Abstract
The dispersal mechanism of the two-spotted spider mite Tetranychus urticae Koch (Acari: Tetranychidae) could affect predator–prey population dynamics and the spread of acaricide resistance. To investigate the propensity for spider mite migration in the field, the genetic structure of spider mite populations was studied in two apple orchards using five microsatellite markers. Adult female mites were collected from trees separated by approximately 10–24 m along a line covering a distance of about 100 m. The genetic data suggested that a high population density increased the migration rate among the breeding colonies within a single tree. Spatial autocorrelation analysis suggested a positive genetic structure in the first distance class within the two orchards, which might have been caused by crawling or short-distance aerial dispersal. Meanwhile, mites may also have a large-scale migration system that could cause a high level of gene flow and constrained isolation-by-distance or genetic clines within the approximately 100-m range of the study sites. Therefore, mites might aerially disperse over long distances on a scale of <100 m while also taking shorter trips among nearby trees within a distance of 10–24 m in the apple orchards.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
A key component in the population dynamics of herbivorous arthropods is the dispersal ability among available host plants. Many herbivorous arthropods have winged adult stages that can fly long distances in search of suitable host plants, but the wingless spider mites generally must rely on crawling for their dispersal (Mitchell 1973; Alves et al. 2005). Crawling dispersal dictates shorter-scale migrations that are primarily adapted to the location of food and for breeding colony expansion into uninfested areas with the same plant (Hussey and Parr 1963; Kielkiewicz 1996). However, in addition to crawling, spider mites can be carried by the wind to great heights (Coad 1931) and for long distances (Smitley and Kennedy 1985, 1988; Lawson et al. 1996; Osakabe et al. 2005). Aerial dispersal accelerates migration to uninfested crops (Smitley and Kennedy 1988; Grafton-Cardwell et al. 1991) and may affect population dynamics within large-scale prey–predator systems (Pels and Sabelis 1999). Aerial dispersion has also been associated with the spreading of the acaricide resistance genes (Grafton-Cardwell et al. 1991).
The dispersal process used by a particular mite will typically depend on habitat conditions. Degradation of habitat conditions such as an increase in population density or a decrease in host quality increases the proportion of individuals that exhibit aerial dispersal behavior under laboratory conditions (Li and Margolies 1993a). In field populations, degradation conditions, increase in predator density, and application of acaricide accelerated aerial dispersal (Smitley and Kennedy 1988; Grafton-Cardwell et al. 1991; Margolies 1995). Therefore, migration rates and mean dispersal distances associated with both crawling and aerial mechanisms are expected to vary over time. Neutral genetic makers are useful tools for investigation of complicated dispersal processes such as those observed in spider mites.
The two-spotted spider mite Tetranychus urticae Koch (Acari: Tetranychidae) is an important cosmopolitan species that damages a large variety of plants including fruit trees, vegetables, and flowers. Additionally, the two-spotted spider mite has acquired resistance to many types of acaricides (Croft and van de Baan 1988), making this species a major pest in greenhouses, crop fields, and fruit orchards (Bolland et al. 1998). A better understanding of dispersal could be helpful for the successful management of mite populations by providing reliable estimates of population dynamics and the potential risk of spreading the acaricide resistance genes (Grafton-Cardwell et al. 1991; Dunley and Croft 1992).
Using microsatellite markers, Uesugi et al. (2009) discovered that the level of genetic differentiation was high both between and within rose beds in T. urticae populations in a greenhouse (22 or 27 m length). Gene flow was most likely to occur between neighboring breeding colonies and the level of gene flow was too low, even within a rose bed, to act against genetic drift and homogenize allele frequencies. Hinomoto and Takafuji (1994) also found limited of gene flow on strawberries among leaf populations, colonies recognized by a patch of injured plants, and populations sub-divided by sections of a greenhouse. These studies suggest that large-scale dispersal such as aerial dispersal contributes very little to gene flow in a semi-enclosed greenhouse. However, gene flow due to mite dispersal has not been well characterized in open field populations, despite the substantial numbers of airborne mites seen in such locations (Boykin and Campbell 1984; Smitley and Kennedy 1988).
