Abstract
Endophytic microorganisms inhabit internal plant tissues in the host plant without causing any symptoms or negative effects. Although the diversity of endophytes has been evaluated by both culture-dependent and culture-independent methods, less information is available on yeast communities. Therefore, in this study a culture-independent method was used to examine endophytic yeasts associated with rice leaves based on the large subunit of ribosomal DNA using a semi-nested PCR technique. Sequence analysis indicated that the colonization frequency and the relative species frequency (RF) of endophytic yeast phylotypes were 0.41 and 0.06, respectively, and the majority of the yeast phylotypes were basidiomycetous yeasts. The phylotypes were designated as five known species (Cryptococcus victoriae, Debaryomyces hansenii, Debaryomyces vindobonensis, Meyerozyma guilliermondii and Pseudozyma antarctica), together with seventeen phylotypes closest to Candida metapsilosis, Cryp. foliicola, Cryp. laurentii, Pseudozyma abaconensis, Pseudozyma aphidis and Trichosporon asahii, among which some could be novel species. The most prevalent phylotypes were those closest to Cryp. foliicola (47.5 % RF) followed by D. hansenii (22.8 % RF) and P. antarctica (16.8 % RF). The presence of the phylotypes related to species known for their potential applications as biocontrol agents and plant growth promoting hormone producers suggests that they may have valuable applications. In addition, our findings revealed the occurrence of novel phylotypes at high frequency, which should encourage extensive studies to discover novel yeast species and to understand their roles in the rice leaves.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Endophytic microorganisms comprising bacteria, filamentous fungi and yeasts inhabit internal tissues of the host plant without causing any symptoms or negative effects (Petrini 1991). Endophytes have been receiving attention due to their ability to produce various beneficial bioactive compounds which are involved in plant health and adaptation to both abiotic and biotic stress and thus have potential agricultural, industrial and pharmaceutical applications (Trindade et al. 2002; Arnold et al. 2003; Tian et al. 2004; Higginbotham et al. 2013). At present it is commonly accepted that almost every plant species is inhabited by endophytic fungi and that more than one type of fungal endophyte may be found within a single plant (Petrini et al. 1992). Endophytes associated with crop plants, including rice have been reported. The reports have focused on bacterial and filamentous fungal community structures and their biological activities, but only a few reports have described the isolation, localization or diversity of endophytic yeasts. For instance, Fusarium and Streptomyces were respectively predominant endophytic fungi and actinomycetes in rice leaves and roots from Guangdong province in southern China while yeasts were detected only in leaves and only at low frequency (3.7 %); no yeasts were detected in roots (Tian et al. 2004). Yuan et al. (2010) characterized the fungal communities in the roots of rare wild rice (Oryza granulate) using both culture-dependent and culture-independent approaches. The results demonstrated that most of the fungal isolates belonged to Ascomycota with 34.5 % being of undescribed species, whereas the frequently detected clones were classified as Basidiomycota, 60.2 % being of unknown taxa. Among the detected clones, only one yeast phylotype was found; this was closely related to Trichosporon mucoides. To date endophytic yeasts have been isolated from a few plant species; for example Cryptococcus sp. and Rhodotorula sp. in leaves of tomato (Larran et al. 2001), Cyberlindnera saturnus, producing indole-3-acetic acid (IAA) and indole-3-pyruvic acid (IPYA), in roots of maize (Nassar et al. 2005), Rhodotorula graminis and Rhodotorula mucilaginosa, producing IAA, in stems of wild cottonwood and hybrid poplar (Xin et al. 2009).
Although microbial strains have been isolated by conventional methods, it has not been possible to isolate numerous microorganisms in the environment due to the fact that their cultivation conditions are unknown and that there is a possibility that some species might be unculturable. Culture-independent methods are increasingly employed to analyze the microbial diversity directly from tissue or environmental samples via DNA extraction, along with PCR amplification of nuclear ribosomal gene (rDNA) and separation techniques such as random cloning, amplified rDNA restriction analysis (ARDRA), amplified ribosomal intergenic spacer analysis (ARISA), denaturing gradient gel electrophoresis (DGGE), temperature gradient gel electrophoresis (TGGE), and terminal restriction fragment length polymorphism (T-RFLP) (Anderson and Cairney 2004; Nielsen et al. 2007; Yuan et al. 2010; Singh et al. 2011). These techniques are subject to biases and their own limitations because of the DNA quality, the primers used, copy numbers of the target gene and the length of amplified products (Jones and Richards 2011). To reduce biases, several primers have been developed as universal primers (Toju et al. 2012) and specific primers (White et al. 1990; Gardes and Bruns 1993; Harrison et al. 2011) together with the optimization of amplification programs such as nested PCR or semi-nested PCR which increases the sensitivity and/or specificity of PCR (Ibeas et al. 1996; Oros-Sichler et al. 2006; Chae et al. 2012; Díaz et al. 2013).
Rice is a major grain food crop and its production for both local consumption and export is the main agricultural activity in Thailand. Aware that endophytes possess beneficial biological properties and that attention has not yet been focused on endophytic yeasts, we aimed to evaluate the biodiversity of endophytic yeast in rice leaves using a culture-independent approach via semi-nested PCR for construction of a D1/D2 library followed by ARDRA and sequencing.
Materials and methods
Sample collection
Forty-six samples of healthy leaves of rice (Oryza sativa) were collected from rice fields in five provinces in Thailand between October 2011 and December 2011 (Table 1). Leaf samples were kept in ice boxes during collection and then at 4 °C in the laboratory. DNA extraction was conducted within 24 h after returning to the laboratory and not longer than 1 week after collection.
