Abstract
The rate of human health care-associated infections caused by Acinetobacter baumannii has increased significantly in recent years for its remarkable resistance to desiccation and most antibiotics. Phospholipases, capable of destroying a phospholipid substrate, are heterologous group of enzymes which are believed to be the bacterial virulence determinants. There is a need for in silico studies to identify potential vaccine candidates. A. baumannii phospholipase D (PLD) role has been proved in increasing organism’s resistance to human serum, destruction of host epithelial cell and pathogenesis in murine model. In this in silico study high potentials of A. baumannii PLD in elicitation of humoral and cellular immunities were elucidated. Thermal stability, long half-life, non-similarity to human and gut flora proteome and non-allergenicity were in a list of A. baumannii PLD positive properties. Potential epitopic sequences were also identified that could be used as peptide vaccines against A. baumannii and various other human bacterial pathogens.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The most significant clinical representative of Acinetobacter spp. in hospitals is Acinetobacter baumannii. According to the Centers for Disease Control (CDC) reports, A. baumannii is cause of about 80 % of documented Acinetobacter infections. A. baumannii has appeared as a globally important multidrug-resistant (MDR) trouble maker over the last two decades. Considerable potency to upregulate or obtain antibiotic resistance genes even to forceful antibiotics, resistance to drying and disinfectants, having ability to airborne transmission and patient-to-patient transmission account for long-term persistence and shedding of these microorganisms into their surrounding environment A. baumannii potential to survive in clinical environment has been reported especially in Asian and Middle Eastern hospitals and from war zones in Iraq and Afghanistan (Camp and Tatum 2010; Peleg et al. 2008; Towner 2009). A. baumannii relevant problematic infections are ventilator-associated pneumonia, skin, central nervous system, urinary tract, bone, blood-stream and soft tissue infections, wound infections and secondary meningitis. Given the aforesaid reasons, it is obvious that many of these infections occur in intensive care units (ICUs), where the severely ill patients are treated with broad-spectrum antibiotics or require mechanical ventilation, or bear wound or burn injuries (Peleg et al. 2008; Towner 2009). Mortality rates of A. baumannii infections range from 19 to 54 % (Russo et al. 2013). Pathogenic bacteria produce various substances like capsule, slime, proteins, toxins, and enzymes to aid in the survival, propagation and invasion in the host, and also in eluding defense mechanisms of the host. Requisite of host invasion by microorganisms is production of hydrolytic enzymes which destroy the outer cell envelope. The main targets of these enzymes are membrane lipids and proteins (Braun 2008; Ghannoum 2000).
Membrane ester linkage in glycerophospholipids could be cleaved by a heterogeneous group of enzymes namely “phospholipases” (PLs) depending on specific ester bond targeted, A, B, C and D letters are used in the nomenclature of phospholipase (Ghannoum 2000; Ivanovska 2003).
Phospholipase D (PLD) is a signal-activated enzyme that can catalyze two reactions: (1) cleavage of the phosphate side to yield phosphatidic acid and free polar head group of choline, inositol and ethanolamine, depending on the primary substrate; (2) generation of different phospholipid by primary or secondary alcohols transphosphatidylation (Yang and Roberts 2002). Several other phosphatidyl transferase enzymes involved in lipid metabolism such as cardiolipin and phosphaidylserine synthases hydrolyze equivalent bond, too (Liscovitch et al. 1999; Ponting and Kerr 1996; Schmiel and Miller 1999).
The existence of PLD has been proven in both eukaryotes, including animals, plants, fungi, protozoa, and prokaryote bacteria (Exton 1997). PLD is one of Corynebacterium pseudotuberculosis major exotoxins which contributes to spread of bacteria by hydrolyzing ester linkages in host cell membrane (McKean et al. 2007). This microorganism causes caseous lymphadenitis (CLA), a globally distributed disease of sheep and goats (Dorella et al. 2006). Commercial combined vaccines based on cell-culture supernatant of this bacterium, containing PLD as a main component, and other pathogens are available in some countries. Glanvac is one example of these kinds of vaccines (Stanford et al. 1998). Also significant protection against C. pseudotuberculosis has been reported by vaccination of animal models based on PLD with different strategies such as attenuated vaccine via constructing PLD-negative (Hodgson et al. 1992) or genetically inactive PLD mutants of C. pseudotuberculosis (Hodgson et al. 1999), DNA vaccine expressing some antigens along with genetically manipulated PLD (Chaplin et al. 1999) and subunit vaccine by producing recombinant inactive PLD (Fontaine et al. 2006; Young 2011). In the case of A. baumannii, studies are indicative of PLD role in increasing organism’s ability to prosper in the human serum, epithelial cell invasion and pathogenesis (Cerqueira and Peleg 2011; de Leseleuc and Chen 2012; Jacobs et al. 2010). Several methods using whole cell, capsular polysaccharide, outer membrane proteins, autotransporter, biofilm protein were tested to produce a therapeutic antiserum against A. baumannii and indicated the possibility of their vaccination potential (Bentancor et al. 2012; Fattahian et al. 2011; McConnell et al. 2011; McConnell and Pachon 2010; Russo et al. 2013). However, some of them have several disadvantages such as high costs of preparation, limited protection against some isolates. The prevalence, variability and solubility of single antigen candidates barricade delivery of an ideal effective vaccine. Since no vaccine has been licensed for A. baumannii, attempts should be made to arrive at better vaccine candidates with least drawbacks. Better recognition of the molecules and mechanisms involved in pathogenicity of this microorganism would help the researchers arrive at their objectives of finding efficient targets for new therapies. The present work focuses on in silico analysis of one of the most neglected molecules of A. baumannii described as PLD.
Similar in silico methods were reported to have been used in identification of vaccine candidates in Salmonella Typhi (Prabhavathy et al. 2011), Mycobacterium tuberculosis (Somvanshi et al. 2008), Neisseria meningitides (Chandra et al. 2010) and Neisseria gonorrhoeae (Barh et al. 2010). The peptide vaccine in question can be used against some other pathogenic bacteria too. To the best of our knowledge, this is the first in silico study designed to address important characteristics of PLD as a suitable vaccine candidate and is the first attempt made to develop peptide vaccine against A. baumannii. Our results identified PLD as a vaccine candidate owing to its intrinsic advantages.
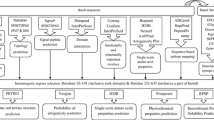
2 Methods
2.1 Primary Sequence Analysis
The sequence of A. baumanii ATCC19606 PLD with accession number WP_001985196.1 was retrieved from NCBI protein database (http://www.ncbi.nlm.nih.gov/protein), in FASTA format.