Therefore, in this study, we analyzed genetic structure using microsatellite markers associated with individual tree populations of the two-spotted spider mite in apple orchards. From our gene flow estimates, we discuss the migration of T. urticae among trees in an orchard.
Materials and methods
Study sites and mite sampling
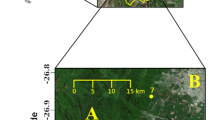
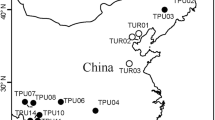
During September 10–11, 2006, T. urticae female adults were collected from two apple orchards, one in Susaka Town (site A: 36°40′N, 138°55′E) and the other in Azumino Town (site B: 36°14′N, 137°51′E), Nagano Prefecture, Japan (Fig. 1). Mite damage to apple trees was more serious at site B as evidenced by the brown color of the abaxial surfaces of most leaves caused by the heavy infestation. In contrast, at site A, such changes were not found and the population density was relatively low (<1 female adult per leaf). Other potential host plants for the mites were scarce in the orchards. Graminaceous weeds covered site A and rice straw was spread at site B. However, stocks of white clover, Trifolium repens L. (Fabaceae), which is a potential host, were scattered at site B. Other apple orchards with similar environments were located around these sties.
At site A, trees were planted at intervals of 5–8 m in line and their branches did not contact neighboring trees (Fig. 2). Therefore, mites were expected to migrate between trees by crawling on the ground or by aerial dispersal. At site B, trees were planted at intervals of 2 and 7.5 m within and between lines, respectively, and their branches were in contact with neighboring trees in the same line (Fig. 2). Therefore, mites could also crawl along the branches to the next tree. To exclude effects on gene flow of direct crawling dispersal between trees, we selected trees for sampling along a diagonal transect (Fig. 2). Mites were collected from trees separated by distances of 10–24 m along straight lines designated A–1 through A–7 at site A and B–1 through B–9 at site B (Fig. 2).
Approximately 50 leaves were randomly collected from shoots of each of a total of 16 trees. From these leaves 1–2 adult female mites per leaf were collected using a fine brush. The mites were placed on kidney bean leaf disks of Phaseolus vulgaris L. (Fabaceae) prior to the extraction of DNA.
Microsatellite analysis
DNA was extracted by the procedure of Goka et al. (2001). Five microsatellite loci were chosen to analyze population structure. The primer pairs used for DNA amplification using polymerase chain reaction (PCR) were Tu1b16 (Navajas et al. 1998), TkMS015 (Nishimura et al. 2003), TuCT04, TuCA18, and TuCA25 (Uesugi and Osakabe 2007). PCR amplifications were carried out with a TaKaRa PCR Thermal Cycler TP600 (TaKaRa Bio, Kyoto, Japan) as follows: 3 min at 96°C, 32–38 cycles for 1 min at 96°C, 1 min at 52°C, and 2 min at 72°C, followed by 10 min at 72°C. The 20 μl reaction volume contained 0.4 μM of each primer, 0.25 mM of dNTPs, 10 mM Tris–HCl (pH 8.4), 50 mm KCl, 1.5 mM MgCl2, 1 U of Taq polymerase (TaKaRa Ex Taq), and 1 μl of the DNA template solution (approximately 1 ng/μl) prepared by the method of Osakabe et al. (2005). PCR products were analyzed on an ABI 3130-200 Genetic Analyzer with Gene Mapper (Applied Biosystems, Foster City, CA, USA).
