Abstract
A segment of DNA was isolated that complemented several poorly characterised sporulation-defective white-colony mutants of Streptomyces coelicolor A3(2) from an early collection (Hopwood et al., J Gen Microbiol 61: 397–408, 1970). Complementation was attributable to a gene, SCO4543, named whiJ, encoding a likely DNA-binding protein. Surprisingly, although some mutations in whiJ had a white colony phenotype, complete deletion of the wild-type or mutant gene gave a wild-type morphology. The whiJ gene is a member of a large paralogous set of S. coelicolor genes including abaAorfA, which regulates antibiotic production; and genes flanking whiJ are paralogues of other gene classes that are often associated with whiJ-like genes (Gehring et al., Proc Natl Acad Sci USA 97: 9642–9647, 2000). Thus, the small gene SCO4542 encodes a paralogue of the abaAorfD gene product, and SCO4544 encodes a paralogue of a family of likely anti-sigma factors (including the product of abaAorfB). Deletion of SCO4542 resulted in a medium-dependent bald- or white-colony phenotype, which could be completely suppressed by the simultaneous deletion of whiJ. A model is proposed in which WhiJ binds to operator sequences to repress developmental genes, with repression being released by interaction with the WhiJ-associated SCO4542 protein. It is suggested that this activity of SCO4542 protein is prevented by an unknown signal.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Streptomyces coelicolor A3(2) has been widely used as the model for this genus of antibiotic-producing, developmentally complex bacteria. Early studies of sporulation-deficient white colony mutants identified seven loci, whiA, whiB, whiC, whiD, whiG, whiH and whiI, that appeared to be specifically required for sporulation of aerial hyphae in S. coelicolor (Hopwood et al. 1970; Chater 1972). All of these genes except whiC have since been cloned and sequenced, and found to encode proteins playing likely regulatory roles that are essential for normal sporulation (Chater and Chandra 2006; Flärdh and Buttner 2009). Later studies of further, often phenotypically less well-defined, white colony mutants revealed some further genes (Ryding et al. 1999), including one gene (bldN) for a sigma factor that is involved in both aerial growth and the sporulation of aerial hyphae (Bibb et al. 2000). In addition, reverse genetic approaches have revealed further developmentally significant genes, such as the ssg genes that appear to be organisers of cell-division (Noens et al. 2005; Traag and van Wezel 2008; Xu et al. 2009).
Among the initial collection of whi mutants, three with somewhat poorly defined oligosporogenous phenotypes (C53, C77, C193) were found to have mutations close to strA (Chater 1972; Chater and Merrick 1976). Because of the difficulties of scoring these phenotypes, it was difficult to establish whether the mutations in these strains mapped to a single locus, so only one mutation was allocated a gene identifier: whiC193. Further studies of strains carrying this mutation were cut short when it turned out that all of the stocks had died. Here we describe the isolation and characterisation of a DNA fragment, containing intact genes SCO4541-SCO4545, that complemented the strA-linked mutations in strains C53 and C77, leading to the recognition of the whiJ gene (SCO4543). Following our deposition of the DNA sequence of the complementing clone, work done in parallel elsewhere showed that a transposon insertion in SCO4543 gave a similar (but more severe) phenotype (Gehring et al. 2000). As noted by Gehring et al. (2000) and Chater and Chandra (2006), whiJ is a member of a large family of paralogous genes present in the S. coelicolor genome, and the genes flanking it are likewise members of paralogous families, with the three kinds of genes often being found clustered together. We here describe the further analysis of whiJ and flanking sequences both by targeted mutagenesis and by computer analysis of the genome sequences of several streptomycetes. The results indicated that WhiJ protein is a repressor of sporulation that responds to a signal transmitted via at least one of the adjacent genes.
Materials and methods
Bacterial strains and microbiological techniques
S. coelicolor A3(2) derivatives C51, C53, C77, C141, C151, C246, C248, C249 and C253 were all from the collection of mutagen-induced developmental mutants described by Hopwood et al. (1970). Derivatives of strain M145, the plasmid-free version of A3(2) whose genome sequence has been determined (Bentley et al. 2002), were also constructed in this work (Table 1). The general conditions for maintenance and growth of these strains, and the media YEME (Yeast extract, Malt extract medium), TSB (tryptone soy broth), SMMS (supplemented minimal medium), DNA (Difco Nutrient Agar), MM (minimal medium, with mannitol as carbon source), SFM (soy flour mannitol medium), R2 (a hypertonic medium) and R2YE (a yeast extract-enriched version of R2) were as in Kieser et al. (2000). For work done in E. coli, maintenance and growth on medium LB were as in Sambrook and Russell (2001). The general hosts for cloning in E. coli were DH5α (Sambrook and Russell 2001) and XL1-Blue (Stratagene), and the non-methylating strain ET12567 was used for the preparation of non-methylated DNA for introduction into S. coelicolor (McNeil 1988).