Selected sequence was subjected to blastp against non redundant protein sequence (nr) Database with A. baumanii as a selected organism in NCBI server (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The resulted sequence with a similarity >50 %, coverage > 50 % and E-value < 10−4 were selected and aligned with multiple sequence alignment tool PRALINE (http://ibivu.cs.vu.nl/programs/pralinewww/) (Simossis and Heringa 2005). It has high quality output files for publication and presentation purposes (Kumar and Srivastava 2012). The aim was to choose the longest sequence to continue working on it.
The primary structure and the basic physico-chemical properties of the selected PLD sequences were computed using ProtParam tool (http://web.expasy.org/protparam/).
Subcellular localization of PLD in gram-negative bacteria was predicted by PSLpred (http://www.imtech.res.in/raghava/) a hybrid approach-based method with an overall high accuracy of 91.2 % (Bhasin et al. 2005) and CELLO at cello.life.nctu.edu.tw which uses multiple Support Vector Machines (SVMs) to assign a query protein to one of the five Gram-negative localization sites (Yu et al. 2006).
Presence of signal peptide sequence was tested by Signal-3L software at http://www.csbio.sjtu.edu.cn/bioinf/Signal-3L/ (Shen and Chou 2007) and SignalP at http://www.cbs.dtu.dk/services/SignalP-3.0/ (Dyrlov Bendtsen et al. 2004).
Signal-3L predictor software consisting of three prediction engines is known by high success prediction rates and short computational time (Shen and Chou 2007). According to a benchmark study SignalP 3.0 was found to be the best method of signal peptide prediction (Choo et al. 2009).
Solubility of PLD was evaluated by Recombinant Protein Solubility Prediction (http://biotech.ou.edu/#rt) by discriminated analysis of charge average, molecular weight, amino acid fractions, aliphatic, alpha-helix propensity, beta-sheet propensity, average pI and hydrophilicity parameters (Diaz et al. 2010). Surface accessibility of PLD was estimated by IEDB surface accessibility prediction (http://tools.immuneepitope.org/tools/bcell/iedb_input) at default threshold (Emini et al. 1985).
Algpred sever (http://www.imtech.res.in/raghava/algpred/submission.html) allowed prediction of allergens based on combining different approaches of SVMs, MEME/MAST, IgE epitope mapping and blast search against 2,890 allergen-representative peptides (ARPs). The protein sequence is identified as allergen if any one of the methods predicts it as allergen (Saha and Raghava 2006a, b). Algpred has the highest specificity compared with other predictors (Dimitrov et al. 2013).
The PLD protein close homologues in the human proteome were identified through human blast p using a BLOSUM62 matrix (with abovementioned cutoff values). The same analysis was performed against mouse proteome, because it is most widely used animal model in studying diseases.
Similarity of PLD protein to gut flora proteins was searched through blast (with abovementioned cutoff values) against the proteome of 95 organisms microbes comprising the human gut flora, obtained from a report by Raman et al. (2008), that inhabit the healthy human gut.
2.2 Secondary and Tertiary Structure Prediction
The secondary protein structure was predicted by self-optimized prediction align method, SOPMA (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html) (Geourjon and Deleage 1995) using default parameters (Window width: 17, similarity threshold: 8 and number of states: 4) and PSIPRED v 3.3 (http://bioinf.cs.ucl.ac.uk/psipred/). The PSIPRED assembles several structure prediction methods, data to analyze protein sequences (McGuffin et al. 2000).
3D protein structure predictions were performed by online I-Tasser (iterative threading assembly refinement) software at http://zhanglab.ccmb.med.umich.edu/I-TASSER/. It combines ab initio folding and threading modeling methods to generate tertiary structure. It has been ranked as the best method for the automated protein structure prediction in the recent CASP experiments because of using composite approaches in protein structure prediction (Roy et al. 2010).
2.3 Model Evaluation and Refinement
The 3D model with the best score of I-Tasser was selected and the structure was evaluated qualitatively by PROSESS (Protein Structure Evaluation Suite and Server) (http://www.prosess.ca) (Berjanskii et al. 2010) and Qmean (Qualitative model energy analysis) (http://swissmodel.expasy.org/qmean/cgi/index.cgi?page=help) (Benkert et al. 2008).
PROSESS is a comprehensive web server, with ability to unify previously tested methods to evaluate predicted structures (Berjanskii et al. 2010). Qmean describes the major geometrical aspects of protein structures by scoring function to distinguish good models from bad (Benkert et al. 2008).
In addition, the Ramachandran diagram was plotted via Rampage (http://mordred.bioc.cam.ac.uk/~rapper/rampage.php).
Ramachandran plot can determine which torsional angles can obtain insight into the structure of peptides. The selected PLD 3D structure was corrected by ModReiner (http://zhanglab.ccmb.med.umich.edu/ModReiner/) which improves models by minimization of atomic-level energy (Xu and Zhang 2011).
To prove model refinement, the model evaluated by PROSESS, Qmean. The Ramachandran plot was also depicted. The refined model was used at discontinuous B cell epitopes predictions.
2.4 Antigenicity Prediction
Sequence was analyzed for antigencity using IEDB B cell antigenic prediction site (http://tools.immuneepitope.org/tools/bcell/iedb_input) (Zhang et al. 2008), at default threshold value of 1.0, which predicts antigenic determinants on protein sequence, using a semi-empirical method of Kolaskar and Tongaonkar (1990). To ensure accuracy of antigenic areas, Antigenic Peptide Prediction tool available at http://imed.med.ucm.es/Tools/tools.html was employed too. The VaxiJen v2.0 server (http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html) (Doytchinova and Flower 2007) was used to analyze the antigenicity of PLD. VaxiJen is alignment-free approach for antigen prediction. It allows antigen classification based on the physicochemical properties of proteins for vaccine development.
2.5 Identification of B-Cell Epitopes
B-cell epitopes identification plays an important role in vaccine design and antibody production. For prediction of B-cell epitopes, full-length PLD sequence was subjected to B-cell epitope prediction using BCPreds (http://ailab.cs.iastate.edu/bcpreds/index.html) (EL-Manzalawy et al. 2008). Both the BCP and AAP prediction methods with specificity of 75 % and epitope length of 20aa were used. Possibility of BCPreds determined sequences to be epitope through ABCpred (http://www.imtech.res.in/raghava/abcpred/) (Saha and Raghava 2006b) and LBtope (www.imtech.res.in/raghava/lbtope/) (Singh et al. 2013) were investigated.
Spatial Epitope Prediction of Protein Antigens (Seppa) (http://lifecenter.sgst.cn/seppa) (Sun et al. 2009) and BEpro at http://pepito.proteomics.ics.uci.edu/cgi-bin/BEpro.cgi (Sweredoski and Baldi 2008) were used for predicting conformational B-cell epitopes with three-dimensional protein structures. The default threshold of the server is 1.80 to help specify the epitope residues.