DNA extraction from leaf samples
The leaves were cut into fragments (3 cm) and surface-sterilized. The leaf fragments were submerged in 70 % (w/v) ethanol for 2.5 min, followed by 5 % (w/v) sodium hypochlorite for 2.5 min, and then washed 5 times by soaking in 50 ml sterile water and shaking for 2 min. The effectiveness of the surface sterilization procedure was verified by spreading 0.1 ml of the final rinse water onto yeast extract-malt extract (YM) agar (3 g l−1 yeast extract, 3 g l−1 malt extract, 5 g l−1 peptone, 10 g l−1 glucose and 20 g l−1 agar) and placing of a few leaf fragments directly onto YM agar. Furthermore, the efficiency of the removal of DNA from leaf surfaces was examined by the semi-nested PCR amplification (described below in PCR Amplification of D1/D2 domain of large subunit ribosomal DNA section) using DNA extracted from the pellets collected from the rinse water as well as washed leaves and unwashed leaves, that were submerged in washing buffer (1X phosphate buffered saline, Tween 20), and sonicated for 7 min using an ultrasonic bath (Yang et al. 2001), as DNA template. The quantity of the amplified D1/D2 fragment was calculated based on the approximate mass of 100 bp DNA ladder (NEB, USA).
Total genomic DNA from fresh surface-sterilized rice leaves was extracted according to the method of Kurtzman and Robnett (1998) with modification. The leaf sample was ground with mortar and pestle under liquid nitrogen and then treated with 2 × CTAB extraction buffer (2 % (w/v) CTAB, 100 mM Tris–HCl pH 8.4, 1.4 M NaCl, 25 mM EDTA) at 65 °C for 1 h with occasional gentle swirling. DNA was extracted with phenol:chloroform:isoamyl alcohol (25:24:1), followed by chloroform:isoamyl alcohol (24:1). Then, DNA was precipitated by adding 0.54 volume of isopropanol and washed once with 70 % (w/v) ethanol and finally resuspended in 100 μl of sterile distilled water.
PCR Amplification of D1/D2 domain of the large subunit ribosomal DNA
The total genomic DNA extracted from rice leaves was used as a DNA template for the amplification of the D1/D2 domain of the large subunit ribosomal DNA (LSU rDNA) with primers, NL1 and NL4 (Kurtzman and Robnett 1998). However, considerable amplification of rice LSU rDNA was detected. To exclude this disturbance, semi-nested PCR was carried out. Forward primers for amplifying the ITS-D1/D2 domain as an outer fragment were sought out from fungal ITS primers previously published, NSI1 (Martin and Rygiewicz 2005), ITS1-F (Gardes and Bruns 1993), ITS1-F_KYO1 and ITS1-F_KYO2 (Toju et al. 2012). ITS1-F_KYO1 was selected due to its amplification performance in vitro as well as the high percentage of fungal taxa and low percentage of plant taxa amplified in silico (Toju et al. 2012). The primer pairs used for semi-nested PCR were ITS1-F_KYO1 and NL4 as the outer set to amplify the ITS-D1/D2 domain (~1300 bp) and NL1/NL4 as the inner set to amplify only the D1/D2 domain (~600 bp). Although the ITS-D1/D2 domain contained the D1/D2 domain, the amplified D1/D2 domain was used to construct the clone library due to its smaller size and higher yield. The sequences of the primers used are shown in Table 2. Amplification of the ITS-D1/D2 domain was carried out in 25 μl of a reaction mixture containing 50 ng of genomic DNA, 1X PCR buffer, 1X Q-solution, 200 μM of each dNTP, 10 pmol of each primer of the outer set, and 2.5 U of Taq polymerase (Qiagen, Germany). The reaction was performed as follows: initial DNA denaturation at 94 °C for 5 min, followed by 45 cycles of denaturation at 94 °C for 1 min, annealing at 47 °C for 30 s and extension at 72 °C for 1 min and a final extension at 72 °C for 10 min. For the first amplification, 45 cycles were used to enhance the yield of PCR product. Amplification of the D1/D2 domain was carried out in 25 μl of a reaction mixture containing 1/25 dilution of the ITS-D1/D2 PCR product, 1X PCR buffer, 1X Q-solution, 200 μM of each dNTP, 10 pmol of each primer of the inner set, and 2.5 U of Taq polymerase (Qiagen, Germany). The reaction was performed as follows: initial DNA denaturation at 94 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 1 min, annealing at 52 °C for 30 s and extension at 72 °C for 30 s and a final extension at 72 °C for 10 min. Visualization of the amplified DNA was performed by electrophoresis using 0.8 % agarose in 1X TBE buffer (0.09 M Tris–borate, 0.001 M EDTA; pH 8.0); it was then stained with ethidium bromide and observed under a UV illuminator. The expected amplified DNA fragment (~600 bp) was excised and purified with QIAquick® Gel Extraction Kit (Qiagen, Germany) according to the manufacturer’s instructions.
Construction of clone libraries and phylogenetic analysis
The purified PCR products were ligated into pGEM-T ®easy (Promega, USA) and used to transform into Escherichia coli JM109. The randomly selected transformants containing the insert were confirmed by the colony PCR method with NL1 and NL4 primers and screened by amplified ribosomal DNA restriction analysis (ARDRA). PCR products having the correct size of insert were digested with the restriction enzymes, CfoI, HaeIII, and HinfI (Guillamón et al. 1998) and then separated in 2 % agarose gel electrophoresis for 45 min. Clones were grouped into operational taxonomic units (OTUs) according to the RFLP pattern and the representative OTUs were sequenced by First Base Laboratories Sdn Bhd (Malaysia) with NL1 primer using BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, USA). The sequences of the D1/D2 domain of the LSU rDNA were submitted to BLASTn search (Altschul et al. 1997) at the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/) to determine their similarity and phylogenetic affiliation. The sequences were aligned with related species by Multiple Alignment using Fast Fourier Transform (MAFFT) (Katoh and Standley 2013) at the European Bioinformatics Institute (EBI; http://www.ebi.ac.uk/) and the phylogenetic tree was constructed from the evolutionary distance data according to Kimura (1980) by the maximum likelihood method (Felsenstein 1981) using the MEGA version 6.0 (Tamura et al. 2013) with 1000 bootstrap replicates.