Phase contrast microscopy and scanning electron microscopy were as described by Ryding et al. (1998).
DNA procedures and genetic manipulation
Basic procedures for DNA work were as in Sambrook and Russell (2001) for E. coli and Kieser et al. (2000) for Streptomyces. A previously described conjugatable library of SauIIIAI-partially digested fragments of DNA from S. coelicolor M145, cloned in a conjugation-proficient vector (pIJ698, which confers thiostrepton resistance), was maintained as an array of clone-containing S. coelicolor J1501 patches on master-plates (MM containing thiostrepton) (Ryding et al. 1998, 1999; Aínsa et al. 2000). The library was replicated to R2YE plates spread with spores and fragments of C53, a sporulation mutant. After incubation for 5 days, during which the plasmids containing cloned DNA were transferred by natural mating to the C53 recipient, the plates were further replicated to MM lacking the histidine and uracil required by J1501, but containing thiostrepton to select recipients that had received DNA associated with pIJ698. Potential complementation was indicated when exconjugants were grey and sporulating. To confirm complementation, plasmid DNA isolated from the corresponding patch in the library was used in direct transformation of C53 protoplasts (Kieser et al. 2000). The conjugative vectors pSET152 (Bierman et al. 1992) and pDH5 (Hillemann et al. 1991) were used to introduce subcloned DNA fragments into S. coelicolor mutants to test complementation. For routine subcloning, pIJ2925 (Kieser et al. 2000), pDH5 (Hillemann et al. 1991) and pBluescript vectors (Stratagene) were used. Plasmids constructed in this work are listed in Table 1.
To disrupt SCO4543, the entire 3.8–kb complementing insert from pIJ6205 was excised using XbaI and introduced in the XbaI site of a derivative of vector pDH5 so that the EcoRI site internal to SCO4543 was unique. Then, an EcoRI fragment containing a hygromycin resistance cassette was inserted at the EcoRI site of SCO4543, resulting in plasmid pIJ6411. pIJ6411 was used to replace the wild-type allele in S. coelicolor M145, generating strain J2452. For complete gene deletion-replacement mutations, cosmid 2D4 from the library of Redenbach et al. (1996), which contains a DNA insert that includes the entire whiJ locus, was used in PCR-targeted mutagenesis with the apramycin-resistance cassette (Gust et al. 2003, 2004) and primers listed in Table 1. All mutants were confirmed by PCR.
DNA isolated from putative whiJ mutants was used as template in PCR sequencing reactions, using primers 2StD4_14 T1 and 2StD4_14 T2 (Table 1). The sequencing was carried out by the Genome Centre of the John Innes Centre.
In BLAST searches, different cut-off expect values were used to search for paralogues of proteins encoded by the three genes of the whiJ locus, based on the levels at which proteins clearly belonging to different classes began to be found.
Results
Complementation of several poorly characterised whi mutants by a single cloned DNA fragment
Among the white colony mutants of S. coelicolor A3(2) isolated by Hopwood et al. (1970 and unpublished) were many with a poorly defined oligosporogenous phenotype, such as C53 and C77 (Fig. 1). After conjugation of a library of DNA from S. coelicolor M145 (Ryding et al. 1998, 1999) with C53, three exconjugant patches were found to have grey aerial mycelium. The corresponding plasmid clones in the original library were isolated and used to transform C53 and C77. One of them (named pIJ6205) complemented both mutants. The other two complemented neither: possibly they contained inserts of DNA from close to the site of the mutation, which could mobilise nearby chromosomal DNA containing wild-type sequences corresponding to the mutation in C53. The 3.8-kb insert in pIJ6205 was sequenced (Accession number AF106004). Later, when the genome sequence of S. coelicolor M145 became available (Bentley et al. 2002), the sequence was found to correspond to genes SCO4541-4546 (Fig. 2; below we discuss a single nucleotide difference between the two deposited sequences).