Analysis of genetic structure
We analyzed genotypic disequilibrium among five microsatellite loci for each tree population using FSTAT software (Goudet 2001). The heterozygosities were determined within each site (H T) and within each tree (H S). In addition, we calculated average allelic richness (r g; El Mousadik and Petit 1996), and the number of different alleles found when sampling ‘g’ genes using POPULATIONS version 1.2.28 (Langella 2002). We fixed the sample number in each population to 20 genes, namely ten mite samples, when calculating allelic richness (r 20). The fixation index (F IS) of each tree population was calculated and the deviation of F IS from Hardy–Weinberg equilibrium was tested by applying 1,000 permutations of alleles among individuals using FSTAT software (Goudet 2001).
FSTAT version 2.9.3 (Goudet 2001) was also used to evaluate heterozygosity deficiency (f) to determine whether mites were panmictic within a tree (Weir and Cockerham 1984). The level of genetic differentiation among tree populations θ was quantified using FSTAT version 2.9.3 (Goudet 2001). The 95% confidence intervals for f and θ were computed by applying 1,000 permutations of the genotypes using FSTAT version 2.9.3 (Goudet 2001).
We used a cluster analysis to investigate the genetic relationships among mite populations on individual trees. The genetic distances between pairs of mite populations from the same tree were estimated by the Cavalli-Sforza and Edwards’ (1967) chord distance, D C. The resulting distance matrix was used to construct dendrogram with the UPGMA algorithm. These were computed using POPULATIONS version 1.2.28 (Langella 2002).
For analysis of isolation-by-distance (Wright 1943), we compared F ST/(1 − F ST) with the spatial distance of each quadrat population (Rousset 1997) using a Mantel test (1,000 permutations) in GENEPOP (Raymond and Rousset 1995). The isolation-by-distance means decreased with genetic similarity among quadrat populations as the spatial distance between them increased.
Pair-wise genetic distance among individuals was calculated using the method described by Smouse and Peakall (1999) with GENALEX version 6 (Peakall and Smouse 2006). The pairwise spatial distance was also calculated among individuals from different trees. Using the genetic distances, spatial autocorrelation analysis was performed with the correlation value r using GENALEX version 6 (Smouse and Peakall 1999; Peakall and Smouse 2006) to examine the effect of gene flow on genetic structure within each site.
Results
We determined the genotypes of 164 mites collected from seven trees at site A and 217 mites collected from nine trees at site B. Significant genotypic disequilibria were detected only between TkMS015 and TuCT04 in population A–1, and between Tu1b16 and TuCA25, and TkMS015 and TuCT04 in population A–7, (P < 0.05 after Bonferroni correction). Therefore, the effect of genotypic disequilibrium among the five microsatellite loci on genetic structure was minor in these populations. Allelic richness (r 20), heterozygosity within a site (H T) and heterozygosity within a tree (H S) were similar between sites (Table 1). The mite population within a tree was not panmictic because the 95% confidence intervals of heterozygote deficiency (f) did not include zero at both sites (Table 1). This was probably caused by the Wahlund effect because spider mites rarely crawl over a structured habitat such as branches (Hussey and Parr 1963; Osakabe and Komazaki 1999) and tend to create numerous small breeding colonies on a single plant (Hinomoto and Takafuji 1995). Significant (P < 0.05) heterozygote deficits were found in all of tree populations of site A and 4 of 9 populations of site B (Table 2). In addition, the f value was higher at site A (0.328) than at site B (0.078; Table 1). Therefore, the level of genetic differentiation among breeding colonies within a tree was probably higher at site A than at site B.
Genetic differentiation among trees (θ) was positive; θ = 0.028 (0.012–0.047, 95% CI) at site A, θ = 0.012 (0.000–0.020, 95% CI) at site B, and θ = 0.039 (0.035–0.045, 95% CI) among all 16 tree populations (Table 1), suggesting that there was a certain level of limitation in mite migration and complete genetic homogenization was prevented within a scale of studied sites.
The relationship between spatial location and place in the UPGMA dendrogram of mite population from on individual trees was not clear within sites A and B, although the dendrogram identified distinct genetic differences between sites (Fig. 3). The correlation between F ST/(1 − F ST) and spatial distance was not significant at either site A (Mantel test, P = 0.191) or site B (Mantel test, P = 0.307). Therefore, a genetic cline or isolation-by-distance was not detected among mite populations at either site.