2.6 T-Cell Epitope Prediction
Prediction of human HLA restricted determinants within this antigen was detected by the first prediction server using the matrix-based TEPITOPE algorithm, ProPred (http://www.imtech.res.in/raghava/propred) (Singh and Raghava 2001), and ProPredI (http://www.imtech.res.in/raghava/propred1) (Singh and Raghava 2003) servers to predict HTL (MHC class II binding peptides) for 51 alleles and CTL (MHC class I binding peptides(for 47 alleles. ProPred analysis was performed at the default setting with threshold value of 3 %. In the case of PropPred, 4 % default threshold level with 5 % proteasome filter was selected. The 50 % inhibitory concentration (IC50) of common epitopes of both of servers was calculated using quantitative prediction of binding affinity of peptide–MHC binding site, MHCPred (http://www.ddg-pharmfac.net/mhcpred/MHCPred/) (Guan et al. 2003). Epitopes with IC50 value <1,000 nM for allele DRB1*0101 were selected as a good epitope.
2.7 Identification of Common Epitope for Multiple Pathogens
B-cell predicted epitopes by both modules of BCPreds (BCPred and AAP) having cutoff score of >0.8 and antigenic sites predicted by IEDB (cutoff score > 1) were aligned to get common overlapping amino acid sequences that possess both the B-cell binding sites as well as antigenic sequences. Selected sequence(s) was then analyzed with ProPred and ProPredI using parameters as mentioned before, to find whether this common sequence(s) elicit T cell response. The sequence(s) was/were further checked using VaxiJen v2.0 server.
To identify a single epitope against multiple pathogens, full length sequence of A. baumannii PLD was subjected to blastp [cutoff values: bit score (>100), E-value (<10−10), and percentage of identity at amino acid level (>35 %)] against proteome of CDC reported important pathogens associated with antimicrobial resistance (Clostridium difficile, Klebsiella spp., Escherchia coli, N. gonorrhoeae, M. tuberculosis, Campylobacter spp., Enterococcus spp., Pseudomonas aeruginosa, non-typhoidal Salmonella, S. Typhi, Shigella spp., Streptococcus pneumonia and Staphylococcus aureus) (Centers for Disease Control and Prevention 2013).
The essentiality in survival of these microorganisms harboring PLD homologous proteins was checked by blastp against Database of Essential Genes (DEG) at http://tubic.tju.edu.cn/deg/ (Zhang et al. 2004).
PLD homologue sequences of the qualified pathogens were checked for their similarity to human proteome via blastp. Non-human homologues were aligned using PRALINE to determine similarity at common sequence(s) position(s). The antigenicity of these sequences was analyzed using VaxiJen. Their Tcell eliciting ability was predicted by ProPred and ProPredI.
3 Results
3.1 Primary Sequence Analysis
Multiple sequence alignment showed the selected PLD sequence to be the longest one (543aa) as against a protein from A. baumannii ATCC 17978 (accession number ABS90299.1) as the shortest one (77aa). ProtParam parameters shown in Table 1 reveal the physicochemical properties of the protein. The number of positively charged residues are slightly higher than negatively charged residues. Both subcellular localization prediction sites viz., CELLO and PSLpred identified PLD as a periplasmic protein with the reliability index of 2.683 and 1 respectively. A potential signal peptide comprising 36 amino acids was predicted in the N-terminal of the protein by signal-3L and SignalP software. The PLD sequence has 95.5 % chance of solubility when overexpressed in E. coli. The average score of surface accessibility predicted by Emini Surface Accessibility Prediction was 1.00 and the maximum and minimum scores were 5.763 and 0.052, respectively. Predicted peptides are given in Table 2. Based on different allergen prediction approaches in AlgPred, the PLD protein was not detected as an allergen. PLD was not similar to any human or murine protein. The most similar human and/murine proteins had 22/34 % identity, query coverage of 31/36 %, E-value of 0.088/0.36 and total score of 38.5/36.2 respectively. PLD protein Blastp against proteome of gut flora showed that just one out of the 95 bacteria namely Citrobacter sp. ATCC 29220 had the cutoff prerequisites. PLD domain protein of this bacterium had 54 % similarity and 88 % query coverage with A. baumannii PLD. E-value of the resulting output was 3−120.
3.2 Secondary and Tertiary Structure Prediction
The secondary structure prediction defined each residue into either beta sheet, alpha helix or random coil structures. SOMPA analysis revealed alpha helix more than random coil or beta strand (Table 3). The secondary structure prediction of PSIPRED shown in Fig. 1 indicates the abundance of alpha helix parts was more than beta sheet segments. Five 3D model structures for protein were generated by I-TASSER. The best confidence score (C-score) of the models was −1.25. In addition, the expected TM-score for this model was 0.56 ± 0.15. The expected RMSD was 10.4 ± 4.6 Å. This model was used for evaluation and refinement.
3.3 Model Evaluation and Refinement
The refined 3D structure of PLD is shown in Fig. 2. Comparison of the validation indices of modeled structure before and after refinement showed that Qmean and PROSESS scores vary as follows: score was improved from 0.441 to 0.458. PROSESS overall and torsion angel quality scores were improved from 3.5 to 4.5 and 2.5 to 3.5, respectively. Covalent and non-covalent band quality remained unchanged. Ramachandran plot revealed that before refinement 87.1, 9.4 and 3.5 % of amino acid residues of modeled 3D structure were incorporated in the favored, allowed and outlier regions respectively (Fig. 3a) while 0.2 % was located in the outlier area after refinement with the increased percentage of favored region to 90.6 % (Fig. 3b).
3.4 Antigenicity Prediction
IEDB B-Cell antigenic predicted sites listed in Table 4 show the shortest sequence of 6 amino acids at the start position of 460 and the longest one consisting of 50 residues with the start position of 5. High similarity of Antigenic Peptide Prediction results ensured the accuracy of antigenic areas. VaxiJen classified PLD as probable antigen, because it has a value of 0.4768 which is above the normal threshold value of 0.4.
3.5 Identification of B-Cell Epitopes
B-cell epitopes prediction by BCpreds generated 8 epitopes with Bcpred model 2 of which possessed scores below 0.8 with start position of 259 and 401. It was also presented 9 epitopes with another model, all of which owned scores above 0.9. Qualified epitope of both models is listed in Table 4. Discontinuous B-cell epitopes predicted by Seppa and BEpro were specified in Fig. 4. The PLD have several surface exposed conformational epitopes. Antigenic B-cell epitopes of 53 and 26 mers sequences were identified and analyzed.
