Abstract
A novel strain of Planctomycetes, designated JC670T, was isolated from a high altitude (~ 2900 m above sea level) soil sample collected from Garhwal region in the Western Himalaya. Colonies of this strain were observed to be light pink coloured with spherical to oval shaped cells having crateriform structures distributed all over the cell surface. The cells divide by budding. Strain JC670T was found to grow well at pH 7.0 and pH 8.0 and to tolerate up to 2% NaCl (w/v). MK6 was the only respiratory quinone identified. The major fatty acids of strain JC670T were identified as C18:1ω9c, C18:0 and C16:0, and phosphatidylcholine, two unidentified phospholipids and six unidentified lipids are present as the polar lipids. The polyamines putrescine and sym-homospermidine were detected. Strain JC670T shows high 16S rRNA gene sequence identity (95.4%) with Paludisphaera borealis PX4T. The draft genome size of strain JC670T is 7.97 Mb, with G + C content of 70.4 mol%. Based on phylogenetic analyses with the sequences of ninety-two core genes, low dDDH value (20.6%), low gANI (76.8%) and low AAI (69.1%) results, differential chemotaxonomic and physiological properties, strain JC670T (= KCTC 72850T = NBRC 114339T) is recognised as the type strain of a new species of the genus Paludisphaera, for which we propose the name Paludisphaera soli sp. nov.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The phylum Planctomycetes is an important member of the Planctomycetes/Verrucomicrobia/Chlamydiae (PVC) superphylum, which is well-known for its biotechnological relevance (Wagner and Horn 2006). Though its members were initially considered as an intermediate between prokaryotes and eukaryotes (McInerney et al. 2011) they have now been recognised as bacteria due to the presence of peptidoglycan cell walls and their cell structure is similar to that of Gram-stain-negative bacteria (Wiegand et al. 2018). Members of Planctomycetes are ubiquitous in distribution and are found in diverse habitats with most species reported from aquatic environments (Wiegand et al. 2018). Though members of Planctomycetes can be the abundant phylum in soil microbial communities (Borneman et al. 1996; Borneman and Triplett 1997; Neef et al. 1998), relatively few have been cultivated so far (Buckley et al. 2006; Dedysh and Ivanova 2019; Dedysh et al. 2020; Ivanova and Dedysh 2012; Wang et al. 2002), although progress has recently been made in describing taxa cultivated by Wiegand et al. (2018) such as Tautonia plasticadhaerens (Jogler et al. 2020).
Taxonomically, the phylum Planctomycetes is subdivided into three classes: Planctomycetia, Phycisphaerae and Candidatus Brocadiae (Wiegand et al. 2018). At present, class Planctomycetia contains the majority of the species in the phylum and consists of four orders with validly published names viz., Planctomycetales, Gemmatales, Pirellulales and Isosphaerales (Dedysh et al. 2020). The families Planctomycetaceae and Pirellulaceae contain the majority of the species with validly published names. The family Isosphaeraceae is currently represented by six genera (Paludisphaera, Tundrisphaera, Isosphaera, Aquisphaera, Singulisphaera and Tautonia), which are generally monospecific taxa except the genera Singulisphaera and Tautonia. Recently, one more member has been added to the genus Tautonia, i.e. T. plasticadhaerens (Jogler et al. 2020). Members of the family Isosphaeraceae are stalk-free planctomycetes with spherical cells that divide by budding and are known to colonise a wide range of habitats such as hot springs, peat moss and aquatic sediments (Kulichevskaya et al. 2016). Genomic attributes of the strains belonging to the phylum Planctomycetes indicate genome sizes of 3–12 Mb and a DNA G + C content of 40–71% (Wiegand et al. 2018). Within the family Isosophaeraceae, the genome size ranges from 5.4–10.4 Mb while DNA G + C content ranges from 62.2–71.1% (Bondoso et al. 2011; Wiegand et al. 2018; Jogler et al. 2020).
The genus Paludisphaera was described by Kulichevskaya et al. (2016) to accommodate a new planctomycete of the family Isosphaeraceae isolated from a boreal Sphagnum peatbog. So far, the genus Paludisphaera has a single species, namely Paludisphaera borealis. The species is characterised by bright-pink colonies, small (1.5–2.5 μm) spherical cells that occur singly, in pairs or in short chains containing up to 10 cells and reproduce by budding (Kulichevskaya et al. 2016). Genome-based investigation of P. borealis PX4T indicates high glycolytic properties and production of a diverse range of secondary metabolites (Ivanova et al. 2017).