The multiallelic correlograms for spatial analysis demonstrated a low level of positive autocorrelation in the first distance class at both sites (Fig. 4). In other words, individual mites inhabiting trees separated by 10–24 m were more likely to share similar alleles than individual mites on trees separated by greater distances. In addition, correlation values (r) decreased from the first to the second distance class at both sites, suggesting significant genetic structure at less than the distance of the second class. These results suggest that the migration rate was relatively high among nearby trees while the range was less than the distance of second class. Negative r values were detected at more than the distance of second class, particularly in site A. Therefore, mites could also have different migration system in more than the distance class. However, the spatial genetic autocorrelations were low for all distance classes (r < 0.02) including the first distance class at both sites; the 95% confidence intervals of the r values did not exclude the null hypothesis of no genetic structure (Fig. 4). Therefore, a level of gene flow might be relatively high over long distances of <100 m, resulting in no genetic structure among mite populations of distant trees on the scale of studied sites.
Multiallelic correlograms (solid lines) with 95% null hypothesis confidence regions (dotted lines) of no genetic structure in tree populations of Tetranychus urticae in site A and site B. Distance measure was by order of tree location (10–24 m). Vertical bar indicates 95% confidence intervals of the r values that were determined by bootstrapping
Discussion
In our study, T. urticae had a significant positive autocorrelation within a short range (first distance class, 10–24 m) in the two apple orchards, which could be the result of frequent the migration between nearby apple trees. In a semi-enclosed greenhouse habitat, the typical crawling dispersal mechanism characteristic of most mites, which results in the short range migration and consequently the fine-scale genetic structure such as isolation-by-distance or genetic cline, appeared to be dominant (Uesugi et al. 2009). At the study sites, fine-scale genetic structure could have also been the result of crawling among the ground or short trips by aerial dispersal between apple trees, or both. However, we did not detect isolation-by-distance or a genetic cline among tree populations, suggesting frequent migration over distant trees on the scale of studied sites (<100 m). These patterns may indicate the use of different dispersal mechanisms or different patterns of aerial dispersal; that is, mites may frequently migrate long distances while also taking relatively short trips. However, large-scale migration cannot be explained by typical crawling dispersal.
One possibility is that an aerial dispersal mechanism largely contributed to the gene flow among apple tree populations in the two orchards. Aerial dispersal of T. urticae has been observed by aerial traps in open field habitat (Boykin and Campbell 1984; Smitley and Kennedy 1988; Grafton-Cardwell et al. 1991). Substantial numbers of airborne mites could successfully migrate between trees via wind in an orchard and could contribute to gene flow among apple trees on a large scale. Another possibility is that the mite populations of individual trees we analyzed could have been colonized by the same population source from an alternative host. In fact, the two-spotted spider mite often moves from alternative adjacent host plants (Grafton-Cardwell et al. 1991; Morishita 1992, 1997). However, this explanation appears to be less likely because we did not find an alternative host within and around the orchards that could have provided a habitat for a source population. Further study of population genetics is necessary to confirm which dispersal mechanisms contributed to high levels of gene flow among mite populations of individual trees.
The large-scale gene flow in the open field habitats could be the result of adequate wind velocity required for takeoff and subsequent aerial dispersal. In fact, the mite appears to require some minimal wind speed, 1.5 (Smitley and Kennedy 1985) or 3 m/s (Osakabe et al. 2008), for aerial dispersal, which is rare in semi-closed habitats such as greenhouses. Additionally, other factors such as humidity, migration of predators from surrounding vegetation, selection pressure by acaricides, and seasonal changes in host quality might be responsible for the difference in frequency of large-scale dispersal between semi-closed and open habitats (Margolies and Kennedy 1988; Li and Margolies 1993a, b; Margolies 1995).