3.6 T-Cell Epitope Prediction
Full length protein was subjected to T-cell epitope prediction. Number of ProPred predicted epitopes which bind MHC class II were 95. One of them bound 42 alleles and 11 other bound just one allele. The number of ProPred I predicted epitopes binding MHC class I were 44. The number of alleles that they can bind varies from 1 to 19. There were 9 common epitopes generated both TCL and HCL mediated immune response (Table 5). “LLNDPLEAL” sequence with the binding ability to 25 MHC alleles was the best common epitope. Lower IC50 is equal to higher affinity. All the common epitopes possessed expected IC50 value (<1,000 nM).
3.7 Identification of Common Epitope for Multiple Pathogens
Two common antigenic B-cell epitopes were identified using IEDB and BCPreds which meet all the selection criteria of antigenic B-cell epitope(s). One of them was 53 mers sequence positioned at 5–57 (QSFHSKQLQTHQLAKGFLIKASIVVCSSFAVALTGCSTLPKHSPEPIQYADI) and the other one was 26 mers sequence positioned at 304–329 (FDWVKAEVVKDSPDKIRSKAKKEEHL) with acceptable VaxiJen scores (>0.4) of 0.4340 and 0.5426 (Table 4). Both sequences were analyzed with two ProPred and ProPredI servers. 53 mers sequence consisted of 8 MHC class II binding epitopes and 4 MHC class I binding epitopes. Two of these epitopes were common, represented by bold characters in the Table 6. Four MHC class II and one MHC class I binding epitopes existed within 26 mers sequence with no common epitope.
Blastp of A. baumannii PLD against multiple pathogens showed that C. difficile, M. tuberculosis, Campylobacter spp., P. aeruginosa, S. pneumonia and S. aureus did not have any specified PLD homologues. According to DEG blastp analysis PLD homologous proteins in S. Typhi, S. Typhimurium, and E. coli were not considered essential. Since DEG includes limited number of human pathogens data, essentiality of PLD homologous proteins for other pathogens (Table 7) could not be checked by this server. The PLD homologues sequences of the rest of the pathogens did not show any similarity to human proteome. Multiple alignments of PLD homologues using PRALINE indicated that there was not any sequence similarity at the position of the first selected sequence (5–57). Relative similarity existed at the position of the second selected sequences (304–329) (Fig. 5).
The multiple alignments of PLD homologues of other human pathogens using PRALINE Sequence similarity at the sequence “FDWVKAEVVKDSPDKIRSKAKKEEHL” position. Microorganisms from top to bottom of aligning rows are: A. baumannii, N. gonorrhoeae, S. flexneri, S. dysenteriae, S. sonnei, S. boydi, S. Enteritidis and K. pneumonia
The similar sequences of different pathogens are given in Table 7. Antigenicity analysis of these sequences by VaxiJen showed S. flexneri score was below the threshold. These homologous peptide sequences were capable of stimulating T-cells.
4 Discussion
PLD affects host cell penetration and lysis ability of microorganism. A. baumannii may avoid the host’s immune response and contribute to antibiotic tolerance with the help of this natural property (Antunes et al. 2011). In this study, PLD was taken as an appropriate target for prevention of infectious processes in order to recognize the immune responses elicited by this enzyme. The selected PLD sequence was the longest of its homologues in A. baumannii strains. A long sequence with common similar PLD protein parts is assumed to be useful in elicitation of strong immune response against most of A. baumannii strains. Computed isoelectric point (pI) of greater than 7 (pI = 9.08) indicates that this protein is basic in character. The extinction coefficient of protein at 280 nm was as high as 78,270 M−1 cm−1 representing a high concentration of Cys, Trp and Tyr. High (91.44) relative protein volume occupied by aliphatic side chains (A, V, I and L), aliphatic index, was a positive factor implied stability of PLD for a wide temperature range (Nazarian et al. 2012). The low range Grand Average hydropathy (GRAVY) value (−0.42) indicates better interaction of protein with water (Sahay and Shakya 2010). ProtParam classifies the PLD protein as stable (Instability index < 40). The estimated half-life of greater than 10 h and the protein stability are attractive features for a vaccine candidate. CELLO and PSLpred predicted PLD as a periplasmic protein. The localization of a protein is correlated with its biological function. Extracellular secretion of proteins such as proteases, phospholipases and toxins are regarded as a major virulence mechanism in bacterial infections. These proteins usually are secreted by the type II pathway and perform periplasmic pool and then fold into a translocation competent conformation before secreting across channels to extracellular environments (Sandkvist 2001). This is further supported by Signal-3L that predicted PLD to have signal sequence. In the secretion process signal peptides function as a tag that directs proteins to the periplasm (Filloux 2010). In a recent in silico study to identify vaccine antigens of A. baumannii (Moriel et al. 2013), PLD was not listed as a good vaccine candidate. The reason was that they selected outer membrane, extracellular or unknown proteins only while neglecting the others in particular the periplasmic proteins.
The high solubility of protein (95.5 %) implies that it could be purified under native condition when expressed in E. coli. In spite of the fact that a protein with its native tertiary structure could be a good candidate for immunogenic studies, this protein should be purified with denaturants like guanidium hydrochloride or urea. During this process, protein loses its bioactive conformational folding reversibility of which is difficult on the basis of thermodynamic rules even after removing this chaotropic denaturants (Dobson 2004; Timasheff and Xie 2003). This denaturation process is required for the prevention of functional toxicity in the body. This goal can be achieved by site directed mutagenesis in the sequences coding protein active site prior to its cloning as carried out on PLDs of Streptomyces chromofuscus and C. pseudotuberculosis (McNamara et al. 1994; Yang and Roberts 2002). The PLD was not predicted as an allergen. Although many of allergen clusters are present in a limited number of protein families, none of the developed techniques has produced a reliable prediction of IgE epitopes du to the limited number of known epitopes (Davies and Flower 2007).
Since inhibition of proteins of normal bacterial flora results in adverse side effects and in colonization of the gut by pathogens (Mai and Morris 2004; Raman et al. 2008), hence, PLD was compared with the proteins of the gut flora by blastp analysis. Citrobacter genus was the only gut flora that possessed a homologous protein. Citrobacter species exist in soil, water, waste water and human intestine. Their presence is not common in the intestinal tract as the most predominant species of Bacteroides, Prevotella, Clostridium, Fusobacterium and Eubacterium (Dethlefsen et al. 2006; Mai and Morris 2004; Suau et al. 1999).
Unlike majority of the protein secondary structures which include mainly β-sheet and α-helix, PLD contains higher percentage of α-helix and random coils. Composition of the secondary structures influences protein tertiary structure, protein quality, availability and digestive behavior (Yu 2005). Different amino acids have distinct tendency to form helical strand and random coils (Malkov et al. 2005). In PLD, the number of helix admirers including (Ala, Leu, Glu, Gln, Arg Met, and Lys) was higher than strand and coil admirers.