In the present study, soil samples were collected from Gangotri, in Uttarkashi district of Uttarakhand state in India. Gangotri is located on the banks of the river Bhagirathi, the main tributary of the Ganga. Being part of the Great Himalayan range, the area exhibits a distinctive diversity of flora and fauna that are adapted to cold environment at high altitude. Several studies have reported the bacterial diversity from high altitude Gangotri soil (Baghel et al. 2005; Kumar et al. 2019) and the presence of taxonomically unclassified sequences indicates the untapped novel bacterial diversity of these ecosystems. During our survey of soil bacterial diversity, we isolated strain JC670T from a soil sample collected nearly 30 km from the town of Gangotri. Combining a polyphasic taxonomic approach together with genomic information, we conclude strain JC670T should be classified as the type strain of a new species of the genus Paludisphaera. To the best of our knowledge, this is the first report of planctomycetes from the Himalaya. This study not only extends the studies on collection of axenic strains of planctomycetes from the Indian subcontinent, their characterisation and description of novel taxa (Kumar et al. 2020a, b), but also represents one of the first attempts to isolate and perform polyphasic characterisation of a Planctomycetes strain of the family Isosphaeraceae from a soil sample.
Materials and methods
Home habitat and isolation of novel strain
Strain JC670T was isolated from a soil sample collected from the bank of the river Bhagirathi (2900 m above sea level, 31° 00′ 3.2″ N 78° 54′ 32.7″ E) at a depth of 10 cm from soil surface. The pH and salinity of the soil were 7.2 and 0.00 ppt, respectively. Soil samples were subjected to enrichment and cultivation in a medium containing (g l−1 in distilled water; pH 7.0): N-acetylglucosamine, 2.0; KH2PO4, 0.1; peptone, 0.1; yeast extract, 0.1; vitamin solution, 10 ml l−l; Hutner’s basal salts, 20 ml l−l prepared in distilled water. The antibiotics (g l−1) streptomycin, 0.4, ampicillin, 0.2 and cycloheximide, 0.025 were added to the media. Vitamin solution contained (mg l−1): vitamin B12, 0.2; biotin, 4; thiamine-HCl.2H2O, 10; Calcium pantothenate, 10; folic acid, 4.0; riboflavin, 10; nicotinamide, 10.0; p-aminobenzoic acid, 10; pyridoxine HCl, 20. Hunter’s basal salts contain (g l−1): nitrilotriacetic acid, 10; MgSO4.7H2O, 30; CaCl2.2H2O, 3.5; (NH4)6MoO7O24.4H2O, 0.01; FeSO4.7H2O, 0.1; and metals stock solution 50 ml. Metal stock solution contain (g l−1): Na-EDTA, 0.25; ZnSO4.7H2O, 1.1; FeSO4.7H2O, 0.5; MnSO4.H2O, 0.15; CuSO4.5H2O, 0.04; Co(NO3)2.6H2O, 0.025; Na2B4O7.10H2O, 0.018. The soil sample (100 mg) was mixed with 10 ml medium in a serum vial of 20 ml capacity and the vial was sealed with butylated rubber stoppers. The serum vial was then incubated for two months at 25 °C to enrich planctomycetes. After two months of incubation, a light pink globular bacterial colony was observed at the bottom of the serum vial. The pink globular colony was further streaked on an agar plate containing the same medium. After three weeks of incubation, pink colonies appeared along with white colonies on the agar plates. The pink colonies were purified through repeated streaking. Pure cultures were maintained on agar plates by repeated sub-culturing and preserved at 4 °C. Purified cultures were grown in the above medium without antibiotics, unless otherwise mentioned. The pink coloured culture isolated from the soil sample was designated strain JC670T.
DNA isolation, PCR, 16S rRNA gene sequencing and BLAST analysis
DNA was isolated from the pure cultures using a commercial DNA isolation kit (Nucleo-pore gDNA Fungal Bacterial Mini Kit, from M/s. Genetix Biotech Asia Pvt. Ltd, India) which was used for PCR amplification and genome sequencing. PCR was performed for 16S rRNA gene amplification using planctomycetes specific primers F40 (Kohler et al. 2008) and R1388 (Stackebrandt et al. 1993). Purified PCR products were sent to AgriGenomePvt. Ltd. (Kochi, India) for 16S rRNA gene sequencing.