Nucleotide sequence accession number
The nucleotide sequence data reported in this work were deposited in GenBank under the accession numbers KM872031 to KM872060.
Data analyses
The colonization frequency (CF) was calculated as number of leaf samples colonized by endophytic yeasts divided by the number of total leaf samples collected. The relative frequency (RF) was calculated as the number of clones in each yeast phylotype, species, genus or phylum divided by the total number of clones in the library, depended on the group involved. Diversity of the clone library was investigated by rarefaction analysis and the rarefaction curve was calculated using the RarefactWin program (Holland 2003).
Results
Diversity of endophytic yeasts by a culture-independent approach
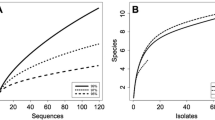
Diversity of the endophytic yeast species in rice leaves was investigated by a culture-independent method in this study. Prior to DNA extraction, the sterility of the washed leaf surface was verified and the results showed that there was no growth of microorganisms. In case of the removal of DNA on the leaf surface, the quantity of PCR products of DNA obtained from the rinse water, washed leaves and unwashed leaves showed a significant reduction of the epiphytic yeast DNA since the quantity of amplified band at each washing step reduced gradually from 100 % in unwashed leaves (R0) to 0 % in the final rinse water (R5) and washed leaves (R6) (Fig. 1). The results indicated that most of the epiphytic yeast DNA was removed from the washed leaves. Due to the long distance from the sampling sites to the laboratory, total DNA from rice leaf samples collected in the Central Region (n = 24), the Eastern Region (n = 5) and the Northeastern Region (n = 17) was extracted within 1, 3 and 5 days of collection. The extracted DNA was used as template for the amplification of the D1/D2 domain of the LSU rDNA of endophytic yeasts. The D1/D2 domain of the LSU rDNA was successfully amplified by semi-nested PCR using ITS1-F_KYO1 and NL4 as the outer set and NL1/NL4 as the inner set. Therefore, 46 D1/D2 libraries were constructed and a total of 1826 randomly picked clones were grouped into 124 operational taxonomic units (OTUs), according to the ARDRA patterns obtained from the independent digestion by three restriction endonucleases: CfoI, HaeIII and HinfI. Representative OTUs from each library were sequenced and analyzed. Through the BLAST analysis, 12 OTUs had sequences typical of yeasts (101clones), whereas 112 OTUs belonged to filamentous fungi (1725 clones) and no OTU matched the rice sequence. Moreover, the number of yeast phylotypes in rice leaves ranged from 0 to 3 phylotypes per sample. Among 46 leaf samples, yeast phylotypes were found in 19 leaf samples, whereas all leaf samples contained filamentous fungal phylotypes. The results revealed that the primer sets used in this study effectively amplified the fungal D1/D2 domain of the LSU rDNA and that yeasts were in a minority among endophytic fungi in rice leaves. The CF and the RF of endophytic yeast phylotypes in rice leaves were 0.41 and 0.06, respectively. The rarefaction curve did not reach a horizontal asymptote (Fig. 2), indicating that more species could be retrieved. Since the objective of this work is the assessment of the biodiversity of endophytic yeast in rice leaves, only yeast clones (101 clones) were further analyzed and are discussed.
Relative efficiency of the removal of DNA from rice leaf surface by the sterilization protocol. Values are mean ± standard deviation and N = 3 for each treatment; R0 is the unwashed leaves, R1–R5 are the 1st–5th rinse water and R6 is washed leaves. The relative efficiency of the removal of DNA was calculated as a percentage of the quantity of amplified D1/D2 fragment of each sample divided by the quantity of amplified D1/D2 fragment of unwashed leaf sample
Based on sequence analysis of the D1/D2 domain of the LSU rDNA, the D1/D2 clone sequences of endophytic yeasts derived from rice leaves were identified and affiliated by their similarity and phylogenetic relationships to the sequences of the closest related strains in NCBI and related type strains. The criteria described by Kurtzman and Robnett (1998), namely, that ascomycetous yeast strains with 0 to 3 nucleotide differences in the D1/D2 domain (~600 bp) are either conspecific or sister species and yeast strains showing nucleotide substitutions greater than six are usually different species and the criteria of Fell et al. (2000), namely, that the majority of basidiomycetous yeast can be identified using the D1/D2 region and strains that differed by two or more nucleotides represent different taxa, were used as the guidelines. A total of 101 sequences of yeast clones showed that 12 OTUs were composed of 28 different phylotypes (Table 3). According to the sequence analysis by BLAST search, three OTUs had diverse phytotypes; ten phylotypes closest to Cryptococcus foliicola in OTU1, one phylotype closest to Pseudozyma antarctica and one phylotype closest to P. aphidis in OTU8, and six phylotypes closest to Debaryomyces hansenii and one phylotype closest to Debaryomyces vindobonensis in OTU11. The closest related species of the clone sequences showed nine (26 clones, 25.7 % RF) and nineteen (75 clones, 74.3 % RF) phylotypes belonged to the Ascomycota and Basidiomycota, respectively. The