Three-dimensional structure of protein prediction has been useful in biomedicine like drug candidate selection. Protein structure precision is often measured by root mean squared deviation (RMSD) of all the equivalent atom pairs and template modeling score (TM-score) of all the residue pairs. Low accuracy of RMSD values (>3 Å) indicates protein does not have solved template structures. Therefore, RMSD value greater than 3 Å would no longer be a good marker of modeling quality or accuracy. Under such circumstances TM-score is more applicable. TM-score value >0.5 indicates a model with an approximately correct topology (Xu and Zhang 2010; Zhang 2009). C-score, based on convergence parameters of the structure assembly simulations and the consensus significance score of multiple threading, ranges from −5 to 2. Greater C-score, higher is the confidence (Roy et al. 2010). These explain correctness of the predicted PLD 3D structure.
B-cell epitope characterization is necessary for understanding the interactions in humoral immune response (Rubinstein et al. 2008). For prediction of B-cell epitopes, BCPreds was used as the main predictor. The results presentation of other available software for predicting B-cell epitopes such as IGpred were of no or limited use for our purpose. Rich output and easy use of prediction results were main features of the used software over other related servers. None of the important measures like accuracy, specificity, sensitivity, and correlation coefficients can be used alone to evaluate the performance of the predictors. These metrics are threshold-dependent and are useful to evaluate performance of the predictor by the area under receiver operator curves (AUC). It is defined as the probability that a randomly chosen positive sample will be ranked higher than a randomly chosen negative sample. AUC value of any predictor with a performance better than random will be between 0.5 and 1.0. AUC greater than 0.7 is indicative of acceptable performance of software. AUC value of BCPred (0.758) and AAP (0.7) is greater than AUC of some B-cell epitope predictors. However AUC has some limitations and can yield misleading conclusions in comparing different predictors, and there is a need for better metrics for performance comparison of different predictors. Therefore it is not easy to state which predictor is better (EL-Manzalawy et al. 2008; Yasser and Honavar 2010). Lysine was the most abundant amino acid in epitope sequences whereas leucine was overrepresented in non-epitopic parts. These findings are in support of previous reports claiming that charged residues are preferred in epitope sequences (Chandra and Singh 2012). Specific amino acid pairs of epitope sequences Y: Y, Y: N, Y: G, Y: R and P: D (Rubinstein et al. 2008) existed in the predicted sequences. Adaptation of secondary structure prediction results and linear epitope sequences revealed that epitopes were significantly enriched with (irregular) random coil and turn structures compared to non epitopic sequences. These structures tend to be more flexible than the other secondary structures. The secondary structure content is important property of protein–protein interfaces. The flexible secondary structures affect conformational adjustment of epitopes upon antibody binding (Chandra and Singh 2012). Comparison of linear epitope sequences and surface accessibility predictions showed almost half of the epitopic sequences in surface regions. In fact, unexposed sequences, capable of stimulating production of specific antibodies, and a few exposed ones are required for generation of antibodies, because they have higher potential to be recognized by the immune systems (Rubinstein et al. 2008).
In this study T-cell epitope prediction was performed using ProPred and ProPredI. Despite development of many computational methods and the focus of many studies on evaluating the success of various MHC-peptide binding prediction methods, there is no consensus on the ideal method (Lafuente and Reche 2009). If sufficient peptide quantities of a pathogen bind to a major histocompatibility complexes (MHC) on the surface of an antigen-presenting cell, the T cell can elicit the cellular immunity (Davies and Flower 2007). This study manifested a lot of PLD epitopic parts which could trigger strong immune response. It is worth mentioning that awareness about distribution of HLA-ABC (MHC class I) and HLA-DR (MHC class II) has clinical application in adaptive immunity and vaccine development (Shankarkumar 2010). Therefore a good vaccine candidate should have T-cell epitope parts with ability to bind most frequent HLA alleles of area used for. No HLA allele was detected by ProPred or ProPredI that PLD could not bind.
An epitope evoking both the B-cell and T-cell (MHC I and MHC II) mediated immunity is highly useful in developing peptide based vaccines. Two peptide sequences of PLD, “QSFHSKQLQTHQLAKGFLIKASIVVCSSFAVALTGCSTLPKHSPEPIQYADI”, and “FDWVKAEVVKDSPDKIRSKAKKEEHL” possessed these properties.
In this analysis, homologous PLD sequences from N. gonorrhoeae, Shigella sp. (flexeneri, sonnei, dysenteriae and boydi), S. enterica subsp. enterica serovar (Typhi, Typhimurium and Enteritidis), E. coli, K. pneumonia and E. gallinarum qualified the cut off value. Hence, it is presumed that their homologous proteins have similar epitopic sequences; therefore, inhibitors for A. baumannii PLD may inhibit these proteins, too. The genus Salmonella consists of two species viz, S. enterica (medically important salmonellae) and S. bongori. Typhoidal serotypes (e.g., S. enterica var Typhi and S. enterica var Paratyphi, and non-typhoidal Salmonella serotypes (NTS serotypes) of S. enterica subsp. Enterica can cause human diseases. S. Typhimurium and S. Enteritidis account for 80 % of NTS serotypes induced infections (Feasey et al. 2012; San Roman et al. 2013). Thus, we chose these two serotypes to find homologous PLD sequences. Enterococci are frequent causes of nosocomial urinary tract infections and nosocomial bacteremia. E. galinarum contribution to these infections is very negligible as compared with E. faecalis and E. faecium. A low level of Vancomycin resistance is found in E. galinarum (Arias and Murray 2012; Teixeira and Merquior 2013). Since the aim of this study was to find a target sequence for antiobiotic resistance species, this species was omitted. All the above-mentioned species of Shigella can produce shigellosis. Because the CDC report did not specify any particular species, we surveyed all of these four species.
S. Typhi, S. Typhimurium and E. coli of CDC reported microorganisms did not require PLD homologous proteins for their survival. In addition to CDC list, Helicobacter pylori and Mycoplasma pulmonis had homologous ones playing key roles for their life. Alignment analysis revealed the first sequence did not exist in most of the pathogen studied. Signal-3L analysis of A. baumannii revealed the first 36 of total 53 PLD amino acids are signal peptide sequence which under natural conditions most part (36aa) of these peptide would be omitted. So, triggering antibody and cell immunity against this part would not have any influence on the inhibition of pathogen unless antibody reacts with the bacterial components from lysed bacteria. The lysed components influence the pathogenesis of infectious diseases (Chen et al. 2009). There were similar sequences in the second peptide sequence position in all the qualified pathogens (Fig. 5; Table 7). In order to demonstrate the possibility of these sequences to be used as peptide vaccine, VaxiJen value was determined. S. flexneri did not qualify VaxiJen threshold. All the homologues peptides had T-cell epitopic sites. A PLD based vaccine for N. gonorrhoeae (Apicella et al. 2007) is in agreement with our in silico results.