Genomic information and in-silico metabolic characterisation
To further investigate the taxonomic position of strain JC670T, whole genome sequencing was carried out using an Illumina HiseqX10 platform and a paired end library generated with sequence coverage of 100x. Genome sequencing was outsourced to AgriGenome Pvt. Ltd, Kochi, India. De novo assembly was performed using Unicycler (Wick et al. 2017) assembly software. The default k-mer sizes were used for Unicycler assembly. Unicycler assembly was used for all further downstream analyses. The genome sequence was checked for any possible contamination using the ContEst service of EZBiocloud (https://www.ezbiocloud.net/tools/contest16s; Yoon et al. 2017) which yielded 16S rRNA gene sequence of only one organism indicating the draft genome sequence was free from sequences of other organisms.The genome was further annotated using the NCBI Prokaryotic Genome Annotation Pipeline and also through the online freely available RAST server (https://rast.theseed.org/FIG/rast.cgi) (Aziz et al. 2008). In silico metabolic characterisation of strain JC670T and P. borealis PX4T was carried out on the basis of its genome information through the online tool PATRIC 3.6.2 (https://www.patricbrc.org/) (Wattam et al. 2017) and using Interactive pathway explorer (iPath3) (https://pathways.embl.de/login.cgi) (Darzi et al. 2018). In silico identification for the presence of gene clusters responsible for the biosynthesis of secondary metabolites in strain JC670T and P. borealis PX4T were carried out using the online freely available tool antiSMASH5.1 (https://antismash.secondarymetabolites.org) (Blin et al. 2019).
OrthoANI score was calculated from the genome sequences using the online service in the EzBioCloud (https://www.ezbiocloud.net/tools/ani) (Yoon et al. 2017). dDDH estimate between strain JC670T and P. borealis PX4T was calculated based on the genome sequences using the Genome-to-Genome Distance Calculator (GGDC 2.1) online service (https://ggdc.dsmz.de/distcalc2.php) (Auch et al. 2010). Average amino acid identity (AAI) was calculated between the genomes of strains JC670T and P. borealis PX4T using the AAI calculator developed by the Konstantinidis lab (Rodriguez and Konstantinidis 2014).
Phylogenetic analysis
The 16S rRNA gene sequence of strain JC670T extracted from the genome using ContEst16S has a sequence length of 1524 nt and analysis of identity was performed using NCBI BLAST (Johnson et al. 2008). The full length 16S rRNA gene sequences of strain JC670T, P. borealis PX4T, clone sequences related to Paludisphaera sp. (obtained from NCBI) along with all other closely related type strains of Isosphaeraceae (obtained from EZBioCloud) were aligned using MUSCLE implemented in MEGA7.0 (Kumar et al. 2016) and the distances were calculated by using the Kimura 2-parameter (Kimura 1980) in a pairwise deletion procedure. Neighbor-joining (NJ), minimum evolution (ME) and maximum likelihood (ML) methods in the MEGA7 software were used to construct phylogenetic trees having Bootstraps of 1000 replication (Felsenstein 1985).
The genome sequence of P. borealis PX4T from the NCBI database (accession number: NZCP019082) was used. The phylogenomic tree was constructed using 92 core genes from publicly available genomes mainly representing the families Pirellulaceae, Planctomycetaceae and Isosphaeraceae. These 92 core genes were retrieved using the Up-to-date Bacterial Core Gene (UBCG) tool (Na et al. 2018). A concatenated sequence of 92 genes was used to construct the RAxML based phylogenomic tree. The tree was generated with random samples of 50% of the homology groups used for the main tree, in a process known as gene-wise jackknifing. Hundreds of these 50% gene-wise jackknife trees were made using RAxML, and the support values at the nodes indicate the number of times a particular branch was observed in the support tree.
Physiological analysis
For organic substrates and nitrogen source utilisation of strain JC670T, liquid basal medium was used as described by Bondoso et al. (2011) with slight modifications. Medium was supplemented with 0.05% w/v yeast extract. (NH4)2SO4 (0.1% w/v) and glucose (0.1% w/v) were used as nitrogen and organic carbon sources respectively, unless otherwise mentioned. For organic substrate utilisation, (NH4)2SO4 (0.1% w/v) was used as a nitrogen source and cell growth was tested with different organic substrates at a concentration of 0.1% (w/v). For nitrogen source utilisation, glucose (0.1% w/v) was used as a carbon source and cell growth was tested with different nitrogen substrates at 0.1% (w/v). To examine the hydrolysis of phytagel, strain JC670T was streaked onto the same medium solidified with 2% phytagel (Sigma-Aldrich) and incubated for 4 weeks (Kulichevskaya et al. 2016). The phytagel hydrolysing property was also checked on media supplemented with N-acetylglucosamine as a sole source of carbon and nitrogen. Both organic and nitrogen substrate utilisation were tested in test tubes (25 × 250 mm) containing 10 ml of liquid basal medium as described above. Requirement of vitamin B12 was tested by growing the cells in liquid media with and without vitamin B12 (0.02 mM). The requirement for vitamin B12 was confirmed by repeated transfer into media lacking vitamin B12. NaCl tolerance (0–7% w/v; final salt concentration in the media) was tested at 25 °C and at pH 7. The optimum temperature (5, 10, 15, 20, 25, 30, 35, 40, 45 °C) required for cell growth was tested at pH 7. pH tolerance (4.0, 5.0, 6.0, 7.0, 8.0, 9.0, 10.0) for cell growth of strain JC670T was tested at 25 °C in buffered broth, as described earlier (Bondoso et al. 2015). Nitrate reduction was tested using conventional methods (MacFaddin et al. 1985). Enzymatic activities were assayed using the API ZYM kit (Biomerieux, France) following the manufacturer’s protocol.