As population density increases, the level of population subdivision caused by crawling dispersal decreases, because female adults are apt to disperse more frequently (Mitchell 1973). In addition, breeding colonies that were initially isolated become connected by increased population density (Hinomoto and Takafuji 1995; Uesugi et al. 2009). Such a relationship between genetic differentiation and population density has been observed in confined greenhouse environments (Hinomoto and Takafuji 1994; Tsagkarakou et al. 1997, 1998, 1999; Navajas et al. 2002). Our data showed that the heterozygote deficiency within an individual tree was lower at site B, which was characterized by much more overall mite damage. This result may also provide the evidence that higher population densities are correlated with higher migration rates among breeding colonies within an individual apple tree.
The high frequency of migration can result in spreading of pesticide resistance genes and accelerate the development of resistance over a large area (e.g., Caprio and Tabashnik 1992; Chevillon et al. 1999; Endersby et al. 2006; Martinelli et al. 2007). In this study, gene flow was frequent over a range of at least 100 m, in which acaricide resistance will easily expand. In the future, we propose to investigate the limitations for large-scale migration of the spider mite and its contribution to gene flow of acaricide resistance in orchards. Such knowledge could provide successful management strategies pertaining to acaricide resistance.
References
Alves EB, Casarin NFB, Omoto C (2005) Dispersal mechanisms of Brevipalpus phoenicis (Geijskes) (Acari: Tenuipalpidae) in citrus groves. Neotrop Entomol 34:89–96
Bolland HR, Gutierrez J, Flechtmann CHW (1998) World catalogue of the spider mite family (Acari: Tetranychidae), with references to taxonomy, synonymy, host plants and distribution. Brill Academic Publishers, Leiden
Boykin LS, Campbell WV (1984) Wind dispersal of the two-spotted spider mites (Acari: Tetranychidae) in North Carolina peanut fields. Environ Entomol 13:212–227
Caprio MA, Tabashnik BE (1992) Gene flow accelerates local adaptation among finite populations—simulating the evolution of insecticide resistance. J Econ Entomol 85:611–620
Cavalli-Sforza LL, Edwards AWF (1967) Phylogenic analysis: models and estimation procedures. Am J Hum Genet 19:233–257
Chevillon C, Raymond M, Guillemaud T et al (1999) Population genetics of insecticide resistance in the mosquito Culex pipiens. Biol J Lin Soc 68:147–157. doi:10.1111/j.1095-8312.1999.tb01163.x
Coad BR (1931) Insects captured by airplane are found at surprising heights. US Dept Agric Yearbk 1931:320–323
Croft BA, van de Baan HE (1988) Ecological and genetic factors influencing evolution of pesticide resistance in teranychid and phytoseiid mites. Exp Appl Acarol 4:277–300. doi:10.1007/BF01196191
Dunley JE, Croft BA (1992) Dispersal and gene flow of pesticide resistance traits in phytoseiid tetranychid mites. Exp Appl Acarol 14:313–325. doi:10.1007/BF01200570
El Mousadik A, Petit RJ (1996) High level of genetic differentiation for allelic richness among populations of the argan tree (Argania spinosa L. Skeels) endemic of Morocco. Theor Appl Genet 92:832–839
Endersby NM, McKechnie SW, Ridland PM et al (2006) Microsatellites reveal a lack of structure in Australian populations of the diamondback moth, Plutella xylostella (L.). Mol Ecol 15:107–118. doi:10.1111/j.1365-294X.2005.02789.x
Goka K, Okabe K, Yoneda M et al (2001) Bumblebee commercialization will cause worldwide migration of parasitic mites. Mol Ecol 10:2095–2099. doi:10.1046/j.0962-1083.2001.01323.x
