on TLR2-mediated recognition. J Immunol 173(5):3392–3397
Fremond CM, Togbe D, Doz E et al (2007) IL-1 receptor-mediated signal is an essential component of MyD88-dependent innate response to Mycobacterium tuberculosis infection. J Immunol 179(2):1178–1189
Bordón J, Plankey MW, Young M et al (2011) Lower levels of interleukin-12 precede the development of tuberculosis among HIV-infected women. Cytokine 56(2):325–331
Cooper AM, Mayer-Barber KD, Sher A et al (2011) Role of innate cytokines in mycobacterial infection. Mucosal Immunol 4(3):252–260
Huang Y, Wang Y, Bai Y et al (2010) Expression of PE_PGRS 62 protein in Mycobacterium smegmatis decrease mRNA expression of proinflammatory cytokines IL-1beta, IL-6 in macrophages. Mol Cell Biochem 340(1–2):223–229
Li J, Chai QY, Zhang Y et al (2015) Mycobacterium tuberculosis Mce3E suppresses host innate immune responses by targeting ERK1/2 signaling. J Immunol 194(8):3756–3767
Muttucumaru DGN, Smith DA, McMinn EJ et al (2011) Mycobacterium tuberculosis Rv0198c, a putative matrix metalloprotease is involved in pathogenicity. Tuberculosis (Edinb) 91(2):111–116
Acknowledgements
This work was supported by National Natural Science Foundation (Grant Nos. 81371851, 81071316, 81271882, 81301394), National Key Research and Development Program (2016YFC0502304), New Century Excellent Talents in Universities (Grant No. NCET-11-0703), The Fundamental Research Funds for the Central Universities (Grant No. XDJK2016E093), The Chongqing Municipal Committee of Education for postgraduates innovation program (Grant No. CYS16073). The Fundamental Research Funds for the Central Universities (Grant Nos. XDJK2016E093, XDJK2016D055, XDJK2012D007), The Chongqing Municipal Committee of Education for postgraduates innovation program (Grant No. CYS16073).
Author information
Authors and Affiliations
Corresponding author
Additional information
Wenmin Yang and Wanyan Deng have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Yang, W., Deng, W., Zeng, J. et al. Mycobacterium tuberculosis PE_PGRS18 enhances the intracellular survival of M. smegmatis via altering host macrophage cytokine profiling and attenuating the cell apoptosis. Apoptosis 22, 502–509 (2017). https://doi.org/10.1007/s10495-016-1336-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10495-016-1336-0