genera belonging to the Ascomycota consisted of Candida (one phylotype in the Lodderomyces–Spathaspora clade), Debaryomyces (seven phylotypes) and Meyerozyma (one phylotype), whereas the genera belonging to the Basidiomycota were Cryptococcus (two phylotypes in the Bulleromyces clade and twelve phylotypes in the victoriae clade), Pseudozyma (two phylotypes) and Trichosporon (one phylotype). The phylotypes, most closely related to type strains of ascomycetous yeasts, which were either identical or had only two to three nucleotides differences (Kurtzman and Robnett 1998) and the phylotypes most closely related to basidiomycetous yeasts, having fewer than two nucleotides difference (Fell et al. 2000), were designated as five known species (Cryp. victoriae, D. hansenii, D. vindobonensis, Meyerozyma guilliermondii and P. antarctica), whereas the others may be novel species closest to Candida metapsilosis, Cryp. foliicola, Cryp. laurentii, Pseudozyma abaconensis, Pseudozyma aphidis and Trichosporon asahii (Table 4). The majority of the endophytic yeast phylotypes in rice leaves were basidiomycetous yeasts and the most prevalent phylotypes were the ones closest to Cryp. foliicola (47.5 % RF) followed by D. hansenii (22.8 % RF) and P. antarctica (16.8 % RF). The phylotypes close to C. metapsilosis, Cryp. laurentii, D. vindobonensis, M. guilliermondii, P. aphidis, P. abaconensis and T. asahii were found at the low frequency of 1.0–5.0 % (Table 4). The phylogenetic tree showed that the analyzed sequences were distributed in four taxonomic orders, namely, Ustilaginales, Trichosporonales and Tremellales in Basidiomycota (Fig. 3) and Saccharomycetales in Ascomycota (Fig. 4). The majority of phylotypes were affiliated in the order Tremellales and most of them were located in the victoriae clade. The tree also revealed the clusters of the novel sequences (15R19, 3R33 and 43R8), which did not include any reference sequences as their phylogenetic neighbors in the victoriae clade, Tremellales. Moreover, the other phylotypes were affiliated in the genera Candida in the Lodderomyces-Spathaspora clade, Debaryomyces, Meyerozyma, Pseudozyma and Trichosporon. Remarkably, the basidiomycetous phylotypes with two or more nucleotides differences were grouped in different clusters of the closest related type strains, implying that they may belong to novel yeast species.
Phylogenetic tree, based on the sequences of the D1/D2 region of the LSU rDNA, showing the placement of basidiomycetous phylotypes with respect to closely related species. The phylogenetic tree was constructed from evolutionary distance data with Kimara’s two-parameter correction, using the maximum likelihood method and MEGA version 6.0. Numbers at nodes indicate percentages of bootstrap sampling derived from 1000 replicates and bootstrap percentages higher than 50 % are shown. Bar 0.02 Knuc. Sequences obtained in the present study are printed in bold letters and the sequences of type strains are indicated by superscript T
Phylogenetic tree, based on the sequences of the D1/D2 region of the LSU rDNA, showing the placement of ascomycetous phylotypes with respect to closely related species. The phylogenetic tree was constructed from evolutionary distance data with Kimara’s two-parameter correction, using the maximum likelihood method and MEGA version 6.0. Numbers at nodes indicate percentages of bootstrap sampling derived from 1000 replicates and bootstrap percentages higher than 50 % are shown. Bar 0.01 Knuc. Sequences obtained in the present study are printed in bold letters and the sequences of type strains are indicated by superscript T
Discussion
Investigation of endophytic yeast species in rice leaves was carried out by a culture-independent method. Therefore, the removal of viable cells and their DNA on the leaf surface should be performed to avoid the contamination of the endophytic population with epiphytes. After treating the leaves with 70 % (w/v) ethanol for microbial decontamination and 5 % (w/v) sodium hypochlorite for DNA decontamination together with several times washing with sterile water, no growth of microorganisms and no PCR products of the D1/D2 domain from the washed leaves were detected, indicating adequate sterilization and DNA removal of the leaf surface, respectively. The inoculation of Saccharomyces cerevisiae as a control organism on the leaf surface of wheat was proposed for the evaluation of the surface sterilization efficiency in culture-independent fungal endophyte studies (Burdorf et al. 2014); however, a known species used as control organism must not be an endophyte of the plant and must produce amplified products that are different in size from the native endophytes. Therefore, we used epiphytic microorganisms as control organisms to elucidate the reduction in the epiphytic DNA and the comparison of the PCR products with and without sterilization revealed that the sterilization process could significantly reduce the contamination of epiphytic yeast DNA and sufficiently prevent the amplification of the D1/D2 domain of the epiphytic yeast DNA. However, a small amount of residual DNA may remain on the leaf surface. Furthermore, it is possible that epiphytic yeasts colonized the inner tissues of the leaves prior to surface sterilization due to the long storage period of leaf samples collected in the Eastern and Northeastern regions.