Chemotaxonomic characterisation
For fatty acid analysis, growing cells were harvested by centrifugation (10,000 g for 15 min at 4 °C) at a cell density of 70% of the maximum optical density (100% = 0.9 OD660). Cellular fatty acids were methylated, separated and identified according to the instructions of the Microbial Identification System (Microbial ID; MIDI 6.0 version; method, RTSBA6) (Sasser 1990), which was carried out by Royal Research Labs, Secunderabad, India. Polar lipids of strain JC670T were extracted, separated and characterised as described previously (Kates 1972; Oren et al. 1996). Quinones were extracted with a chloroform/methanol (2:1, v/v) mixture, purified by TLC and analysed by HPLC (Imhoff et al. 1984). Polyamines of strain JC670T were extracted and identified according to a recent method (Kumar et al. 2020b).
Microscopy
Cell morphology, cell size, cell shape and cell division were observed under transmission and scanning electron microscope (Carl Zeiss LSM880 /Philips XL3O). Transmission electron microscopy (H-7500 Hitachi) was used to observe cross sections of the cells.
Results and discussion
Phylogenetic inference
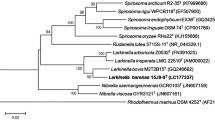
The 16S rRNA gene sequence of strain JC670T extracted from the draft genome has a sequence length of 1524 nt and a sequence similarity of 95.4% with that of P. borealis PX4T as determined by BLAST and ClustalW analyses. The 16S rRNA gene sequence based phylogenetic tree with combined bootstrap values obtained from NJ, ME, ML trees (Fig. 1) and 92 core genes based phylogenomic tree (Fig. 2) confirmed the clustering of strain JC670T with the type species of the genus Paludisphaera, P. borealis.
Phylogenetic tree based on 16S rRNA gene sequences showing the phylogenetic relationship of strain JC670T with P. borealis PX4T and other closely related species. The tree was made with MEGA 7 software and Gemmata obscuriglobus DSM 5831 T was used as out-group. The GenBank accession numbers for 16S rRNA gene sequences are shown in parentheses. Numbers at nodes indicate Bootstrap values from 1000 repetitions corresponding in the NJ/ME/ML analysis. Bar, 0.02 nucleotide substitution per position. *Clone sequences; $Axenic culture
POCP (Table S2) and AAI (Table S1) values between the strain JC670T and P. borealis PX4T were 67.3% and 69.1%, respectively. A POCP threshold below 50% was proposed to determine new genera (Qin et al. 2014) and the value of 67.3% observed between the two strains clearly indicates that they belong to the same genus. Further, AAI value of 69.1% between the strains fell close to the recommended cut-off of 80% used for the genus delineation (Rodriguez and Konstantinidis 2016). The OrthoANI value between JC670T and P. borealis PX4T was 76.8%. The dDDH value between strain JC670T and P. borealis PX4T was 20.6%. The ANI and dDDH values between the strain JC670T and P. borealis PX4T are far below the recommended cut off values (95–96% cut off for ANI and 70% cut off for dDDH) for prokaryotic species delineation (Meier-Kolthoff et al. 2014; Chun et al. 2018). Thus, strain JC670T is concluded to represent a novel species of the genus Paludisphaera.
Genomic characteristics
Based on the NCBI Prokaryotic Genome Annotation Pipeline, the genome size of strain JC670T is 7.97 Mb with N50 value of 217, 617, while the genome size of P. borealis PX4T is 7.65 Mb. The genome of strain JC670T has 6,453 predicted genes of which 6,213 are protein coding genes, 3 rRNA operons, 55 tRNA coding and179 genes are pseudogenes; and no CRISPR repeats were found. The genome of P. borealis PX4T was predicted to contain a total of 5877 genes of which 5642 are protein coding, 150 pseudogenes, 73 tRNA genes, 9 rRNA operons, 3 genes coding for other RNAs and one CRISPR repeat.