Goudet J (2001) FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). Available at http://www2.unil.ch/popgen/softwares/fstat.htm. Accessed 29 Apr 2009
Grafton-Cardwell EEJ, Granett J, Normington SM (1991) Influence of dispersal from almonds on the population-dynamics and acaricide resistance frequencies of spider mites infesting neighboring cotton. Exp Appl Acarol 10:187–212. doi:10.1007/BF01198650
Hinomoto N, Takafuji A (1994) Studies on the population structure of the two-spotted spider mite, Tetranychus urticae Koch, by allozyme variability analysis. Appl Entomol Zool (Jpn) 29:259–266
Hinomoto N, Takafuji A (1995) Genetic changes in the population structure of the two-spotted spider mite, Tetranychus urticae Koch (Acari: Tetranychidae), on vinyl-house strawberry. Appl Entomol Zool (Jpn) 30:521–528
Hussey NW, Parr WJ (1963) Dispersal of the glasshouse red spider mite Tetranychus urticae Koch (Acari: Tetranychidae). Entomol Exp Appl 6:207–214. doi:10.1007/BF00300527
Kielkiewicz M (1996) Dispersal of Tetranychus cinnabarinus on various tomato cultivars. Entomol Exp Appl 80:254–257. doi:10.1007/BF00194769
Langella O (2002) POPULATIONS 1.2.28. Population genetic software (individuals or populations distances, phylogenetic trees). Available from http://bioinformatics.org/~tryphon/populations/. Accessed 29 Apr 2009
Lawson DS, Nyrop JP, Dennehy TJ (1996) Aerial dispersal of European red mites (Acari: Tetranychidae) in commercial apple orchards. Exp Appl Acarol 20:193–202. doi:10.1007/BF00054511
Li J, Margolies DC (1993a) Effects of mite age, mite density, and host quality on aerial dispersal behavior in the two-spotted spider mite. Entomol Exp Appl 68:79–86. doi:10.1007/BF02380584
Li J, Margolies DC (1993b) Quantitative genetics of aerial dispersal behavior and life-history traits in Tetranychus urticae. Heredity 70:544–552. doi:10.1038/hdy.1993.78
Margolies DC (1995) Evidence of selection on spider mite dispersal rates in relation to habitat persistence in agroecosystems. Entomol Exp Appl 76:105–108. doi:10.1007/BF02382315
Margolies DC, Kennedy GG (1988) Fenvalerate-induced aerial dispersal by the two-spotted spider mite. Entomol Exp Appl 46:233–240. doi:10.1007/BF00364194
Martinelli S, Clark PL, Zucchi MI et al (2007) Genetic structure and molecular variability of Spodoptera frugiperda (Lepidoptera: Noctuidae) collected in maize and cotton fields in Brazil. Bull Entomol Res 97:225–231. doi:10.1017/S0007485307004944
Mitchell R (1973) Growth and population dynamics of a spider mite (Tetranychus urticae K., Acarina: Tetranychidae). Ecology 54:1349–1355. doi:10.2307/1934198
Morishita M (1992) Movement of two species of tetranychid mites (Acarina: Tetranychidae) from border vegetation to watermelon fields. Jap J Appl Entomol Zool 36:25–30 (in Japanese with English summary)
Morishita M (1997) Intercrop movement of the two-spotted spider mite, Tetranychus urticae Koch (Acari: Tetranychidae) from chrysanthemum to pea field. Jap J Appl Entomol Zool 41:33–38 (in Japanese with English summary)
Navajas MJ, Thistlewood HMA, Lagnel J et al (1998) Microsatellite sequences are under-represented in two mite genomes. Insect Mol Biol 7:249–256. doi:10.1111/j.1365-2583.1998.00066.x
Navajas M, Perrot-Minnot ML, Lagnel J et al (2002) Genetic structure of a greenhouse population of the spider mite Tetranychus urticae: spatio-temporal analysis with microsatellite markers. Insect Mol Biol 11:157–165. doi:10.1046/j.1365-2583.2002.00320.x