Although internal transcribed spacer (ITS) has been proposed as the official fungal barcode, a culture-independent method based on LSU rDNA analyses was used because LSU rDNA is primarily used for standard identification of yeasts and has less variation in length than ITS. Also, there is an extensive reference database for taxonomic study. The primers: NL1 and NL4, have been commonly used for amplification of the D1/D2 domain of pure cultures, but have been applied to mixed DNA samples in only a few instances such as investigations of fruit-associated fungal communities (Taylor et al. 2014), of microorganisms associated with mosquitoes (Gusmão et al. 2010) and of yeast diversity associated with invasive bark beetle (Lou et al. 2014). Although there was no report about biased amplification of other eukaryotic sequences, our results showed that these universal primers could also amplify the LSU rDNA of rice, and this interfered with the amplification of fungal LSU rDNA. Nested PCR or semi-nested PCR, involving two sets of primers for two successive runs of PCR reaction, the second set intended to amplify a secondary target within the first run product, is used to increase the sensitivity and/or specificity of PCR products. Furthermore, PCR primers used in the exploration of microbial diversity in environmental samples should have high specificity for the representative target genes. The ITS1-F_KYO1 primer designed by Toju et al. (2012) showed enhanced coverage across diverse fungal taxa and the in silico analysis revealed that the primer matched 99 % of ascomycete and basidiomycete ITS taxa without significant taxonomic biases whilst inhibiting the amplification of plant sequences. Therefore, semi-nested PCR combined with the highly specific primers (ITS1-F_KYO1) was carried out to enhance the specific amplification of small amounts of fungal DNA compared to rice DNA in the reaction. The primer sets of ITS1-F_KYO1 and NL4 as the outer set and NL1/NL4 as the inner set used for semi-nested PCR confirmed their specificity to fungal sequences in the presence of many copies of rice LSU rDNA and the products of the D1/D2 domain were successfully obtained. However, the competition of yeast DNA with the large amount of filamentous fungal DNA resulted in numerous filamentous fungal sequences in the clone libraries. Accordingly, only 12 OTUs (101 clones) out of 124 OTUs (1826 clones) were identified to be yeast phylotypes by the ARDRA technique followed by sequencing, homology search and phylogenetic analyses. The OTUs were clustered by the restriction patterns with different restriction enzymes for rapid identification; however, this technique was unable to distinguish the difference among the phylotypes related to Cryp. foliicola (OTU1), P. antarctica and P. aphidis (OTU8), and D. hansenii and D. vindobonensis (OTU11). Therefore, the sequence analysis was needed for the identification of these phytotypes. The yeast phylotypes found in 19 rice leaf samples ranged from 0 to 3 phylotypes per sample and among these phylotypes belonging to the Basidiomycota were dominant, while all samples contained filamentous fungi phylotypes (86–99 % similarity to closely related species) ranging from 2 to 11 phylotypes per sample. Moreover, the leaf samples collected from the Northeastern region, except for the sample No. R3, contained fewer endophytic phylotypes than those from Central and Eastern regions and only the filamentous fungal phylotypes related to Passalora janseana were observed. On the other hand, various genera of the filamentous fungal phylotypes; for instance Bipolaris, Cladosporium, Passalora, Ramularia, Setosphaeria and Xylaria, were found in the leaf samples collected from the Central and Eastern regions (data not shown), suggested that the sampling site may affect the distribution of fungal endophytes in rice leaf. The endophytic distribution in the plant is affected by various ecological factors such as geographic location, sampling site, season and type of plant tissue (Collado et al. 1999; Gai et al. 2009; Naik et al. 2009; Solis et al. 2015). However, the rarefaction curve indicated that additional sampling is needed to retrieve the remaining yeast species. This finding confirmed that filamentous fungi were relatively much more abundant than yeasts in rice leaves, corresponding to the culture-dependent study reported by Tian et al. (2004) that endophytic yeasts could be isolated only from leaves but not roots of rice plants at the isolation frequency of 3.7 %. Likewise, endophytic fungi of wild rice (Oryzae granulate) analyzed by a culture-independent approach based on ITS sequences showed that the phylotype accumulation curve did not reach saturation level. The frequently detected clones were classified as Basidiomycota and only one yeast phylotype, closely related to T. mucoides, was found (Yuan et al. 2010). On the culture-independent basis, the biases of clone library construction, PCR amplification condition and/or ARDRA screening may limit the sampling saturation (Jones and Richards 2011).
The endophytic yeast community in rice leaves was dominated by phylotypes that belonged to the Basidiomycota. The prevalent phylotypes were Cryptococcus spp. located in the victoriae clade, Tremellales. Interestingly, the novel phylotypes were found at high frequency (56.4 %), suggesting much more exploration of endophytic yeasts in rice leaves is needed. Our findings demonstrated that the endophytic yeast phylotypes consisted of five known species and seventeen novel phylotypes, indicating high proportion of novel yeast species in rice leaves. Although the contamination by epiphytic yeast DNA in this study is possible, it could be noted that the phylotypes stated in this study are endophytic yeasts in rice leaves according to the following reasons. The phylotypes related to Cryp. foliicola, Cryp. victoriae, C. metapsilosis, D. hansenii, D. vindobonensis, P. abaconensis, and T. asahii are endophytic yeasts in rice leaves because the assessment of epiphytic yeasts from rice leaves in Thailand by the enrichment technique (Limtong and Kaewwichian 2015) and culture-independent approach (Nasanit et al. 2015) showed that none of these yeast species were detected. In relation to the species reported as rice epiphytic yeast, the phylotypes related to Cryp. laurentii and P. aphidis may be novel phylotypes due to the high number of nucleotide substitutions (3–51 nucleotides), whereas P. antractica and M. guilliermondii, are probably endophytic phylotypes since their DNA was present in the leaf samples (37R and 44R), from which DNA was extracted within 24 h of collection. Hence, the colonization in the inner tissues of rice leaves probably occurred in the paddy field. Nonetheless, it should be noted that a culture-independent approach may not depict the actual yeast diversity in rice leaves due to the presence of large quantity of fungal DNA and the limitation of primer sets for specific annealing with the yeast LSU rDNA. The direct amplification method could remarkably enhance detection of endophytic yeasts that may be novel species and/or unculturable. Furthermore, the development of specific primers designed for amplification of yeast rDNA and the use of a multi primer approach might make it possible to recover diverse phylotypes from the environmental samples of interest (Nakano et al. 2010; Singh et al. 2011; Singh et al. 2012).
