Abstract
Saitoella coloradoensis sp. nov. (NRRL YB-2330, CBS 12360, type strain, MycoBank accession number 563858) is described. This new member of the phylum Ascomycota, subphylum Taphrinomycotina was isolated from insect frass occurring in an Engelmann spruce (Picea engelmannii) that was growing in Colorado, USA. Multigene sequence analysis showed that S. coloradoensis is distinct from Saitoella complicata, the only other known species of Saitoella. The two species may be separated phenotypically from growth reactions on d-xylose, ribitol and methyl-α-d-glucoside. Asexual reproduction is by budding and both species produce thick-walled, spherical cells that appear morphologically similar to the ascogenous cells formed in plant host tissue by species of Protomyces and some species of Taphrina. The thick-walled cells did not form ascospores but did produce buds when placed on fresh growth media.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The monotypic yeast genus Saitoella was described by Goto et al. (1987) with the type species S. complicata based on two strains that had been isolated from soil collected in Laya, Bhutan. The strains were initially classified from phenotype as Rhodotorula glutinis (Goto and Sugiyama 1970). Later, it was discovered that the isolates gave a negative diazonium blue B color reaction, typical of ascomycetous yeasts (Sugiyama et al. 1985), and that the cell wall ultrastructure was characteristic of the Ascomycota (Goto et al. 1987). Because S. complicata synthesizes carotenoid pigments, forms coenzyme Q 10 and lacks xylose in cellular hydrolysates, it resembles the plant pathogenic yeast-like genera Protomyces and Taphrina (Sugiyama et al. 1985; Sugiyama and Hamamoto 2011).
Phylogenetic analysis of genes for nuclear small subunit rRNA (Nishida and Sugiyama 1993), domains D1 and D2 of large subunit rRNA (Kurtzman and Robnett 1998) and multigene sequence analysis (Sugiyama et al. 2006) placed Saitoella, Protomyces and Taphrina in the same clade, which also includes Schizosaccharomyces, Pneumocystis and Neolecta. Sugiyama et al. (2006) noted from multigene sequence analysis that the position of Saitoella in the subphylum Taphrinomycotina is not strongly resolved, but the genus may be most closely related to Protomyces and Taphrina. Recently, the new genus Archaeorhizomyces was described for strains isolated from coniferous ectomycorrhizal root tips, and multigene sequence analysis placed this new taxon in the Taphrinomycotina (Rosling et al. 2011). The preceding studies and that of James et al. (2006) resolved the phylum Ascomycota into three major clades or subphyla. The subphylum Taphrinomycotina encompasses the above noted seven genera and is basal to the subphyla Saccharomycotina and Pezizomycotina, which are sister taxa. Budding yeasts such as Saccharomyces are placed in the Saccharomycotina, whereas “filamentous” species, such as Aspergillus and Neurospora are members of the Pezizomycotina (Hibbett et al. 2007).
The present widespread practice of identifying yeasts newly isolated from nature by DNA sequence analysis has resulted in the discovery of many new species, but additional strains of Saitoella have not been isolated and the genus is still known only from the original two cultures obtained from Himalayan soil (Goto and Sugiyama 1970) that formed the basis for description of S. complicata (Goto et al. 1987). During characterization of unidentified yeasts maintained in the ARS Culture Collection, a new species of Saitoella was recognized from D1/D2 LSU rRNA gene sequence analysis. The genetic separation between this species and S. complicata was supported by additional gene sequences. This new species, which was isolated from insect frass in an Engelmann spruce (Picea engelmannii) growing in Colorado, USA, is described here.
Materials and methods
The proposed new species of Saitoella, NRRL YB-2330, was isolated by L. J. Wickerham from insect frass collected in 1950 by staff of the U.S. Forest Service from an Engelmann spruce (Picea engelmannii) growing in the White River National Forest, Meeker, CO, USA. The composition of the culture media used and procedures for conducting growth and sugar fermentation tests were by standard methods (Kurtzman et al. 2011). Growth tests were conducted in liquid media and incubated at 25°C for four weeks. Cultures of NRRL YB-2330 and S. complicata NRRL Y-17804 were tested for ascosporulation on YM, 5% malt extract, RG and water agar media, as well as on water agar containing 1.5 cm2 sections of leaves from mulberry (Morus rubra) and lily-of-the-valley (Convallaria majalis) and branch tips with needles from a Colorado blue spruce (Picea pungens). Freshly collected leaf sections and spruce bud tips were placed in tubes of water agar and autoclaved for 20 min at 121°C. The agar surface was inoculated with cells near the protruding leaf sections.