The difference (5.8 mol%) in G + C content between strain JC670T and P. borealis PX4T (Table S3) further indicates that these two are distinct species since strains of a species will have less than 3% difference in the G + C content (Meier-Kolthoff et al. 2014). The genome sequences of strain JC670T and P. borealis PX4T were aligned using PATRIC software to identify the multiple maximal matches and local collinear blocks. The genome sequence of strain JC670T was used as a reference against P. borealis PX4T. The alignments of the local collinear blocks in both strains differ from each other significantly (Fig. S1). The alignment facilitates detection of homologous regions that are shuffled or inverted due to DNA rearrangement or recombination. Comparison of the genome map based on the protein sequence identity shows that majority of the proteins of strain JC670T share 30–90% similarity with those of P. borealis PX4T (Fig. S2). Both strain JC670T and P. borealis PX4T share 10–20% protein sequence similarity with other members of the family Isosphaeraceae (Fig. S2). These results suggest a clear dissimilarity between strain JC670T with P. borealis PX4T and between the two strains and other taxa of the family Isosphaeraceae.
In silico metabolic characterisation
In silico metabolic characterisation showed that strain JC670T and P. borealis PX4T have the 2-C-methyl-D-erythritol 4-phosphate/1-deoxy-D-xylulose 5-phosphate pathway for the biosynthesis of five carbon isoprene units (isopentenyl pyrophosphate), which is eventually the precursor for the synthesis of carotenoids and quinones (Eisenreich et al. 2004; Zhao et al. 2013). Genes encoding for carotenoid biosynthetic enzymes such as lycopene β-cyclase (5.5.1.19), β-carotene hydroxylase (1.14.13.129), zeaxanthin epoxidase (1.14.15.21) and neoxanthin synthase (5.3.99.9) were found in both the strains. Hence, both strains are putatively capable of forming neoxanthin from lycopene via intermediates such as γ-carotene, β-carotene, zeaxanthin and violaxanthin. Gene clusters for the biosynthesis of neomycin, puromycin, kanamycin, tetracycline, novobiocin and gentamycin antibiotics are present in both of the strains. The streptomycin biosynthetic pathway gene cluster was exclusively predicted in P. borealis PX4T. Both strains are capable of purine and pyrimidine metabolism. P. borealis PX4T is putatively capable of degradation and metabolism of xenobiotic compounds such as xylene, atrazine, benzoate, toluene, ethyl benzene and caprolactam (Fig. S3). Due to the complex life cycle of planctomycetes, they are believed to produce bioactive secondary metabolites (Wiegand et al. 2020). There were six putative secondary metabolite gene clusters predicted in strain JC670T and P. borealis PX4T. Both strains have gene clusters for mixed heterocyst-glycolipid synthase-like and Type I polyketide synthases (Mixed hgIE-KS-Type I PKS) and terpenes biosynthesis. Type I and Type III polyketide synthase gene clusters were exclusively predicted in strain JC670T.
Morphological and physiological analyses
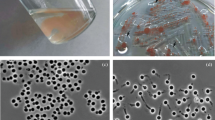
SEM imaging showed that cells of strain JC670T are present singly or in tissue-like aggregates (Fig. 3a). Cells of strain JC670T were observed to be spherical to oval shaped (1.6–1.7 × 1.3–1.5 μm; Fig. 3b), having crateriform structures distributed all over the surface. TEM images of JC670T cells showed the presence of inner and outer membranes, invaginations of the cytoplasmic membrane, inclusion bodies, cytoplasm and nucleoid region (Fig. 3c). Cells of strain JC670T multiply by budding and the daughter cells are smaller than the parent cells (Fig. 3d).
Scanning (a, b) and transmission (c, d) electron micrographs of cell of strain JC670T. a Spherical cells occur singly or in tissue-like aggregates. Bar, 2 μm. b Cells having crateriform structures (CR). Bar, 1 μm. c Ultrathin section showing invagination of cytoplasmic membrane (ICM), Inner membrane (IM), outer membrane (OM), inclusions (IN), cytoplasm (CP) and nucleoid region (N). d Cells of strain JC670T multiply by budding (BD) wherein daughter cells are smaller than parent cells
NaCl is not essential for growth of strain JC670T, which tolerates up to 2% (w/v) NaCl with an optimum at 0%. The optimum temperature for the growth of strain JC670T was determined to be 22–25 °C and no cell growth occurs above 30 °C or below 4 °C. The optimum pH for the growth of strain JC670T was found to be 7.0 and cell growth was observed at pH 7.0 and pH 8.0. Strain JC670T is unable to reduce nitrate and vitamin B12 is not required for its growth. Growth observations with different organic substrates/nitrogen sources of strain JC670T are given in the species description and Table 1.
Enzymatic activities determined for strain JC670T using the API ZYM kit indicate positive results for alkaline phosphatase, esterase (C4), esterase lipase (C8), leucine arylamidase, valine arylamidase, acid phosphatase and naphthol-AS-BI-phosphohydrolase, with negative results for lipase (C14), cysteine arylamidase, trypsin, α-chymotrypsin, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase and α-fucosidase. Strain JC670T can hydrolyse phytagel (Fig. S4) only in the absence of N-acetylglucosamine in the medium, as also observed previously for P. borealis (Kulichevskaya et al. 2016).