Phenotype of C77, a typical whiJ mutant, after 5 days on SFM. a Petri-dish cultures of C77 and its parent strain, A3(2). C77 grows vigorously, and produces aerial mycelium, but does not develop the grey pigment typical of wild-type spores. b SEM of wild-type sporulating aerial mycelium. Thick arrows, well-developed long spore chains. Thin wavy arrow, spiral aerial hyphae at an early stage of sporulation. Bar, 10 μm. c SEM of C77. Arrows, straight aerial hyphae that have failed to sporulate. C77 sporulates so infrequently that spore chains are seldom seen by SEM, though a few spores are usually found by phase-contrast microscopy. Bar, 10 μm
The whiJ gene cluster. The plasmid pIJ6205, which complemented whiJ mutants, was isolated from a plasmid library of S. coelicolor M145 DNA. Sequencing revealed ORFs SCO4542, SCO4543, SCO4544, SCO4545, and incomplete SCO4541 and SCO4546. Complementation tests using the plasmid subclones shown (see also Table 1), followed by sequencing of mutant alleles and disruption of SCO4543 at the EcoRI site, confirmed that SCO4543 was whiJ
In conjugation-mediated tests for its ability to complement a large number of other white mutants from the collection of Hopwood et al. (1970), pIJ6205 complemented several poorly characterised, mostly oligosporogenous, mutants (C51, C56, C89, C141, C151, C241, C246, C248, C249, C253). C53 and C77 had previously been found to have a whi mutation located just clockwise of strA (Chater and Merrick 1976). Since strA is presumed to encode the ribosomal protein RpsL (gene SCO4659, about 115 genes anticlockwise of SCO4542-4546), the genomic position of the complementing DNA was exactly as predicted from the earlier genetic linkage mapping. The locus corresponding to these mutations was named whiJ.
Identification of whiJ in the complementing DNA
The ability of smaller fragments from the original 3.8-kb clone to complement the whiJ mutation in C77 was tested (Fig. 2). A fragment in which the only intact ORF was SCO4543 (plasmid pIJ6415) complemented all the mutants that had been complemented by the initial 3.8-kb insert, except for C151.
To confirm that SCO4543 corresponded to whiJ, we constructed a mutant (J2452) with a hygromycin resistance gene inserted into the EcoRI site in SCO4543 (leaving codons 1–93 intact). Colonies of J2452 formed aerial mycelium, but remained white on prolonged incubation. Examination by light microscopy showed that the aerial mycelium mostly consisted of straight unbranched hyphae that failed to sporulate. This was similar to the phenotype of whiJ mutants (Fig. 1), including the transposon insertion mutant described by Gehring et al. (2000). The SCO4543-containing plasmid pIJ6415 complemented the SCO4543::hyg disruptant, showing that the white colony phenotype of the insertion mutant was not the result of polar effects on the expression of the downstream gene, SCO4542.
The ability of smaller fragments from the original 3.8-kb clone to complement the whiJ mutation in C77 was tested (Fig. 2). Complementation was observed with plasmids pIJ6416 (containing SCO4543 and SCO4544) and pIJ6415, in which the only intact ORF was SCO4543. Plasmid pIJ6415 further complemented all the other mutants that had been complemented by the initial 3.8-kb insert, except for C151. Sequencing of SCO4543 from C77 revealed one difference from the wild-type sequence in pIJ6205, at codon 104, resulting in a change from an acidic glutamate to a basic lysine residue, in agreement with the complementation of ca. 10% of C77 colonies that had received pIJ6409, an integrative plasmid carrying a truncated SCO4543 gene starting at codon 93 (Fig. 2). Sequencing also revealed SCO4543 mutations in C51 (M43I, G122K) and C248 (W88R, L100M). As mentioned above, the sequence of the 3.8-kb fragment differed at one base from that later determined for the equivalent sequence in the M145 genome (Bentley et al. 2002). This difference would cause a Y215F difference in the WhiJ protein. Since the mutant alleles sequenced all encoded an F residue at this position, we presume that F is the correct residue in the wild-type, as indicated in the full genome sequence (Bentley et al. 2002).
Bioinformatic analysis of whiJ and the flanking genes
SCO4543, termed whiJ for the rest of the paper, is a member of a large family of paralogous genes present in the S. coelicolor chromosome (Table 2): 26 match whiJ in a protein-level BLAST search (expect value of 0.001), all gene products falling in the protein size range 258–306 aa. The genes flanking it are likewise members of paralogous families: there are 17 members of the SCO4542-like family, all in the size range 63–96 aa (the small size of these proteins means that some members of the family may be excluded by the cut-off expect value of 0.1); while SCO4544-related genes are strikingly abundant, with 48 members falling into two size classes: 31 of 120–237 aa, and 17 of 552–916 aa (cut-off expect value 0.1). Genes in the whiJ family are nearly always located either in between members of each of the other classes, or next to one or the other of them (Table 2) (Gehring et al. 2000; Chater and Chandra 2006).