Methods for DNA isolation and sequencing of genes for domains 1 and 2 (D1/D2) of large subunit (LSU) rRNA, mitochondrial small subunit (Mt SSU) rRNA, translation elongation factor (EF-1α) and the internal transcribed spacers 1 and 2 (ITS 1 and 2) were reported earlier (Kurtzman and Robnett 1998, 2003). Briefly, following DNA isolation, each of the regions to be sequenced was amplified by PCR and the resulting amplicons were sequenced using the ABI BigDye Terminator Cycle Sequencing kit (Applied Biosystems) and an ABI 3730 automated DNA sequencer according to the manufacturer’s instructions. For phylogenetic analysis, sequences were visually aligned. Phylogenetic relatedness among species was determined from the maximum parsimony and neighbor-joining programs of PAUP*4.063a (Swofford 1998). Bootstrap support for the phylogenetic trees was determined from 1,000 replicates. The species compared and the GenBank accession numbers for the genes sequenced are given in Table 1.
Results and discussion
NRRL YB-2330 was initially recognized as a member of the genus Saitoella from its D1/D2 LSU rRNA gene sequence. The strain differed from NRRL Y-17804, type strain of S. complicata, by four nucleotide differences in D1/D2 (Table 2). To further determine extent of divergence between the two strains, sequences for ITS, EF-1α and Mt SSU were compared. Surprisingly, the two strains differed by only two ITS substitutions, but differences in EF-1α and Mt SSU were substantially greater (Table 2). Yeast species often differ from one another by 1% or greater substitutions in D1/D2 and ITS, but some closely related species may be unresolved by D1/D2 and/or ITS. For example, Meyerozyma guilliermondii and Candida carpophila, which differ by a single D1/D2 nucleotide, show 55% nuclear DNA relatedness as measured by reassociation (Vaughan-Martini et al. 2005), and this latter assessment of genome similarity suggests that the two taxa represent separate species. In contrast, Trichosporon montevideense and T. domesticum are unresolved by ITS, but are easily separated from their D1/D2 sequences, from which Scorzetti et al. (2002) interpreted the presence of two species. Multi-strain comparisons by Fonseca and Rodrigues (2011) for Taphrina, which is more closely related to Saitoella than species in the preceding examples, have suggested that strains of a Taphrina species may show as many as two nucleotide substitutions in D1/D2 and ITS sequences. Thus, separation of S. complicata and the proposed new species of Saitoella is uncertain from substitutions in D1/D2 and ITS, but analysis of additional gene sequences supports the presence of two species among the Saitoella strains examined. The extent of substitutions between the two taxa for EF-1α and Mt SSU is typical of differences seen for closely related species in the genera Saccharomyces (Kurtzman and Robnett 2003) and Komagataella (Kurtzman 2009).
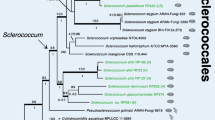
In view of the separation of NRRL YB-2330 from S. complicata by multigene sequence analysis (Fig. 1; Table 2), and the absence of a demonstrable ascosporic state, as described below, the following new species is proposed for the anamorphic genus Saitoella.
Phylogenetic relatedness of Saitoella complicata and S. coloradoensis and their placement near representative species of the neighboring genera Protomyces and Taphrina as represented by the single most parsimonious tree determined from maximum parsimony analysis of concatenated gene sequences from D1/D2 LSU rRNA and EF-1α. The analysis included 1,533 characters of which 266 were parsimony informative. Consistency index = 0.879, rescaled consistency index = 0.612, homoplasy index = 0.121. Bootstrap values are from 1,000 replicates. Schizosaccharomyces pombe (Taphrinomycotina) was the designated outgroup, but an identical tree resulted when Botryozyma nematodophila (Saccharomycotina) was selected as the outgroup species. Neighbor-joining analysis with the Kimura 2-parameter correction gave an identical tree. See text for discussion of phylogenetic placement of Saitoella within the Taphinomycotina
Latin diagnosis of Saitoella coloradoensis Kurtzman et Robnett, sp. nov.