Chemotaxonomic characterisation
The major fatty acids of strain JC670T were identified as C18:1ω9c, C16:0 and C18:0 (Table S4). The polar lipids of strain JC670T were found to include phosphatidylcholine, two unidentified phospholipids and six unidentified lipids (Fig. S5). In addition to phosphatidylcholine, trimethylornithine lipids were observed to be abundant in P. borealis (Kulichevskaya et al. 2016) but these were not detected in strain JC670T. MK6 is the predominant quinone of strain JC670T and polyamines are putrescine and sym-homospermidine (Fig. S6).
Proposal of Paludisphaera soli sp. nov.
Strain JC670T shows similarity with P. borealis PX4T with respect to cell structure, cell arrangement, quinone content, fatty acids and nitrate reduction. Additionally, vitamin B12 is not required for growth of either strain. However, strain JC670T differs from P. borealis PX4T in polar lipid composition, some enzyme activities and utilisation of some carbon and nitrogen sources for growth (Table 1). The phenotypic differences are supported by the molecular differences (16S rRNA gene sequence analysis) and genome relatedness (gANI/AAI, dDDH) values. Cumulatively these differences of strain JC670T with respect to P. borealis PX4T differentiate it into a new species of the genus Paludisphaera, for which we propose the name Paludisphaera soli sp. nov.
Description of Paludisphaera soli sp. nov.
Paludisphaera soli (so'li. L. neut. gen. n. soli of soil, referring to the isolation of the type strain from a soil sample).
Cultures are light pink in colour and strictly aerobic. Cells are spherical to oval shaped and multiply by budding. Non-motile. NaCl is not essential for growth and is tolerated up to 2% (w/v) with optimum at 0%. Vitamin B12 is not required for growth. Utilises α-D-glucose, sucrose, pyruvate, propionate, fructose, D-galactose, mannose, D-xylose and rhamnose as carbon source for growth while starch, ascorbate, acetate, fumarate, mannitol, lactose, inositol, maltose, malic acid, inulin, succinate, sorbitol, benzoic acid or citrate do not support growth. Ammonium sulphate, L-arginine, DL-alanine, L-glycine, L-phenylalanine, L-lysine, L-glutamine, L-proline, L-isoleucine, L-leucine-ornithine, DL-threonine, L-serine, L-tyrosine, Peptone, L-casamino acid, yeast extract, sodium nitrate are used as nitrogen sources for growth. Cysteine, L-methionine, L-aspartic acid, L-tryptophan, L-histidine, L-glutamic acid and urea do not support growth as nitrogen sources. Hydrolyses phytagel. C18:1ω9c, C16:0 andC18:0 are the major fatty acids. MK6 is the only respiratory quinone. Phosphatidylcholine, two unidentified phospholipids and six unidentified lipids are present in the polar lipids. Polyamines are putrescine and sym-homospermidine. The G + C content of the genomic DNA of the type strain is 70.4 mol%.
The type strain JC670T (= KCTC 72850 T = NBRC 114339 T) was isolated from a soil sample from Gangotri region in Uttarakhand, India. The GenBank accession number for the 16S rRNA gene sequence of strain JC670T is LR794334 and the genome (7.97 Mb) sequence has been deposited in GenBank under the accession number JAALJI000000000. The version described in this paper is version JAALJI010000000.