The WhiJ protein, like all its paralogues, has a predicted N-terminal DNA-binding helix-turn-helix domain (ending at residue 56) of the XRE/lambda C1 family. The downstream gene (SCO4542) encodes a 63-aa protein related to the c. 10 kDa acidic products of abaA orfD, one of a cluster of genes that influence secondary metabolism (Fernández-Moreno et al. 2002) and the developmental gene bldB (Pope et al. 1998; Eccleston et al. 2002), though the relationship to bldB was not detected at the cut-off expect value. The diverging upstream gene (SCO4544) encodes a protein with a C-terminal HATPase domain related to that of well-characterised anti-sigma factors, such as SpoIIAB of Bacillus subtilis (at expect values below the applied BLASTP cut-off of 0.1, bona fide anti-sigma factors of this class, and histidine protein kinases, were detected). Indeed, among the many SCO4544 paralogues present in S. coelicolor, one has a gene product that has been shown to interact with both a sigma factor and two anti-anti-sigma factors (Kim et al. 2008). SpoIIAB-like anti-sigmas antagonise members of the class of sigma factors such as σB and σF, in interactions that mediate stress-response and developmental gene expression (Helmann 1998; Mittenhuber 2002; Chen et al. 2003). The 17 larger members of the SCO4544 family all encode proteins with long N-terminal extensions that include putative sensory domains of the PAS or GAF families, and putative phosphorylase domains related to SpoIIE and its paralogues in B. subtilis. In B. subtilis, SpoIIE functions to dephosphorylate (and thereby activate) an anti-anti-sigma factor. No member of this class of large (552–916-aa) SCO4544-family proteins is encoded by a gene adjacent to a whiJ-like gene.
Despite the white colony phenotype of known whiJ mutants, whiJ is not needed for sporulation
The whiJ alleles with white colony phenotypes that were characterised at the DNA sequence level [i.e, a constructed insertion mutant J2452; a transposon insertion mutant (Gehring et al. 2000); and the mutagen-induced whiJ51, −77, −248] retained an intact 5′-end region potentially sufficient to encode a DNA-binding fragment of WhiJ [with the caveat that one of two mutations in whiJ51 results in a conservative change at codon 43 (M43I) that is also seen in several WhiJ paralogues]. To find out whether deletion of the entire gene gave a similar phenotype, we used PCR-targeted mutagenesis to replace the whole of SCO4543 with a cassette encoding apramycin resistance. To our surprise, the resulting mutant had grey, abundantly sporulating colonies that were indistinguishable from the wild-type (Fig. 3a), whether grown on minimal medium (MM), soy flour mannitol medium (SFM) or complex medium (R2YE). This suggested that the mutant alleles giving the white colony phenotype did indeed generate a partially functional WhiJ protein, capable of interfering with normal sporulation. This unexpected result led to the prediction that the introduction of the same deletion into a “standard” whiJ mutant, such as C77, would lead to the recovery of wild-type sporulation. This was confirmed (Fig. 3b).
Phenotypes of deletion mutants (SFM, 5 days). a Deletion of whiJ has no phenotypic effect. b Deletion of a point mutant allele of whiJ restores the wild-type phenotype. c Deletion of SCO4542 confers a bald phenotype. Inset, magnification showing segregation of a bald ΔSCO4542 colony from the heterogenote streakout
From these results, a basic model was developed in which WhiJ normally binds to a cognate operator sequence close to the promoter of one or more genes whose function is essential for sporulation. Sporulation is thereby repressed by wild-type WhiJ, but can be relieved when an unknown checkpoint signal causes WhiJ to detach from operator sites. Mutant alleles giving a white phenotype would retain the ability to bind to operators and repress sporulation, but would have lost the ability to respond to the checkpoint signal. Although such alleles might be expected to be dominant to the wild-type protein, they were found to be recessive (see above). To explain this, we suppose that the mutant forms do not compete well with the wild-type protein in some way—for example, if (like many repressors) WhiJ forms oligomers, the mutant forms may fail to do so, and so be less efficient in DNA-binding; or they might form mixed oligomers with wild-type subunits to give repressors with wild-type responses.
Morphological deficiency of a SCO4542 mutant, and suppression of this phenotype by simultaneous deletion of whiJ
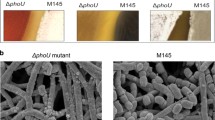
In view of the frequent association of gene pairs in the whiJ and SCO4542 families, it seemed possible that SCO4542 might also play a developmental role. We therefore constructed a cosmid containing a complete deletion of SCO4542, and used the mutated cosmid to replace the whiJ locus of the wild-type strain M145. In constructing the Streptomyces mutant, it was necessary first of all to introduce the mutated cosmid into the chromosome, thereby duplicating the entire chromosomal region contained in the cosmid. These partially diploid strains were selected from intergeneric conjugations by virtue of their resistance to kanamycin, conferred by the neo gene in the vector. Such exconjugants appeared to be morphologically normal. When they were re-plated on SFM medium selecting only for the apramycin resistance conferred by the disruption cassette, a mixture of colony morphologies was invariably seen: most wild-type, and a few bald (Fig. 3c). The latter colonies were kanamycin-sensitive, and were therefore presumptive haploid segregants with a single copy of the whiJ region of the chromosome carrying the SCO4542::AprR mutation. This was verified by PCR.