In agaro YM post dies 3 ad 25°C, cultura est butyrosa et glabra; post dies 10, cultura est butyrosa et salmonea. Cellulae sunt ellipsoideae vel elongatae, 3-4.5 × 5-8.5 μm, singulae et binae. Chlamydosporae formantur globosae, 6-12 μm. In agaro morphologico post dies 7 ad 25°C, nec pseudohyphae nec hyphae formantur. Asci non formantur. Glucosum non fermentatur. Assimilantur glucosum, inulinum, sucrosum, raffinosum, trehalosum, maltosum, melezitosum, amylum solubile, cellobiosum, salicinum, d-xylosum, d-arabinosum (infirme), ethanolum, glycerolum, d-glucitolum (infirme), acidum succinicum (infirme) et potasii nitras. Non assimilantur melibiosum, galactosum, lactosum, methyl-α-d-glucosidum, l-sorbosum, l-rhamnosum, l-arabinosum, d-ribosum, methanolum, erythritolum, ribitolum, galactitolum, d-mannitolum, myo-inositolum, dl-acidum lacticum, acidum citricum, d-gluconatum, d-glucosaminum, N-acetyl-d-glucosaminum, hexadecanum, 2-keto-d-gluconatum, 5-keto-d-gluconatum, et saccharatum. Amylum non formatur. Vitamina externa ad crescentiam non necessaria. Gelatinum non liquescit; esters non fiunt; pellicula non fiunt. Crescit in medio 10% sodii chloridium/5% glucosum. Augmentum in temperatura 37°C non fiunt. Species nova a speciebus aliis sequentibus nucleotiditis nucleus submonas largus D1/D2 rRNA gene, ITS rDNA, mitochondrial submonas parvus rRNA gene et factore elongationis-1α gene distinguenda. Typus: NRRL YB-2330 (CBS 12360) designat stirpem typicam. Isolata a dejectus coleopterorum in Picea engelmannii, Meeker, Colorado, USA. Depositata in Collectione Culturarum ARS (NRRL), Peoria, Illinois, USA.
Description of Saitoella coloradoensis Kurtzman & Robnett, sp. nov.
Growth on YM agar
After 3 days at 17°C, cells divide by budding, which is polar or nearly polar (Fig. 2). Cells are ellipsoid to elongate, 3–4.5 × 5–8.5 μm, and occur singly or as budded pairs. Colony growth is white, butyrous and glistening. After 7–10 days, the growth becomes light pinkish–red, which then darkens to a salmon color. Thick-walled cells, which we have termed chlamydospores, are 6–12 μm in diameter and form within 3–7 days (Fig. 2).
Cell morphology of Saitoella coloradoensis and S. complicata. S. coloradoensis NRRL YB-2330. a Budding cells, YM agar, 3 days, 17°C. b Chlamydospores, 5% malt extract agar, 5 weeks, 17°C. c Budding chlamydospores, 1 day, YM agar, 17°C, after transfer of chlamydospores from 5% malt extract agar, 14 weeks, 17°C. Buds from chlamydospores have given rise to additional budding cells. S. complicata NRRL Y-17804. d Budding cells. e Chlamydospores. f Budding chlamydospores. Conditions for d, e, f, are the same as for a, b, c, respectively. Bar = 5 μm for all photographs
Cells of S. complicata are similar to S. coloradoensis in size, 3–4 × 6–7 μm, and morphology (Fig. 2). Colony growth is also similar, but cultures of S. complicata tend to form the reddish color a few days earlier. Thick-walled chlamydospores, 7–15 μm in diameter, are also formed by S. complicata within 3–7 days (Fig. 2).
Dalmau plate culture on morphology agar
After 7 days at 17°C, growth under the coverglass showed neither hyphae nor pseudohyphae.
Tests for ascospore formation
Saitoella coloradoensis and S. complicata were examined for production of ascospores. Cultures of NRRL YB-2330 and NRRL Y-17804 were incubated at 5°, 17° and 25°C on YM, 5% malt extract, RG and water agar for 7 months with initial examination twice weekly for 2 weeks followed by biweekly examination for the remainder of the incubation time. Ascospore formation was not detected in either species. The two taxa were also mixed and incubated at 5°, 17° and 25°C on YM, 5% malt extract, RG and water agar media for 2 months but there was no evidence of conjugation or ascospore formation.