Abbreviations
- NCBI:
-
National Center for Biotechnology Information
- ANI:
-
Average Nucleotide Index
- AAI:
-
Average Nucleotide Index
- dDDH:
-
Digital DNA-DNA Hybridization
- HPLC:
-
High-Pressure Liquid Chromatography
- KCTC:
-
Korean Collection for Type Cultures
References
Auch AF, Klenk HP, Göker M (2010) Standard operating procedure for calculating genome-to-genome distances based on high-scoring segment pairs. Stand Genomic Sci 2:142–148
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M et al (2008) The RAST server: rapid annotations using subsystems technology. BMC genomics 9:75
Baghel VS, Tripathi RD, Ramteke PW, Gopal K, Dwivedi S, Jain RK, Rai UN, Singh SN (2005) Psychrotrophic proteolytic bacteria from cold environment of Gangotri glacier, Western Himalaya, India. Enzyme Microb Tech 36:654–659
Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Lee SY, Medema MH, Weber T (2019) antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res 47:W81–W87
Bondoso J, Albuquerque L, Nobre MF, Lobo-da-Cunha A, da Costa MS, Lage OM (2011) Aquisphaera giovannonii gen. nov., sp. nov., a planctomycete isolated from a freshwater aquarium. Int J Syst Evol Microbiol 61:2844–2850
Bondoso J, Albuquerque L, Nobre MF, Lobo-da-Cunha A, da Costa MS, Lage OM (2015) Roseimaritima ulvae gen. nov., sp. nov. and Rubripirellula obstinate gen. nov., sp. nov. two novel planctomycetes isolated from the epiphytic community of macroalgae. Syst Appl Microbiol 38:8–15
Borneman J, Skroch PW, O'Sullivan KM, Palus JA, Rumjanek NG, Jansen JL, Nienhuis J, Triplett EW (1996) Molecular microbial diversity of an agricultural soil in Wisconsin. Appl Environ Microbiol 62:1935–1943
Borneman J, Triplett EW (1997) Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl Environ Microbiol 63:2647–2653
Buckley DH, Huangyutitham V, Nelson TA, Rumberger A, Thies JE (2006) Diversity of planctomycetes in soil in relation to soil history and environmental heterogeneity. Appl Environ Microbiol 72:4522–4531
Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, da Costa MS, Rooney AP, Yi H, Xu XW, Meyer SD, Trujillo ME (2018) Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68:461–466
Darzi Y, Letunic I, Bork P, Yamada T (2018) iPath3. 0: interactive pathways explorer v3. Nucleic Acids Res 46:W510–W513
Dedysh SN, Ivanova AA (2019) Planctomycetes in boreal and subarctic wetlands: diversity patterns and potential ecological functions. FEMS Microbiol Eco 95:fiy227
Dedysh SN, Kulichevskaya IS, Beletsky AV, Ivanova AA, Rijpstra WIC, Damsté JSS, Ravin NV (2020) Lacipirellula parvulagen. nov., sp. nov., representing a lineage of planctomycetes widespread in low-oxygen habitats, description of the family Lacipirellulaceae fam. nov. and proposal of the orders Pirellulalesord. nov., Gemmatalesord. nov. and Isosphaerales ord. nov. Syst Appl Microbiol 43:126050
Eisenreich W, Bacher A, Arigoni D, Rohdich F (2004) Biosynthesis of isoprenoids via the non-mevalonate pathway. Cell Mol Life Sci 61:1401–1426
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Imhoff JF (1984) Quinones of phototrophic purple bacteria. FEMS Microbiol Lett 25:85–89
Ivanova AO, Dedysh SN (2012) Abundance, diversity, and depth distribution of planctomycetes in acidic northern wetlands. Front Microbiol 3:5
Ivanova AA, Naumoff DG, Miroshnikov KK, Liesack W, Dedysh SN (2017) Comparative genomics of four Isosphaeraceae planctomycetes: a common pool of plasmids and glycoside hydrolase genes shared by Paludisphaera borealis PX4T, Isosphaera pallida IS1BT, Singulisphaera acidiphila DSM 18658T, and strain SH-PL62. Front Microbiol 8:412
Jogler C, Wiegand S, Boedeker C, Heuer A, Peeters SH, Jogler M, Jetten MSM, Rohde M, Kallscheuer N (2020) Tautonia plasticadhaerens sp. nov., a novel species in the family Isosphaeraceae isolated from an alga in a hydrothermal area of the Eolian Archipelago. Antonie van Leeuwenhoek. https://doi.org/10.1007/s10482-020-01424-3
Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL (2008) NCBI BLAST: a better web interface. Nucleic Acids Res 36:W5–W9
Kates M (1972) Isolation, analysis and identification of lipids. Tech Lipidol 3:268–618
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Köhler T, Stingl U, Meuser K, Brune A (2008) Novel lineages of Planctomycetes densely colonize the alkaline gut of soil-feeding termites (Cubitermes spp.). Environ Microbiol 10:1260–1270
Kulichevskaya IS, Ivanova AA, Suzina NE, Rijpstra WIC, Damste JSS, Dedysh SN (2016) Paludisphaera borealis gen. nov., sp. nov., a hydrolytic planctomycete from northern wetlands, and proposal of Isosphaeraceae fam. nov. Int J Syst Evol Microbiol 66:837–844
Kumar D, Gaurav K, Uppada J, Deepshikha G, Ch S, Ch VR (2020a) Roseimaritima sediminicola sp. nov., a new member of Planctomycetaceae isolated from Chilika lagoon. Int J Syst Evol Microbiol 70:2616–2623