When the bald colonies were restreaked from SFM to Minimal Medium, a moderate amount of white aerial mycelium formed, and on the richer and hypertonic medium R2YE the colonies produced abundant aerial mycelium, which remained white and non-sporulating on prolonged incubation, while the substrate mycelium produced blue pigment (as did the wild-type) (Fig. 4). It thus appeared that on SFM, SCO4542 is normally needed for aerial growth and actinorhodin production; while on R2YE that requirement is bypassed, and aerial hyphae can grow, but not differentiate into spores. Thus, the action of SCO4542 shows some sensitivity to environmental cues.
Because of the possibility that SCO4542 protein might interact with WhiJ, we went onto test the phenotype of a SCO4542/SCO4543 double deletion mutant constructed by PCR-targeting. The mutant was morphologically indistinguishable from the wild-type M145 parent on any of the media tested (result not shown). Since the morphological defects resulting from SCO4542 deletion could be bypassed by eliminating the repressor encoded by whiJ (SCO4543), we propose that SCO4542 acts via WhiJ.
Discussion
A cascade of negatively acting steps regulating development and secondary metabolism
The transposon mutagenesis study of Gehring et al. (2000) and our results reported here have identified a further sporulation regulatory gene, whiJ. Unexpectedly, and unlike any of the previously studied whi genes, whiJ is not required for sporulation—a complete deletion of whiJ had no phenotypic effect. Instead, the morphological deficiencies of the whiJ mutants result from alleles that retain partial function.
These results indicate that WhiJ is a repressor of development, whose function in the wild-type is more or less irrelevant to conditions encountered in the laboratory, but which presumably has some adaptive benefit under natural conditions. Genomic analysis suggested that the adjacent gene, SCO4542, might be involved in some interplay with whiJ, and indeed a SCO4542 mutant also had a white colony phenotype on R2YE, though the SCO4542 mutant was devoid of aerial mycelium on SFM medium. Remarkably, the SCO4542 mutant phenotype was entirely suppressed by simultaneous deletion of whiJ.
It appears that sporulation in Streptomyces is subject to many checkpoint controls, particularly involving exposure to stress (e.g. Cho et al. 2001; Kelemen et al. 2001; Sevcikova et al. 2001; Chater 2001; Viollier et al. 2003; Chater and Horinouchi 2003; Lee et al. 2005). The observations on SCO4542 and whiJ are explicable by a model in which SCO4542 protein is involved in transmitting a checkpoint signal to WhiJ (Fig. 5). In the absence of this signal transmission, WhiJ binds to one or more targets in the genome and thereby prevents the expression of genes involved in development. The various phenotypic phenomena that we have described can then be explained as follows. In the laboratory, the checkpoint perceived via SCO4542 protein is constitutively passed, so that SCO4542 protein prevents WhiJ from binding to target DNA. In this situation, WhiJ would play no part in regulating development, hence the absence of phenotypic effects of deleting the whiJ gene. On the other hand, the deletion specifically of SCO4542 would result in the constitutive binding of WhiJ to its target(s), and therefore the repression of development indicated by a bald colony phenotype.
Model for the negatively acting cascade encoded by whiJ and SCO4542. WhiJ is a repressor of unidentified genes needed for aerial growth and/or antibiotic production. WhiJ is sensitive to SCO4542 protein, which interferes with repression. In turn, SCO4542 protein is sensitive to some unknown signal, which results in release of WhiJ from SCO4542 protein, thereby repressing development. This signal is presumed to be absent under laboratory conditions, so development is independent of WhiJ. Under certain conditions (such as growth on R2YE medium), WhiJ is modified to a form (WhiJ*) that cannot repress aerial growth, but does repress sporulation of aerial hyphae. WhiJ* is also sensitive to SCO4542 protein. The forms of WhiJ in mutants such as C53 and C77 are presumed to be insensitive to SCO4542 protein
To explain the conditional white phenotype of the SCO4542 mutant, we suggest that WhiJ may undergo a SCO4542-independent change during growth on certain media, such as R2YE, with the second form (WhiJ*) being ineffective in repressing early development, but effective in repressing later stages. On this model, whiJ mutant alleles that give a white colony phenotype would mimic the WhiJ* form. Since the constructed disruptant J2542 and the transposon-induced mutant of Gehring et al. (2000) would presumably express just the N-terminal helix-turn-helix-containing domain, it is tempting to speculate that WhiJ* is a form in which an interaction between the N-terminal domain and the rest of the protein is prevented. The change to WhiJ* might either be a direct response of WhiJ to a signal or be mediated by some other, converging, signal transduction system that can interact with WhiJ.