Formation of large, thick-walled chlamydospores, which are filled with small lipid droplets, began after 3–10 days incubation at 5°, 17°, 25°, 28° and 32°C (Fig. 2). Chlamydospores were formed by both species, but the description of S. complicata did not report their presence (Goto et al. 1987). Chlamydospores formed most abundantly on 5% malt extract agar followed by YM and RG agars, respectively. Chlamydospores that formed at 17°C appeared unchanged after several months of further incubation, but chlamydospores formed at temperatures of 25°C or greater showed loss of internal structure after a few weeks followed by breakage of the cell wall. The chlamydospores that formed on 5% malt extract agar at 17°C were transferred after 1 month to YM, 5% malt extract and RG agar media and incubated at 5°, 17° and 25°C for 3 months. Ascospores did not form under these conditions. Usually, 12–24 h after transfer of chlamydospores to fresh media, 10–20% of the chlamydospores formed ellipsoid to elongate buds that continued to bud producing typical vigorously growing yeast cultures (Fig. 2). Often, the very small oil droplets in the chlamydospores coalesced into larger droplets, but pressure applied to the coverglass on the microscope slide under observation caused the chlamydospores to rupture and fragment the sometimes cell-like larger lipid droplets. In an additional attempt to stimulate ascospore formation, chlamydospores from 2 month growth on 5% malt extract agar, 17°C, were inoculated onto water agar slants containing sections of autoclaved leaves from mulberry (dicot) and lily-of-the-valley (monocot) and the branch tips of Colorado blue spruce (monocot) because autoclaved plant material such as sections of carrots, cucumbers and leaves stimulate ascospore formation in some yeasts (Kurtzman et al. 2011). These preparations were incubated at 5°, 17° and 25°C for 4 months, but the only change was occasional bud formation by the chlamydospores. However, for species of Taphrina and Protomyces, a specific living host plant is required for ascosporulation (Fonseca and Rodrigues 2011; Kramer 1973), and the same may be true for Saitoella, if it is a plant pathogen.
Fermentation and growth reactions
The growth reactions for S. coloradoensis and S. complicata are given in Table 3. Neither species fermented glucose, galactose, maltose, sucrose, lactose or raffinose. Assimilation reactions for the two species differ somewhat allowing their separation from reactions on methyl-α-d-glucoside, d-xylose, d-arabinose, ribitol, d-glucitol and 10% NaCl/5% glucose. Results for S. complicata are similar to those reported by Sugiyama and Hamamoto (2011), except that in our tests, growth was absent with mannitol and d-arabinose, but present with soluble starch.
Type
NRRL YB-2330, the type strain, is preserved as a lyophilized preparation in the ARS Culture Collection, National Center for Agricultural Utilization Research, Peoria, IL, USA, and as CBS 12630, with the Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands. The MycoBank accession number for this strain is 563858. The strain had been isolated by L. J. Wickerham from insect frass in an Engelmann spruce growing in Colorado, USA, and remained unidentified until the present study.
Etymology
The species name coloradoensis was selected to denote the origin of the type strain from the U.S. State of Colorado.
The genus Saitoella was first shown from single gene phylogenetic analyses to be a member of the Taphrinomycotina (Nishida and Sugiyama 1993; Kurtzman and Robnett 1998; Kurtzman and Sugiyama 2001), and this placement was supported by multigene sequence analysis (Sugiyama et al. 2006). These analyses suggest, as indicated in Fig. 1, that Saitoella appears closely related to Protomyces and Taphrina. The latter two genera are plant pathogens that are only known to form an ascosporic state in the tissue of their hosts, but they produce a budding asexual state that is easily grown on agar media and the colonies become pigmented pinkish or red as also seen for Saitoella (Goto and Sugiyama 1970; Goto et al. 1987). For Taphrina, dikaryotic subcuticular hyphae diploidize and asci are directly produced on the hyphae by some species, but for other species asci arise from thick-walled ascogenous cells (Mix 1949; Kramer 1973; Fonseca and Rodrigues 2011). Species of Protomyces form chlamydospore-like ascogenous cells in host tissue that germinate to form multispored asci (Reddy and Kramer 1975). Pavgi and Mukhopadhyay (1969) reported formation of chlamydospores on agar media by Protomyces macrosporus but ascospore formation did not occur. Whether the chlamydospores formed by Saitoella are environmentally resistant asexual cells or analogous to the morphologically similar ascogenous cells typically produced by Protomyces and some species of Taphrina in host plant tissue is unknown, but their formation raises the possibility that Saitoella may be a plant pathogen. Many of the described species of Protomyces are known only from herbarium specimens and their identity is uncertain, which raises the possibility that some of these taxa may be misidentified species of Saitoella. However, testing this hypothesis will require sequence analysis of all described Protomyces species as well as sequence analysis of the poorly characterized plant pathogenic genera Burenia, Protomycopsis, Taphridium and Volkartia, which are not known from culture, but are morphologically similar to Protomyces (Reddy and Kramer 1975; Kurtzman 2011). Of still further interest concerning the possible habitat of Saitoella is the study of Allison et al. (2010). In this work, DNA was cloned from soil sampled in an Alaskan (USA) boreal forest. Of the 433 fungal sequences obtained, three (GU212336, GQ892426, GQ892425) were identical to the D1/D2 LSU rRNA gene sequence of S. complicata.