Kumar D, Gaurav K, Sreya PK, Shabbir A, Uppada J, Sasikala Ch, Ramana CV (2020b) Gimesia chilikensis sp. nov., a haloalkali-tolerant planctomycete isolated from Chilika lagoon and emended description of the genus Gimesia. Int J Syst Evol Microbiol 70:3647–3655
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Kumar S, Suyal DC, Yadav A, Shouche Y, Goel R (2019) Microbial diversity and soil physiochemical characteristic of higher altitude. PLoS ONE 14:e0213844
Mac Faddin JF (1985) Media for isolation-cultivation-identification-maintenance of medical bacteria. Williams & Wilkins, Philadelphia
Mc Inerney JO, Martin WF, Koonin EV, Allen JF, Galperin MY, Lane N, Embley AJM, TM, (2011) Planctomycetes and eukaryotes: a case of analogy not homology. BioEssays 33:810–817
Meier-Kolthoff JP, Klenk HP, Göker M (2014) Taxonomic use of DNA G+ C content and DNA–DNA hybridization in the genomic age. Int J Syst Evol Microbiol 64:352–356
Na SI, Kim YO, Yoon SH, Ha SM, Baek I, Chun J (2018) UBCG: Up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J Microbiol 56:280–285
Neef A, Amann R, Schlesner H, Schleifer KH (1998) Monitoring a widespread bacterial group: in situ detection of planctomycetes with 16S rRNA-targeted probes. Microbiology 144:3257–3266
Oren A, Duker S, Ritter S (1996) The polar lipid composition of Walsby's square bacterium. FEMS Microbiol Lett 138:135–140
Qin QL, Xie BB, Zhang XY, Chen XL, Zhou BC, Zhou J, Oren A, Zhang YZ (2014) A proposed genus boundary for the prokaryotes based on genomic insights. J Bacteriol Res 196:2210–2215
Rodriguez RLM, Konstantinidis KT (2014) Bypassing cultivation to identify bacterial species. Microbe 9:111–118
Rodriguez-R LM, Konstantinidis KT (2016) The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes (No. e1900v1). Peer J Preprints
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids.
Stackebrandt E, Liesack W, Goebel BM (1993) Bacterial diversity in a soil sample from a subtropical Australian environment as determined by 16S rDNA analysis. FASEB J 7:232–236
Wagner M, Horn M (2006) The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr Opin Biotechnol 17:241–249
Wang J, Jenkins C, Webb RI, Fuerst JA (2002) Isolation of Gemmata-like and Isosphaera-like planctomycete bacteria from soil and freshwater. Appl Environ Microbiol 68:417–422
Wattam AR, Davis JJ, Assaf R, Boisvert S, Brettin T, Bun C, Conrad N, Dietrich EM, Disz T, Gabbard JL, Gerdes S, Henry CS, Kenyon RW et al (2017) Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res 45:D535–D542
Wick RR, Judd LM, Gorrie CL, Holt KE (2017) Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PloS Comput Biol 13:e1005595
Wiegand S, Jogler M, Jogler C (2018) On the maverick Planctomycetes. FEMS Microbiol Rev 42:739–760
Wiegand S, Jogler M, Boedeker C, Pinto D, Vollmers J, Rivas-Marín E, Oberbeckmann S et al (2020) Cultivation and functional characterization of 79 Planctomycetes uncovers their unique biology. Nat Microbiol 5:126–140
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613
Zhao L, Chang WC, Xiao Y, Liu HW, Liu P (2013) Methylerythritol phosphate pathway of isoprenoid biosynthesis. Annu Rev Biochem 82:497–530
Acknowledgements
MS thanks CSIR, RK and KG thank the DBT, New Delhi and JU thanks TEQIP III for the award for research fellowships. Ramana thanks the Department of Biotechnology, Government of India for the award of Tata Innovation fellowship. Sasikala thanks UGC for Mid-Career Award. We thank the staff and the laboratory facilities provided by Centre for Interdisciplinary Studies of Mountain & Hill Environment, University of Delhi.
Funding
This work received grant specifically from Department of Biotechnology, Government of India. Financial support received from Council for Scientific and Industrial Research (CSIR), and AICTE, New Delhi is acknowledged. Infrastructure facilities funded by DST-FIST, UGC-SAP (DRS) and TEQIP, AICTE-RPS and MODROBS are acknowledged. The financial assistance of DST-PURSE Grant to MKP is gratefully acknowledged.
Author information
Authors and Affiliations
Contributions
RK and MS performed sample collection. RK, MS and KG isolated the strain and performed the initial cultivation, strain deposition and strain characterisation, KG performed the electron microscopic analysis and media optimisation, JU performed the genomic and phylogenetic analysis, SA performed and analysed the data for polyamines, RK, MS and KG wrote the manuscript, Ramana, Sasikala and MKP supervised the study and contributed to text preparation and revised the manuscript. All authors read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Ethical approval
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kaushik, R., Sharma, M., Gaurav, K. et al. Paludisphaera soli sp. nov., a new member of the family Isosphaeraceae isolated from high altitude soil in the Western Himalaya. Antonie van Leeuwenhoek 113, 1663–1674 (2020). https://doi.org/10.1007/s10482-020-01471-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-020-01471-w