It seems likely that each of the many pairs of genes paralogous with SCO4543 and SCO4542 may be functioning in a similar manner, with the SCO4542 homologue being involved in transmitting a signal to the SCO4543 homologue and thereby changing its regulatory action.
Implications of the frequent association of whiJ paralogues with SCO4544 paralogues
In this paper we do not report on mutations in SCO4544, which is also a member of a paralogous family frequently represented in whiJ-like clusters, and which will need to be accommodated as the models for whiJ action become increasingly refined. Here we make two observations: a strain in which the three genes SCO4542-4544 are all deleted has a medium dependent phenotype of being bald and overproducing antibiotics (N Bird and KF Chater, unpublished); and the SCO4544 product is a member of a protein family at least one of whose members is able to interact with a sigma factor and with two anti-anti-sigma factors, and is highly likely to act as an anti-sigma factor (Kim et al. 2008). Furthermore, two of these genes (SCO0673 and SCO0868) are next to genes annotated as encoding anti-anti-sigma factors, while a third, SCO7277, is next to a gene encoding a sigma factor of the cognate family. This family of sigmas includes at least two members, σF and σN, whose function is primarily developmental (Potuckova et al. 1995; Dalton et al. 2007), and several that function primarily in stress responses, but which also influence development (Cho et al. 2001; Kelemen et al. 2001; Sevcikova et al. 2001; Viollier et al. 2003; Lee et al. 2005). It has been suggested that there may be highly complex cross-talk in the regulation of activity of sigma factors of this family (Kim et al. 2008). The evidence here that this regulation is also likely to be sensitive to signal input from whiJ-like gene sets appears to complicate matters even further.
The distribution of whiJ-like gene sets
Clusters similar to the whiJ cluster are widespread and numerous in sequenced genomes of other streptomycetes, but they are generally absent from any other bacterial genomes. The only exceptions are a few examples in other mycelial actinomycetes (Chater and Chandra 2006). Strikingly, though, the whiJ cluster is flanked on both sides by genes that are absent from other sequenced Streptomyces genomes, and does not itself appear to have orthologues in those genomes as judged by reciprocal BLAST analysis.
Not all such gene sets are conserved among different Streptomyces genomes, perhaps indicating that different species perceive an incompletely congruent set of signals as part of their adaptation to a particular range of ecological niches. Thus, the nature of the suite of such sets in any one organism may play a part in speciation. A mechanism for the acquisition of whiJ-family clusters by lateral gene transfer is suggested by the presence of such a cluster in the terminal inverted repeats of SCP1, a highly transmissible linear plasmid of S. coelicolor that has been shown to integrate stably into the chromosome (Bentley et al. 2004).
There is little information to indicate the nature of the signals, or the target genes regulated by WhiJ-like proteins, or the areas of physiology whose activity is influenced by these regulatory complexes. However, one whiJ-like cluster, abaA, has been found to influence secondary metabolism (Fernández-Moreno et al. 2002), while mutations in an isolated SCO4542-like gene, bldB, cause severe impairment of development (Merrick 1976; Eccleston et al. 2002). We have also found that mutations in the complex whiJ-like cluster SCO2859-2869 give rise to changes in both morphological differentiation and antibiotic production (M Korberska, K Fowler and KF Chater, unpublished). It therefore seems possible that a significant fraction of such genes may have the potential to repress morphological and physiological development. Thus, the systematic elimination of all the whiJ-like gene sets from a genome might be expected to lead to increased readiness to differentiate. In this connection, it will be of interest to enumerate whiJ-locus-like gene clusters in Streptomyces venezuelae, a remarkably rapidly growing streptomycete that sporulates very quickly even in liquid medium (Flärdh and Buttner 2009).