5 Conclusions
Inhibition of PLD activity by humoral and cellular immune response could have significant therapeutic potential against A. baumannii. The peptide sequence “FDWVKAEVVKDSPDKIRSKAKKEEHL” could serve as a peptide vaccine against A. baumannii. The sequence with slight modifications can find similar application against some other human pathogens.
References
Antunes LÕC, Imperi F, Carattoli A, Visca P (2011) Deciphering the multifactorial nature of Acinetobacter baumannii pathogenicity. PLoS ONE 6:e22674. doi:10.1371/journal.pone.0022674
Apicella MA, Edwards JL, Gibson BW, Scheffler K (2007) Vaccine and compositions for the prevention and treatment of neisserial infections. Google Patents
Arias CA, Murray BE (2012) The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol 10:266–278. doi:10.1038/nrmicro2761
Barh D, Misra AN, Kumar A (2010) In silico identification of dual ability of N. gonorrhoeae ddl for developing drug and vaccine against pathogenic Neisseria and other human pathogens. JPB 3(3):82–90. doi:10.4172/jpb.1000125
Benkert P, Tosatto SC, Schomburg D (2008) QMEAN: a comprehensive scoring function for model quality assessment. Proteins Struct Funct Bioinform 71:261–277. doi:10.1002/prot.21715
Bentancor LV, Routray A, Bozkurt-Guzel C, Camacho-Peiro A, Pier GB, Maira-Litran T (2012) Evaluation of the trimeric autotransporter Ata as a vaccine candidate against Acinetobacter baumannii infections. Infect Immun 80:3381–3388. doi:10.1128/IAI.06096-11
Berjanskii M, Liang Y, Zhou J, Tang P, Stothard P, Zhou Y, Cruz J, MacDonell C, Lin G, Lu P (2010) PROSESS: a protein structure evaluation suite and server. Nucleic Acids Res 38:W633–W640. doi:10.1093/nar/gkq375
Bhasin M, Garg A, Raghava GPS (2005) PSLpred: prediction of subcellular localization of bacterial proteins. Bioinformatics 21:2522–2524. doi:10.1093/bioinformatics/bti309
Braun G (2008) Virulence mechanisms of Acinetobacter. In: Bergogne-Berezin E et al (eds) Acinetobacter biology and pathogenesis. Springer, Berlin, pp 145–154
Camp C, Tatum OL (2010) A review of Acinetobacter baumannii as a highly successful pathogen in times of war. Lab Med 41:649–657. doi:10.1309/LM90IJNDDDWRI3RE
Centers for Disease Control and Prevention (2013) Antibiotic resistance threats in the United States
Cerqueira GM, Peleg AY (2011) Insights into Acinetobacter baumannii pathogenicity. IUBMB Life 63:1055–1060. doi:10.1002/iub.533
Chandra S, Singh TR (2012) Linear B cell epitope prediction for epitope vaccine design against meningococcal disease and their computational validations through physicochemical properties. Netw Model Anal Health Inform Bioinform 1:153–159. doi:10.1007/s13721-012-0019-1
Chandra S, Singh D, Singh TR (2010) Prediction and characterization of T-cell epitopes for epitope vaccine design from outer membrane protein of Neisseria meningitidis serogroup B. Bioinformation 5:155. doi:10.6026/97320630005155
Chaplin PJ, De Rose R, Boyle JS, McWaters P, Kelly J, Tennent JM, Lew AM, Scheerlinck JP (1999) Targeting improves the efficacy of a DNA vaccine against Corynebacterium pseudotuberculosis in sheep. Infect Immun 67:6434–6438
Chen LY, Pan WW, Chen M, Li JD, Liu W, Chen G, Huang S, Papadimos TJ, Pan ZK (2009) Synergistic induction of inflammation by bacterial products lipopolysaccharide and fMLP: an important microbial pathogenic mechanism. J Immunol 182:2518–2524. doi:10.4049/jimmunol.0713933
Choo KH, Tan TW, Ranganathan S (2009) A comprehensive assessment of N-terminal signal peptides prediction methods. BMC Bioinform 10:S2. doi:10.1186/1471-2105-10-S15-S2
Davies MN, Flower DR (2007) Harnessing bioinformatics to discover new vaccines. Drug Discov Today 12:389–395. doi:10.1016/j.drudis.2007.03.010
de Leseleuc L, Chen W (2012) Recent advances in the immunopathogenesis of Acinetobacter Baumannii infection. In: Amal A (ed) Pulmonary infection. In Tech, Rijeka, pp 23–36
Dethlefsen L, Eckburg PB, Bik EM, Relman DA (2006) Assembly of the human intestinal microbiota. Trends Ecol Evol 21:517–523. doi:10.1016/j.tree.2006.06.013
Diaz AA, Tomba E, Lennarson R, Richard R, Bagajewicz MJ, Harrison RG (2010) Prediction of protein solubility in Escherichia coli using logistic regression. Biotechnol Bioeng 105:374–383. doi:10.1002/bit.22537
Dimitrov I, Flower DR, Doytchinova I (2013) AllerTOP—a server for in silico prediction of allergens. BMC Bioinform 14:1–9. doi:10.1186/1471-2105-14-S6-S4
Dobson CM (2004) Principles of protein folding, misfolding and aggregation. Semin Cell Dev Biol 15(1):3–16. doi:10.1016/j.semcdb.2003.12.008
Dorella FA, Pacheco LGC, Oliveira SC, Miyoshi A, Azevedo V (2006) Corynebacterium pseudotuberculosis: microbiology, biochemical properties, pathogenesis and molecular studies of virulence. Vet Res 37:201–218. doi:10.1051/vetres:2005056
Doytchinova IA, Flower DR (2007) VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform 8:4. doi:10.1186/1471-2105-8-4
Dyrlov Bendtsen J, Nielsen H, von Heijne G, Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340:783–795. doi:10.1016/j.jmb.2004.05.028
El-Manzalawy Y, Dobbs D, Honavar V (2008) Predicting linear B-cell epitopes using string kernels. J Mol Recognit 21:243–255. doi:10.1002/jmr.893
Emini EA, Hughes JV, Perlow D, Boger J (1985) Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide. J Virol 55:836–839
Exton JH (1997) New developments in phospholipase D. J Biol Chem 272:15579–15582. doi:10.1074/jbc.272.25.15579
Fattahian Y, Rasooli I, Mousavi Gargari SL, Rahbar MR, Darvish Alipour Astaneh S, Amani J (2011) Protection against Acinetobacter baumannii infection via its functional deprivation of biofilm associated protein (Bap). Microb Pathog 51:402–406. doi:10.1016/j.micpath.2011.09.004