The yeast phylotypes discussed in this study were reported to be associated with plants such as grasses, rice, sugarcane, medicinal plants, fruits, flowers and nectar as both epiphytes and endophytes (Middelhoven 1997; Trindade et al. 2002; Wang et al. 2011; Glushakova et al. 2014, Limtong and Kaewwichian 2015, Limtong et al. 2014) and the ones related to Cryptococcus, especially Cryp. foliicola, were found at high frequency. Moreover, the high frequency of occurrence of novel phylotypes, mainly the basidiomycetous ones, revealed undiscovered yeast species by a culture-dependent approach. Cryp. foliicola inhabits substrates of plant origins, such as leaves (Bakys et al. 2009; Wang et al. 2011) and nectar (Mittelbach et al. 2015), as well as thrives in the extreme environment, such as acidic (Russo et al. 2008) and cold (Arenz et al. 2010; de Garcia et al. 2012) environments. In the present study, the phylotypes related to Cryp. foliicola were found in leaf samples collected from the Northeastern and Central regions, mainly from Jasmine rice (3R, 5R, 10R, 15R, 33R, 34R and 35R). Although it is not possible to specify the variety of rice with which the other phytotypes were associated, the results suggested the possibility that they were associated with Jasmine rice. D. hansenii, the second most common phylotypes in the present study, is widespread in nature, such as the phylloplane, soil, and many habitats with low water activity (Suzuki et al. 2011). Moreover, some phylotypes presented in this study such as M. guilliermondii, and D. hansenii were reported as yeast associated with the brown planthopper (Nilaparvata lugens), an insect pest of rice (Hou et al. 2013), and Cryp. laurentii was isolated from internal organs of beetles (Vega and Dowd 2005). Therefore, it seems likely that insects are a route for the transmission of yeasts into plant tissues. Furthermore, it can be assumed that M. guilliermondii, and P. antractica might inhabit both the leaf surface and internal leaf tissue due to their occurrence as epiphytes (Limtong and Kaewwichian 2015; Nasanit et al. 2015) and endophytes (the present study). The occurrence of yeast species being both epiphyte and endophyte of the same plant species was recently reported by Khunnamwong et al. (2014) that Wickerhamiella siamensis, was a novel endophytic and epiphytic yeast species isolated from sugarcane leaf in Thailand. Furthermore, when the plant growth-promoting traits of epiphytic and endophytic yeasts isolated from rice leaves in Thailand were investigated, 13 strains of nine yeast species, including M. guilliermondii, produced indole-3-acetic acid (IAA) (Nutaratat et al. 2014). IAA production of epiphytic yeasts on sugarcane leaves in Thailand was also determined and the results revealed that 69 strains of 23 yeast species, including Cryp. laurentii, had the capability of producing IAA (Limtong et al. 2014). Cryp. victoriae isolated from fruits had high antagonistic activity against Penicillium expansum, Pen. digitatum and Botrytis cinera (Lutz et al. 2013; Ghosh et al. 2013). Trindade et al. (2002) reported that P. antarctica isolated from pitanga ripe fruits produced mycotoxin, protease, pectinase or β-glucosidase. D. hansenii isolates obtained from citrus fruits were proved to be a potential biocontrol agent against green mold (Pen. digitatum), blue mold (Pen. italicum) and sour rot (Galactomyces candidum) of citrus fruits (Chalutz and Wilson 1990; Hernández-Montiel et al. 2010). Although it is not possible to specify the roles of these phylotypes, the reviewed information suggests, there are potentially beneficial endophytic yeast strains in rice leaves.
References
Altschul SF, Madden TL, Schaffer JZ, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Anderson IC, Cairney JWG (2004) Diversity and ecology of soil fungal communities: increased understanding through the application of molecular techniques. Environ Microbiol 6:769–779
Arenz BE, Held BW, Jurgens JA, Blanchette RA (2010) Fungal colonization of exotic substrates in Antarctica. Fungal Divers 49:13–22
Arnold AE, Mejia LC, Kyllo D, Rojas EI, Maynard Z, Robbins N, Herre EA (2003) Fungal endophytes limit pathogen damage in tropical trees. Proc Natl Acad Sci USA 100:15649–15654
Bakys R, Vasaitis R, Barklund P, Ihrmark K, Stenlid J (2009) Investigations concerning the role of Chalara fraxinea in declining Fraxinus excelsior. Plant Pathol 58:284–292
Burdorf RJ, Laing MD, Morris CD, Jamal-Ally SE (2014) A procedure to evaluate the efficiency of surface sterilization methods in culture-independent fungal endophyte studies. Braz J Microbiol 45:977–983
Chae HS, Park GN, Kim SH, Jo HJ, Kim JT, Jeoung HY, An DJ, Kim NH, Shin BW, Kang YI, Chang KS (2012) Rapid direct identification of Cryptococcus neoformans from pigeon droppings by nested PCR using CNLAC1 gene. Poult Sci 91:1983–1989
Chalutz E, Wilson CL (1990) Postharvest biocontrol of green and blue mold and sour rot of citrus fruit by Debaryomyces hansenii. Plant Dis 74:134–137
Collado J, Platas G, Gonzales I, Pelaez F (1999) Geographical and seasonal influence on the distribution of fungal endophytes in Quercus ilex. New Phytol 143:525–532