References
Aínsa JA, Ryding NJ, Hartley N, Findlay KC, Bruton CJ, Chater KF (2000) WhiA, a protein of unknown function conserved among gram-positive bacteria, is essential for sporulation in Streptomyces coelicolor A3(2). J Bacteriol 182:5470–5478
Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang CH, Kieser T, Larke L, Murphy L, Oliver K, O’Neil S, Rabbinowitsch E, Rajandream MA, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147
Bentley SD, Brown S, Murphy LD, Harris DE, Quail MA, Parkhill J, Barrell BG, McCormick JR, Santamaría RI, Losick R, Yamasaki M, Kinashi H, Chen CW, Chandra G, Jakimowicz D, Kieser HM, Kieser T, Chater KF (2004) SCP1, a 356 023 bp linear plasmid adapted to the ecology and developmental biology of its host, Streptomyces coelicolor A3(2). Mol Microbiol 51:1615–1628
Bibb MJ, Molle V, Buttner MJ (2000) Sigma(BldN), an extracytoplasmic function RNA polymerase sigma factor required for aerial mycelium formation in Streptomyces coelicolor A3(2). J Bacteriol 182:4606–4616
Bierman M, Logan R, O’Brien K, Seno ET, Rao N, Schoner BE (1992) Plasmid cloning vectors for the conjugal transfer of DNA from E. coli to Streptomyces spp. Gene 116:43–49
Chater KF (1972) A morphological and genetic mapping study of white colony mutants of Streptomyces coelicolor. J Gen Microbiol 72:9–28
Chater KF (2001) Regulation of sporulation in Streptomyces coelicolor A3(2): a checkpoint multiplex? Curr Opin Microbiol 4:667–673
Chater KF, Chandra G (2006) The evolution of development in Streptomyces analysed by genome comparisons. FEMS Microbiol Rev 30:651–672
Chater KF, Horinouchi S (2003) Signalling early developmental events in two highly diverged Streptomyces species. Mol Microbiol 48:9–15
Chater KF, Merrick MJ (1976) Approaches to the study of differentiation in Streptomyces coelicolor A3(2) In: Macdonald KD (ed) 2nd international symposium on the genetics of industrial micro-organisms. Academic Press, London, pp 583–593
Chen C-C, Lewis RJ, Harris R, Yudkin MD, Delumeau O (2003) A supramolecular complex in the environmental stress signalling pathway of Bacillus subtilis. Mol Microbiol 49:1657–1669
Cho YH, Lee EJ, Ahn BE, Roe JH (2001) SigB, an RNA polymerase sigma factor required for osmoprotection and proper differentiation of Streptomyces coelicolor. Mol Microbiol 42:205–214
Dalton KA, Thibessard A, Hunter JI, Kelemen GH (2007) A novel compartment, the ‘subapical stem’ of the aerial hyphae, is the location of a sigN-dependent, developmentally distinct transcription in Streptomyces coelicolor. Mol Microbiol 64:719–737
Eccleston M, Ali R, Seyler R, Westpheling J, Nodwell J (2002) Structural and genetic analysis of the BldB protein of Streptomyces coelicolor. J Bacteriol 184:4270–4276
Fernández-Moreno M, Martin-Triana AJ, Martinez E, Niemi J, Kieser H, Hopwood D, Malpartida F (2002) abaA, a new pleiotropic regulatory locus for antibiotic production in Streptomyces coelicolor. J Bacteriol 174:2958–2967
Flärdh K, Buttner MJ (2009) Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat Rev Microbiol 7:36–49
Gehring A, Nodwell J, Beverley S, Losick R (2000) Genomewide insertional mutagenesis in Streptomyces coelicolor reveals additional genes involved in morphological differentiation. Proc Natl Acad Sci USA 97:9642–9647
Gust B, Challis GL, Fowler K, Kieser T, Chater KF (2003) PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci USA 100:1541–1546
Gust B, Chandra G, Jakimowicz D, Yuqing T, Bruton CJ, Chater KF (2004) Lambda red-mediated genetic manipulation of antibiotic-producing Streptomyces. Adv Appl Microbiol 54:107–128
Helmann JD (1998) Anti-sigma factors. Curr Opin Microbiol 2:135–141
Hillemann D, Pühler A, Wohlleben W (1991) Gene disruption and gene replacement in Streptomyces via single-stranded DNA transformation of integration vectors. Nucleic Acids Res 19:727–731
Hopwood DA, Wildermuth H, Palmer HM (1970) Mutants of Streptomyces coelicolor defective in sporulation. J Gen Microbiol 61:397–408
Kelemen GH, Viollier PH, Tenor JL, Marri L, Buttner MJ, Thompson CJ (2001) A connection between stress and development in the multicellular prokaryote Streptomyces coelicolor A3(2). Mol Microbiol 40:804–814
Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA (2000) Practical Streptomyces genetics. The John Innes Foundation, Norwich