References
Allison SD, McGuire KL, Treseder KK (2010) Resistance of microbial and soil properties to warming treatment seven years after boreal fire. Soil Biol Biochem 42:1872–1878
Fonseca Á, Rodrigues MG (2011) Taphrina fries (1832). In: Kurtzman CP, Fell JW, Boekhout T (eds) The yeasts, a taxonomic study, 5th edn. Elsevier Science, Amsterdam, pp 823–858
Goto S, Sugiyama J (1970) Studies on Himalayan yeasts and molds (IV). Several asporogenous yeasts, including two new taxa of Cryptococcus. Can J Bot 48:2097–2101
Goto S, Sugiyama J, Hamamoto M, Komagata K (1987) Saitoella, a new anamorphic genus in the Cryptococcaceae to accommodate two Himalayan yeast isolates formerly identified as Rhodotorula glutinis. J Gen Appl Microbiol 33:75–85
Hibbett DS, Binder M, Bischoff JF, Blackwell M, Cannon PF, Eriksson O, Huhndorf S, James T, Kirk PM, Lücking R, Lumbsch T, Lutzoni F, Matheny PB, McLaughlin DJ, Powell MJ, Redhead S, Schoch CL, Spatafora JW, Stalpers JA, Vilgalys R, Aime C, Aptroot A, Bauer R, Begelow D, Benny GL, Castlebury LA, Crous PW, Dai YC, Gams W, Geiser DM, Griffith GW, Hawksworth D, Hestmark G, Hosaka K, Humber RA, Hyde KD, Ironside JE, Köljalg U, Kurtzman CP, Larsson KH, Lichtwardt R, Longcore J, Miadlikowska J, Miller A, Moncalvo JM, Mozley-Standridge S, Oberwinkler F, Parmasto E, Reeb V, Rogers JD, Ryvarden L, Sampaio JP, Schüßler A, Sugiyama J, Thorn RG, Tibell L, Untereiner WA, Walker C, Wang Z, Weir A, Weiss M, White MM, Winka K, Yao YJ, Zhang N (2007) A higher-level phylogenetic classification of the fungi. Mycol Res 111:509–547
James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, Cox CJ, Celio G, Gueidan C, Fraker E, Miadlikowska J, Lumbsch HT, Rauhut A, Reeb V, Arnold AE, Amtoft A, Stajich JE, Hosaka K, Sung GH, Johnson D, O’Rourke B, Crockett M, Binder M, Curtis JM, Slot JC, Wang Z, Wilson AW, Schüssler A, Longcore JE, O’Donnell K, Mozley-Standridge S, Porter D, Letcher PM, Powell MJ, Taylor JW, White MM, Griffith GW, Davies DR, Humber RA, Morton JB, Sugiyama J, Rossman AY, Rogers JD, Pfister DH, Hewitt D, Hansen K, Hambleton S, Shoemaker RA, Kohlmeyer J, Volkmann-Kohlmeyer B, Spotts RA, Serdani M, Crous PW, Hughes KW, Matsuura K, Langer E, Langer G, Untereiner WA, Lücking R, Büdel B, Geiser DM, Aptroot A, Diederich P, Schmitt I, Schultz M, Yahr R, Hibbett DS, Lutzoni F, McLaughlin DJ, Spatafora JW, Vilgalys R (2006) Reconstructing the early evolution of fungi using a six-gene phylogeny. Nature 443:818–822
Kramer CL (1973) Protomycetales and Taphrinales. In: Ainsworth GC, Sparrow FK, Sussman AS (eds) The fungi—an advanced treatise, vol IVA. Academic Press, New York, pp 33–41
Kurtzman CP (2009) Biotechnological strains of Komagataella (Pichia) pastoris are Komagataella phaffii as determined from multigene sequence analysis. J Ind Microbiol Biotechnol 36:1435–1438
Kurtzman CP (2011) Protomyces Unger (1833). In: Kurtzman CP, Fell JW, Boekhout T (eds) The yeasts, a taxonomic study, 5th edn. Elsevier Science, Amsterdam, pp 725–731