Feasey NA, Dougan G, Kingsley RA, Heyderman RS, Gordon MA (2012) Invasive non-typhoidal salmonella disease: an emerging and neglected tropical disease in Africa. Lancet 379:2489–2499. doi:10.1016/S0140-6736(11)61752-2
Filloux A (2010) Secretion signal and protein targeting in bacteria: a biological puzzle. J Bacteriol 192:3847–3849. doi:10.1128/JB.00565-10
Fontaine MC, Baird G, Connor KM, Rudge K, Sales J, Donachie W (2006) Vaccination confers significant protection of sheep against infection with a virulent United Kingdom strain of Corynebacterium pseudotuberculosis. Vaccine 24:5986–5996. doi:10.1016/j.vaccine.2006.05.005
Geourjon C, Deleage G (1995) SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Computer applications in the biosciences: CABIOS 11:681–684. doi:10.1093/bioinformatics/11.6.681
Ghannoum MA (2000) Potential role of phospholipases in virulence and fungal pathogenesis. Clin Microbiol Rev 13:122. doi:10.1128/CMR.13.1.122-143.2000
Guan P, Doytchinova IA, Zygouri C, Flower DR (2003) MHCPred: a server for quantitative prediction of peptide–MHC binding. Nucleic Acids Res 31:3621–3624. doi:10.1093/nar/gkg510
Hodgson AL, Krywult J, Corner LA, Rothel JS, Radford AJ (1992) Rational attenuation of Corynebacterium pseudotuberculosis: potential cheesy gland vaccine and live delivery vehicle. Infect Immun 60:2900–2905
Hodgson ALM, Carter K, Tachedjian M, Krywult J, Corner LA, McColl M, Cameron A (1999) Efficacy of an ovine caseous lymphadenitis vaccine formulated using a genetically inactive form of the Corynebacterium pseudotuberculosis phospholipase D. Vaccine 17:802–808. doi:10.1016/S0264-410X(98)00264-3
Ivanovska N (2003) Phospholipases as a factor of pathogenicity in microorganisms. J Mol Catal B Enzym 22:357–361. doi:10.1016/S1381-1177(03)00050-X
Jacobs AC, Hood I, Boyd KL, Olson PD, Morrison JM, Carson S, Sayood K, Iwen PC, Skaar EP, Dunman PM (2010) Inactivation of phospholipase D diminishes Acinetobacter baumannii pathogenesis. Infect Immun 78:1952. doi:10.1128/IAI.00889-09
Kolaskar AS, Tongaonkar PC (1990) A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett 276:172–174. doi:10.1016/0014-5793(90)80535-Q
Kumar S, Srivastava G (2012) PRALINE, a protein multiple sequence alignment tool: user perspective analysis. GERF Bull Biosci 3(2):28–29
Lafuente EM, Reche PA (2009) Prediction of MHC-peptide binding: a systematic and comprehensive overview. Curr Pharm Des 15:3209–3220. doi:10.2174/138161209789105162
Liscovitch M, Czarny M, Fiucci G, Lavie Y, Tang X (1999) Localization and possible functions of phospholipase D isozymes. Biochim Biophys Acta (BBA) Mol Biol Lipids 1439:245–263. doi:10.1016/S1388-1981(99)00098-0
Mai V, Morris JG (2004) Colonic bacterial flora: changing understandings in the molecular age. J Nutr 134:459–464
Malkov S, Zivkovic MV, Beljanski MV, Zaric SD (2005) Correlations of amino acids with secondary structure types: connection with amino acid structure. arXiv preprint q-bio/0505046
McConnell MJ, Pachon J (2010) Active and passive immunization against Acinetobacter baumannii using an inactivated whole cell vaccine. Vaccine 29:1–5. doi:10.1016/j.vaccine.2010.10.052
McConnell MJ, Dominguez-Herrera J, Smani Y, Lopez-Rojas R, Docobo-Perez F, Pachon J (2011) Vaccination with outer membrane complexes elicits rapid protective immunity to multidrug-resistant Acinetobacter baumannii. Infect Immun 79:518–526. doi:10.1128/IAI.00741-10
McGuffin LJ, Bryson K, Jones DT (2000) The PSIPRED protein structure prediction server. Bioinformatics 16:404–405. doi:10.1093/bioinformatics/16.4.404
McKean SC, Davies JK, Moore RJ (2007) Expression of phospholipase D, the major virulence factor of Corynebacterium pseudotuberculosis, is regulated by multiple environmental factors and plays a role in macrophage death. Microbiology 153:2203–2211. doi:10.1099/mic.0.2007/005926-0
McNamara PJ, Bradley GA, Songer JG (1994) Targeted mutagenesis of the phospholipase D gene results in decreased virulence of Corynebacterium pseudotuberculosis. Mol Microbiol 12:921–930. doi:10.1111/j.1365-2958.1994.tb01080.x
Moriel DG, Beatson SA, Wurpel DÓJ, Lipman J, Nimmo GR, Paterson DL, Schembri MA (2013) Identification of novel vaccine candidates against multidrug-resistant Acinetobacter baumannii. PLoS ONE 8:e77631. doi:10.1371/journal.pone.0077631
Nazarian S, Mousavi Gargari SL, Rasooli I, Amani J, Bagheri S, Alerasool M (2012) An in silico chimeric multi subunit vaccine targeting virulence factors of enterotoxigenic Escherichia coli (ETEC) with its bacterial inbuilt adjuvant. J Microbiol Methods 90:36–45. doi:10.1016/j.mimet.2012.04.001
Peleg AY, Seifert H, Paterson DL (2008) Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev 21:538. doi:10.1128/CMR.00058-07
Ponting CP, Kerr ID (1996) A novel family of phospholipase D homologues that includes phospholipid synthases and putative endonucleases: identification of duplicated repeats and potential active site residues. Protein Sci 5:914–922. doi:10.1002/pro.5560050513
Prabhavathy K, Perumal P, SundaraBaalaji N (2011) In silico identification of B- and T-cell epitopes on OMPLA and LsrC from Salmonella typhi for peptide-based subunit vaccine design. Indian J Biotechnol 10:440–451
Raman K, Yeturu K, Chandra N (2008) targetTB: a target identification pipeline for Mycobacterium tuberculosis through an interactome, reactome and genome-scale structural analysis. BMC Syst Biol 2:109. doi:10.1186/1752-0509-2-109
Roy A, Kucukural A, Zhang Y (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5:725–738. doi:10.1038/nprot.2010.5
Rubinstein ND, Mayrose I, Halperin D, Yekutieli D, Gershoni JM, Pupko T (2008) Computational characterization of B-cell epitopes. Mol Immunol 45:3477–3489. doi:10.1016/j.molimm.2007.10.016