Kim ES, Song JY, Kim DW, Chater KF, Lee KJ (2008) A possible extended family of regulators of sigma factor activity in Streptomyces coelicolor. J Bacteriol 190:7559–7566
Lee E, Karoonuthaisiri N, Kim H, Park J, Cha C, Kao C, Roe J (2005) A master regulator sigma governs osmotic and oxidative response as well as differentiation via a network of sigma factors in Streptomyces coelicolor. Mol Microbiol 57:1252–1264
McNeil DJ (1988) Characterisation of a unique methyl-specific restriction system in Streptomyces avermitilis. J Bacteriol 170:5607–5612
Merrick M (1976) A morphological and genetic mapping study of bald colony mutants of Streptomyces coelicolor. J Gen Microbiol 96:299–315
Mittenhuber G (2002) A phylogenomic study of the general stress response sigma factor σB of Bacillus subtilis and its regulatory proteins. J Mol Microbiol Biotechnol 4:427–452
Noens EEE, Mersinias V, Traag BA, Smith CP, Koerten HK, Van Wezel GP (2005) SsgA-like proteins determine the fate of peptidoglycan during sporulation of Streptomyces coelicolor. Mol Microbiol 58:929–944
Pope MK, Green B, Westpheling J (1998) The bldB gene encodes a small protein required for morphogenesis, antibiotic production, and catabolite control in Streptomyces coelicolor. J Bacteriol 180:1556–1562
Potuckova L, Kelemen GH, Findlay KC, Lonetto MA, Buttner MJ, Kormanec J (1995) A new RNA polymerase sigma factor, sigma(F), is required for the late stages of morphological differentiation in Streptomyces spp. Mol Microbiol 17:37–48
Redenbach M, Kieser HM, Denapaite D, Eichner A, Cullum J, Kinashi H, Hopwood DA (1996) A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol Microbiol 21:77–96
Ryding NJ, Kelemen GH, Whatling CA, Flärdh K, Buttner MJ, Chater KF (1998) A developmentally regulated gene encoding a repressor-like protein is essential for sporulation in Streptomyces coelicolor A3(2). Mol Microbiol 29:343–357
Ryding NJ, Bibb MJ, Molle V, Findlay KC, Chater KF, Buttner MJ (1999) New sporulation loci in Streptomyces coelicolor A3(2). J Bacteriol 181:5419–5425
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
Sevcikova B, Benada O, Kofronova O, Kormanec J (2001) Stress response sigma factor sigma(H) is essential for morphological differentiation of Streptomyces coelicolor A3(2). Arch Microbiol 177:98–106
Traag BA, Van Wezel GP (2008) The SsgA-like proteins in actinomycetes: small proteins up to a big task. Antonie Van Leeuwenhoek 94:85–97
Viollier PH, Kelemen GH, Dale GE, Nguyen KT, Buttner MJ, Thompson CJ (2003) Specialized osmotic stress response systems involve multiple SigB-like sigma factors in Streptomyces coelicolor. Mol Microbiol 47:699–714
Xu Q, Traag BA, Willemse J, McMullan D, Miller MD, Elsliger MA, Abdubek P, Astakhova T, Axelrod HL, Bakolitsa C, Carlton D, Chen C, Chiu HJ, Chruszcz M, Clayton T, Das D, Deller MC, Duan L, Ellrott K, Ernst D, Farr CL, Feuerhelm J, Grant JC, Grzechnik A, Grzechnik SK, Han GW, Jaroszewski L, Jin KK, Klock HE, Knuth MW, Kozbial P, Krishna SS, Kumar A, Marciano D, Minor W, Mommaas AM, Morse AT, Nigoghossian E, Nopakun A, Okach L, Oommachen S, Paulsen J, Puckett C, Reyes R, Rife CL, Sefcovic N, Tien HJ, Trame CB, van den Bedem H, Wang S, Weekes D, Hodgson KO, Wooley J, Deacon AM, Godzik A, Lesley SA, Wilson IA, van Wezel GP (2009) Structural and functional characterizations of SsgB, a conserved activator of developmental cell division in morphologically complex actinomycetes. J Biol Chem 284:25268–25279
Acknowledgements
We gratefully thank Helen Kieser and David Hopwood for strains recovered from archives, Govind Chandra for carrying out bioinformatic analysis and Kay Fowler for technical advice. This work was supported by grants from the John Innes Foundation (NJR) and the BBSRC grant CAD 04380 (JAA). KFC is a John Innes Foundation Emeritus Fellow.
Author information
Authors and Affiliations
Corresponding author
Additional information
José A. Aínsa, Nick Bird, and N. Jamie Ryding have made equal contributions to the work reported.
Rights and permissions
About this article
Cite this article
Aínsa, J.A., Bird, N., Ryding, N.J. et al. The complex whiJ locus mediates environmentally sensitive repression of development of Streptomyces coelicolor A3(2). Antonie van Leeuwenhoek 98, 225–236 (2010). https://doi.org/10.1007/s10482-010-9443-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-010-9443-3