Kurtzman CP, Robnett CJ (1998) Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie van Leeuwenhoek 73:331–371
Kurtzman CP, Robnett CJ (2003) Phylogenetic relationships among yeasts of the “Saccharomyces complex” determined from multigene sequence analyses. FEMS Yeast Res 3:417–432
Kurtzman CP, Sugiyama J (2001) Ascomycetous yeasts and yeastlike taxa. In: McLaughlin DJ, McLaughlin EG, Lemke PA (eds) The mycota. vol. VII part A (systematics and evolution). Springer-Verlag, Berlin, pp 179–200
Kurtzman CP, Fell JW, Boekhout T, Robert V (2011) Methods for isolation, phenotypic characterization and maintenance of yeasts. In: Kurtzman CP, Fell JW, Boekhout T (eds) The yeasts, a taxonomic study, 5th edn. Elsevier Science, Amsterdam, pp 87–110
Mix AJ (1949) A monograph of the genus Taphrina. Univ Kansas Sci Bull 23:1–167
Nishida H, Sugiyama J (1993) Phylogenetic relationships among Taphrina, Saitoella, and other higher fungi. Mol Biol Evol 10:431–436
Pavgi MS, Mukhopadhyay AN (1969) Artificial culture and in vitro chlamydospore development of Protomyces macrosporus Unger. Pathol Microbiol 33:287–295
Reddy MS, Kramer CL (1975) A taxonomic revision of the Protomycetales. Mycotaxon 3:1–50
Rosling A, Cox F, Cruz-Martinez K, Ihrmark K, Grelet G-A, Lindahl BD, Menkis A, James TY (2011) Archaeorhizomycetes: unearthing an ancient class of ubiquitous soil fungi. Science 333:876–879
Scorzetti G, Fell JW, Fonseca Á, Statzell-Tallman A (2002) Systematics of basidiomycetous yeasts: a comparison of large subunit D1/D2 and internal transcribed spacer rDNA regions. FEMS Yeast Res 2:495–517
Sugiyama J, Hamamoto M (2011) Saitoella S. Goto, Sugiyama, Hamamoto and Komagata (1987). In: Kurtzman CP, Fell JW, Boekhout T (eds) The yeasts, a taxonomic study, 5th edn. Elsevier Science, Amsterdam, pp 1313–1315
Sugiyama J, Fukagawa M, Chiu S-W, Komagata K (1985) Cellular carbohydrate composition, DNA base composition, ubiquinone systems and diazonium blue B color test in the genera Rhodosporidium, Leucosporidium, Rhodotorula and related basidiomycetous yeasts. J Gen Appl Microbiol 31:519–550
Sugiyama J, Hosaka K, Suh SO (2006) Early diverging Ascomycota: phylogenetic divergence and related evolutionary enigmas. Mycologia 98:996–1005
Swofford DL (1998) PAUP*4.0: phylogenetic analysis using parsimony. Sinauer Associates, Sunderland
Vaughan-Martini A, Kurtzman CP, Meyer SA, O’Neill EB (2005) Two new species in the Pichia guilliermondii clade: Pichia caribbica sp. nov., the ascosporic state of Candida fermentati, and Candida carpophila comb. nov. FEMS Yeast Res 5:463–469
Acknowledgments
We thank two anonymous reviewers and Heide-Marie Daniel for helpful comments. The mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. The USDA is an equal opportunity provider and employer.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kurtzman, C.P., Robnett, C.J. Saitoella coloradoensis sp. nov., a new species of the Ascomycota, subphylum Taphrinomycotina. Antonie van Leeuwenhoek 101, 795–802 (2012). https://doi.org/10.1007/s10482-011-9694-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-011-9694-7