Russo TA, Beanan JM, Olson R, MacDonald U, Cox AD, Michael FS, Vinogradov EV, Spellberg B, Luke-Marshall NR, Campagnari AA (2013) The K1 capsular polysaccharide from Acinetobacter baumannii is a potential therapeutic target via passive immunization. Infect Immun 81:915–922. doi:10.1128/IAI.01184-12
Saha S, Raghava GPS (2006a) AlgPred: prediction of allergenic proteins and mapping of IgE epitopes. Nucleic Acids Res 34:W202–W209. doi:10.1093/nar/gkl343
Saha S, Raghava GPS (2006b) Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins Struct Funct Bioinform 65:40–48. doi:10.1002/prot.21078
Sahay A, Shakya M (2010) In silico analysis and homology modelling of antioxidant proteins of Spinach. J Proteomics Bioinform 3:148–154. doi:10.4172/jpb.1000134
San Roman B, Garrido V, Grillo MJ (2013) Non-typhoidal Salmonellosis. In: Giese M (ed) Molecular vaccines. Springer, Berlin, pp 329–342
Sandkvist M (2001) Type II secretion and pathogenesis. Infect Immun 69:3523–3535. doi:10.1128/IAI.69.6.3523-3535.2001
Schmiel DH, Miller VL (1999) Bacterial phospholipases and pathogenesis. Microbes Infect 1:1103–1112. doi:10.1016/S1286-4579(99)00205-1
Shankarkumar U (2010) Complexities and similarities of HLA antigen distribution in Asian subcontinent. Indian J Hum Genet 16:108. doi:10.4103/0971-6866.73397
Shen HB, Chou KC (2007) Signal-3L: a 3-layer approach for predicting signal peptides. Biochem Biophys Res Commun 363:297–303. doi:10.1016/j.bbrc.2007.08.140
Simossis VA, Heringa J (2005) PRALINE: a multiple sequence alignment toolbox that integrates homology-extended and secondary structure information. Nucleic Acids Res 33:W289–W294. doi:10.1093/nar/gki390
Singh H, Raghava GPS (2001) ProPred: prediction of HLA-DR binding sites. Bioinformatics 17:1236–1237. doi:10.1093/bioinformatics/17.12.1236
Singh H, Raghava GPS (2003) ProPred1: prediction of promiscuous MHC Class-I binding sites. Bioinformatics 19:1009–1014. doi:10.1093/bioinformatics/btg108
Singh H, Ansari HR, Raghava GP (2013) Improved method for linear B-cell epitope prediction using Antigen’s primary sequence. PLoS ONE 8:e62216. doi:10.1371/journal.pone.0062216
Somvanshi P, Singh V, Seth PK (2008) In silico prediction of epitopes in virulence proteins of Mycobacterium tuberculosis H37Rv for diagnostic and subunit vaccine design. J Proteomics Bioinform 1:143–153. doi:10.4172/jpb.1000020
Stanford K, Brogden KA, McClelland LA, Kozub GC, Audibert F (1998) The incidence of caseous lymphadenitis in Alberta sheep and assessment of impact by vaccination with commercial and experimental vaccines. Can J Vet Res 62:38
Suau A, Bonnet R, Sutren M, Godon JJ, Gibson GR, Collins MD, Dore J (1999) Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl Environ Microbiol 65:4799–4807
Sun J, Wu D, Xu T, Wang X, Xu X, Tao L, Li YX, Cao ZW (2009) SEPPA: a computational server for spatial epitope prediction of protein antigens. Nucleic Acids Res 37:W612–W616. doi:10.1093/nar/gkp417
Sweredoski MJ, Baldi P (2008) PEPITO: improved discontinuous B-cell epitope prediction using multiple distance thresholds and half sphere exposure. Bioinformatics 24:1459–1460. doi:10.1093/bioinformatics/btn199
Teixeira LM, Merquior VLC (2013) Enterococcus. In: de Filippis I, Mc Kee ML (eds) Molecular typing in bacterial infections. Springer, Berlin, pp 17–26
Timasheff SN, Xie G (2003) Preferential interactions of urea with lysozyme and their linkage to protein denaturation. Biophys Chem 105:421–448. doi:10.1016/S0301-4622(03)00106-6
Towner KJ (2009) Acinetobacter: an old friend, but a new enemy. J Hosp Infect 73:355–363. doi:10.1016/j.jhin.2009.03.032
Xu J, Zhang Y (2010) How significant is a protein structure similarity with TM-score = 0.5? Bioinformatics 26:889–895. doi:10.1093/bioinformatics/btq066
Xu D, Zhang Y (2011) Improving the physical realism and structural accuracy of protein models by a two-step atomic-level energy minimization. Biophys J 101:2525–2534. doi:10.1016/j.bpj.2011.10.024
Yang H, Roberts MF (2002) Cloning, overexpression, and characterization of a bacterial Ca2+-dependent phospholipase D. Protein Sci 11:2958–2968. doi:10.1110/ps.0225302
Yasser EM, Honavar V (2010) Recent advances in B-cell epitope prediction methods. Immunom Res 6:S2. doi:10.1186/1745-7580-6-S2-S2
Young N (2011) Optimizing, purification and inactivation of recombinant Corynebacterium pseudotuberculosis phospholipase D. Dissertation, Occidental College
Yu P (2005) Protein secondary structures (a-helix and b-sheet) at a cellular level and protein fractions in relation to rumen degradation behaviours of protein: a new approach. Br J Nutr 94:655–665. doi:10.1079/BJN20051532
Yu CS, Chen YC, Lu CH, Hwang JK (2006) Prediction of protein subcellular localization. Proteins Struct Funct Bioinform 64:643–651. doi:10.1002/prot.21018
Zhang Y (2009) Protein structure prediction: when is it useful? Curr Opin Struct Biol 19:145–155. doi:10.1016/j.sbi.2009.02.005
Zhang R, Ou HY, Zhang CT (2004) DEG: a database of essential genes. Nucleic Acids Res 32:D271–D272. doi:10.1093/nar/gkh024
Zhang Q, Wang P, Kim Y, Haste-Andersen P, Beaver J, Bourne PE, Bui HH, Buus S, Frankild S, Greenbaum J (2008) Immune epitope database analysis resource (IEDB-AR). Nucleic Acids Res 36:W513–W518. doi:10.1093/nar/gkn254
Acknowledgments
The authors would like to thank Shahed University for supporting this work.
Conflict of interest
We declare no conflict of interests.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zadeh Hosseingholi, E., Rasooli, I. & Mousavi Gargari, S.L. In Silico Analysis of Acinetobacter baumannii Phospholipase D as a Subunit Vaccine Candidate. Acta Biotheor 62, 455–478 (2014). https://doi.org/10.1007/s10441-014-9226-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10441-014-9226-8