Nishimura S, Hinomoto N, Takafuji A (2003) Isolation, characterization, inheritance and linkage of microsatellite markers in Tetranychus kanzawai (Acari: Tetranychidae). Exp Appl Acarol 31:93–103. doi:10.1023/B:APPA.0000005128.70282.a4
Osakabe Mh, Komazaki S (1999) Laboratory experiments on a change in genetic structure with an increase of population density in the citrus red mite population, Panonychus citri (McGregor) (Acari: Tetranychidae). Appl Entomol Zool (Jpn) 34:413–420
Osakabe Mh, Goka K, Toda S et al (2005) Significance of habitat type for the genetic population structure of Panonychus citri (Acari: Tetranychidae). Exp Appl Acarol 36:25–40. doi:10.1007/s10493-005-1672-1
Osakabe Mh, Isobe H, Kasai A et al (2008) Aerodynamic advantages of upside down take-off for aerial dispersal in Tetranychus spider mites. Exp Appl Acarol 44:165–183. doi:10.1007/s10493-008-9141-2
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295. doi:10.1111/j.1471-8286.2005.01155.x
Pels B, Sabelis MW (1999) Local dynamics, overexploitation and predator dispersal in an acarine predator–prey system. Oikos 86:573–583. doi:10.2307/3546662
Raymond M, Rousset F (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Heredity 86:248–249
Rousset F (1997) Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145:1219–1228
Smitley DR, Kennedy GG (1985) Photo-oriented aerial dispersal behavior of Tetranychus urticae (Acari, Tetranychidae) enhances escape from the leaf surface. Ann Entomol Soc Am 78:609–614
Smitley DR, Kennedy GG (1988) Aerial dispersal of the tow-spotted spider mite (Tetranychus urticae) from field corn. Exp Appl Acarol 5:33–46. doi:10.1007/BF02053815
Smouse PE, Peakall R (1999) Spatial autocorrelation analysis of individual multiallele and multilocus genetic structure. Heredity 82:561–573. doi:10.1038/sj.hdy.6885180
Tsagkarakou A, Navajas M, Lagnel J et al (1997) Population structure in the spider mite Tetranychus urticae (Acari: Tetranychidae) from Crete based on multiple allozymes. Heredity 78:84–92. doi:10.1038/hdy.1997.10
Tsagkarakou A, Navajas M, Papaioannou-Souliotis P et al (1998) Gene flow among Tetranychus urticae (Acari: Tetranychidae) populations in Greece. Mol Ecol 7:71–79. doi:10.1046/j.1365-294x.1998.00305.x
Tsagkarakou A, Navajas M, Rousset F et al (1999) Genetic differentiation in Tetranychus urticae (Acari: Tetranychidae) from greenhouses in France. Exp Appl Acarol 23:365–378. doi:10.1023/A:1006293627880
Uesugi R, Osakabe Mh (2007) Isolation and characterization of microsatellite loci in the two-spotted spider mite, Tetranychus urticae (Acari: Tetranychidae). Mol Ecol Notes 7:290–292. doi:10.1111/j.1471-8286.2006.01583.x
Uesugi R, Kunimoto Y, Osakabe Mh (2009) The fine-scale genetic structure of the two-spotted spider mite in a commercial greenhouse. Exp Appl Acarol 47:99–109. doi:10.1007/s10493-008-9201-7
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evol Int J Org Evol 38:1358–1370. doi:10.2307/2408641
Wright S (1943) Isolation by distance. Genetics 28:114–138
Acknowledgments
This research was partially supported by a Grant-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology, Japan, a Grant-in-Aid from the twenty-first Century COE Program for Innovative Food and Environmental Studies Pioneered by Entomomimetic Sciences at Kyoto University, and a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (B; no. 2238-19, 2007).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Uesugi, R., Sasawaki, T. & Osakabe, M. Evidence of a high level of gene flow among apple trees in Tetranychus urticae . Exp Appl Acarol 49, 281–290 (2009). https://doi.org/10.1007/s10493-009-9267-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10493-009-9267-x