de Garcia V, Zalar P, Brizzio S, Gunde-Cimerman N, van Broock M (2012) Cryptococcus species (Tremellales) from glacial biomes in the southern (Patagonia) and northern (Svalbard) hemispheres. FEMS Microbiol Ecol 82:523–539
Díaz C, Molina AM, Nähring J, Fischer R (2013) Characterization and dynamic behavior of wild yeast during spontaneous wine fermentation in steel tanks and amphorae. Biomed Res Int 2013:Art ID 540465. doi:10.1155/2013/540465
Fell JW, Boekhout T, Fonseca A, Scorezetti G, Statzell-Tallman A (2000) Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int J Syst Evol Microbiol 50:1351–1371
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Gai CS, Lacava PT, Maccheroni W Jr, Glienke C, Araújo WL, Miller TA, Azevedo JL (2009) Diversity of endophytic yeasts from sweet orange and their localization by scanning electron microscopy. J Basic Microbiol 49:441–451
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118
Ghosh S, Santra T, Chakarvarty A (2013) Study of antagonistic yeasts isolated from some natural sources of West Bengal. Agric Biol J N Am 4:33–40
Glushakova AM, Kachalkin AV, Chernov IY (2014) Yeasts in the flowers of entomophilic plants of Moscow region. Microbiology 83:125–134
Guillamón JM, Sabaté J, Barrio E, Cano J, Querol A (1998) Rapid identification of wine yeast species based on RFLP analysis of the ribosomal internal transcribes spacer (ITS) region. Arch Microbiol 169:387–392
Gusmão DS, Santos AV, Marini DC, Bacci M Jr, Berbert-Molina MA, Lemos FJA (2010) Culture-dependent and culture-independent characterization of microorganisms associated with Ades aegypti (Diptera: Culicidae) (L.) and dynamics of bacterial colonization in the midgut. Acta Trop 115:275–281
Harrison E, Muir A, Stratford M, Wheals A (2011) Species-specific PCR primers for the rapid identification of yeasts of the genus Zygosaccharomyces. FEMS Yeast Res 11:356–365
Hernández-Montiel LG, Ochoa JL, Troyo-Diéguez E, Larralde-Corona CP (2010) Biocontrol of postharvest blue mold (Penicillium italicum Wehmer) on Mexican lime by marine and citrus Debaryomyces hansenii isolates. Postharvest Biol Technol 56:181–187
Higginbotham SJ, Arnold AE, Ibañez A, Spadafora C, Coley PD, Kursa TA (2013) Bioactivity of fungal endophytes as a function of endophyte taxonomy and the taxonomy and distribution of their host plants. PLoS ONE 8:e73192. doi:10.1371/journal.pone.0073192
Holland SM (2003) Analytic Rarefaction 1.3. http://strata.uga.edu/software/. Accessed 3 Oct 2014
Hou Y, Ma Z, Dong S, Chen YH, Yu X (2013) Analysis of yeast-like symbiote diversity in the brown planthopper (BPH), Nilaparvata lugans Stål, using a novel nested PCR-DGGE protocol. Curr Microbiol 67:263–270
Ibeas JI, Lozano I, Perdigones F, Jimenez J (1996) Detection of Dekkera-Brettanomyces strains in sherry by a nested PCR method. Appl Environ Microbiol 62:998–1003
Jones MDM, Richards TA (2011) Environmental DNA analysis and the expansion of the fungal tree of life. In: Pöggeler S, Wöstemeyer J (eds) Evolution of fungi and fungal-like organisms: the mycota XIV. Springer-Verlag, Berlin, pp 37–54
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780
Khunnamwong P, Surussawadee J, Jindamorakot S, Limtong S (2014) Wickerhamiella siamensis f.a., sp. nov., a novel endophytic and epiphytic yeast species isolated from sugarcane leaf in Thailand. Int J Syst Evol Microbiol 64:3849–3855
Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kurtzman CP, Robnett CJ (1998) Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie van Leewenhoek 73:331–371
Larran S, Monaco C, Alippi HE (2001) Endophytic fungi in leaves of Lycopersicon esculentum mill. World J Microbiol Biotechnol 17:181–184
Limtong S, Kaewwichian R (2015) The diversity of culturable yeasts in the phylloplane of rice in Thailand. Ann Microbiol 65:667–675
Limtong S, Kaewwichian R, Yongmanitchai W, Kawasaki H (2014) Diversity of culturable yeasts in phylloplane of sugarcane in Thailand and their capability to produce indole-3-acetic acid. World J Microbiol Biotechnol 30:1785–1796
Lou QZ, Lu M, Sun JH (2014) Yeast diversity associated with invasive Dendroctonus valens killing Pinus tabuliformis in China using culturing and molecular methods. Microb Ecol 68:397–415
Lutz MC, Lopes CA, Rodriguez ME, Sosa MC, Sangorrín MP (2013) Efficacy and putative mode of action of native and commercial antagonistic yeasts against postharvest pathogens of pear. Intl J Food Microbiol 164:166–172
Martin KJ, Rygiewicz PT (2005) Fungal-specific PCR primers developed for analysis of the ITS region of environmental DNA extracts. BMC Microbiol 5:28–38
Middelhoven WJ (1997) Identity and biodegradative abilities of yeasts isolated from plants growing in an arid climate. Antonie Van Leeuwenhoek 72:81–89
Mittelbach M, Yurkov AM, Nocentini D, Nepi M, Weigend M, Begerow D (2015) Nectar sugars and bird visitation define a floral niche for basidiomycetous yeast on the Canary Islands. BMC Ecol 15:2–14
Naik BS, Shashikala J, Krishnamurthy YL (2009) Study on the diversity of endophytic communities from rice (Oryza sativa L.) and their antagonistic activities in vitro. Microbiol Res 164:290–296
Nakano Y, Nagahama T, Hatada Y, Nunoura T, Yakami H, Miyazaki J, Takai K, Horikoshi K (2010) Fungal diversity in deep-sea sediments—the presence of novel fungal groups. Fungal Ecol 3:316–325
Nasanit R, Krataithong K, Tantirungkij M, Limtong S (2015) Assessment of epiphytic yeast diversity in rice (Oryza sativa) phyllosphere in Thailand by culture-independent approach. Antonie Van Leeuwenhoek 107:1475–1490
Nassar AH, El-Tarabily KA, Sivasithamparam K (2005) Promotion of plant growth by an auxin-producing isolate of the yeast Williopsis saturnus endophytic in maize (Zea mays L.) roots. Biol Fertil Soils 42:97–108
Nielsen DS, Teniola OD, Ban-Koffi L, Owusu M, Andersson TS, Holzapfel WH (2007) The microbiology of Ghanaian cocoa fermentations analysed using culture-dependent and culture-independent methods. Int J Food Microbiol 114:168–186
Nutaratat P, Srisuk N, Arunrattiyakorn P, Limtong S (2014) Plant growth-promoting traits of epiphytic and endophytic yeasts isolated from rice and sugar cane leaves in Thailand. Fungal Biol 118:683–694
Oros-Sichler M, Gomes NCM, Neuber G, Smalla K (2006) A new semi-nested PCR protocol to amplify large 18S rRNA gene fragments for PCR-DGGE analysis of soil fungal communities. J Microbiol Meth 65:63–75
Petrini O (1991) Fungal endophytes of tree leaves. In: Andrews JH, Hirano SS (eds) Microbial ecology of the leaves. Springer-Verlag, New York, pp 179–197
Petrini O, Sieber TN, Toti L, Viret O (1992) Ecology, metabolite production, and substrate utilization in endophytic fungi. Natl Toxins 1:185–196
Porter TM, Golding GB (2012) Factors that affect large subunit ribosomal DNA amplicon sequencing studies of fungal communities: classification method, primer choice, and errors. PLoS ONE 7:e35749. doi:10.1371/journal.pone.0035749
Russo G, Libkind D, Sampio J, van Broock MR (2008) Yeast diversity in the acidic Rio Agrio-Lake Caviahue volcanic environment (Patagonia, Argentina). FEMS Microbiol Ecol 65:415–5424
Singh P, Raghukumar C, Verma P, Shouche Y (2011) Fungal community analysis in the deep-sea sediments of the Central Indian Basin by culture-independent approach. Microb Ecol 61:507–517
Singh P, Raghukumar C, Verma P, Shouche Y (2012) Assessment of fungal diversity in deep-sea sediments by multiple primer approach. World J Microbiol Biotechnol 28:659–667
Solis MJL, Yurkov A, dela Cruz TE, Unterseher M (2015) Leaf-inhabiting endophytic yeasts are abundant but unevenly distributed in three Ficus species from botanical garden greenhouses in Germany. Mycol Progress 14:1019–1028
Suzuki M, Prasad GS, Kurtzman CP (2011) Debaryomyces Lodder & Kreger-van Rij (1952). In: Kurtzman CP, Fell JW, Boekhout T (eds) The yeasts, a taxonomic study. Elsevier, San Diego, pp 361–372
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Taylor MW, Tsai P, Anfang N, Ross HA, Goddard R (2014) Pyrosequencing reveals regional differences in fruit-associated fungal communities. Environ Microbiol 16:2848–5858
Tian XL, Cao LX, Tan HM, Zeng QG, Jia YY, Han WQ, Zhou SN (2004) Study on the communities of endophytic fungi and endophytic actinomycetes from rice and their antipathogenic acitivities in vitro. World J Microbiol Biotechnol 20:303–309
Toju H, Tanabe AS, Yamamoto S, Sato H (2012) High-coverage ITS primers for the DNA-based identification of ascomycetes and basidiomycetes in environmental samples. PLoS ONE 7:e40863
Trindade RC, Resende MA, Silva CM, Rosa CA (2002) Yeast associated with fresh and frozen pulps of Brazillian tropical fruits. Syst Appl Microbiol 25:294–300
Vega FE, Dowd PF (2005) The role of yeasts as insect endosymbionts. In: Vega FE, Blackwell M (eds) Insect-fungal associations ecology and evolution. Oxford University Press, Oxford, pp 211–243
Wang QM, Boekhout T, Bai FY (2011) Cryptococcus foliicola sp. nov. and Cryptococcus taibaiensis sp. nov., novel basidiomycetous yeast species from plant leaves. J Gen Appl Microbiol 57:285–291
White TJ, Bruns TD, Lee S, Taylor JW (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M, Gefand D, Sninsky J, White T (eds) PCR protocols: a guide to methods and applications. Academic Press, London, pp 315–322
Xin G, Glawe D, Doty SL (2009) Characterization of three endophytic, indole-3-acetic acid-producing yeasts occurring in Populus trees. Mycol Res 113:973–980
Yang CH, Crowley DE, Borneman J, Keen NT (2001) Microbial phyllophere populations are more complex than previously realized. Proc Natl Acad Sci USA 98:3889–3894
Yuan ZL, Zhang CL, Lin FC, Kubicek C (2010) Identity, diversity and molecular phylogeny of the endophytic mycobiota in the roots of rare wild rice (Oryza granulate) from a nature reserve in Yunnan, China. Appl Environ Microbiol 76:1642–1652
Acknowledgments
This work was supported by the Thailand Research Fund through a Thailand Research Fund/TRF Research-Team Promotion Grant (RTA54800009) and the Center for Advanced Studies in Tropical Natural Resources, National Research University-Kasetsart University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Rights and permissions
About this article
Cite this article
Tantirungkij, M., Nasanit, R. & Limtong, S. Assessment of endophytic yeast diversity in rice leaves by a culture-independent approach. Antonie van Leeuwenhoek 108, 633–647 (2015). https://doi.org/10.1007/s10482-015-0519-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-015-0519-y