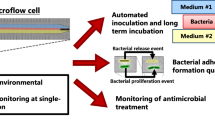
Abstract
We present a real-time microfluidic permeability characterization device for quantifying effects of bacterial deposition on permeability of porous media. Here, we demonstrate a microfluidic approach that allows the effect of bacterial deposition on permeability alteration to be quantified via the measurement of pressure difference, coupled with direct visualization of bacterial distribution. Our experiments reveal three main findings. First, we observe the average percent of cell trapped to be 44.8 ± 9.7%, independent of the average bacterial density at three levels of concentration between 2.05 × 107 and 2.85 × 108 cells/ml. Second, the deposited bacterial cell count appears to follow an exponential reduction in permeability, where the reduction of permeability approaches a pseudo-steady state when a critical number of bacterial cell deposited is achieved. Last, the experiment discerned that bacterial cells tend to preferentially get deposited in regions with larger voids within the porous medium. A theoretical model is developed to determine the relationship between the spatial distribution of voids in the porous medium and the resultant bacterial cell density distribution. The approach described here shows good potential for better understanding the relationship between the spatial distribution of voids and deposited bacteria. More work can be done to further investigate these interactions, such as strategically designed voids to enhance the deposition of bacterial cells, or quantification of inflow bacterial density required to achieve a desired level of pseudo-steady state permeability reduction under various environmental conditions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Soil permeability measures the ability of soil to allow fluid to pass through pores between soil grains (Arora 1987) and is a fundamental physical property in geotechnical engineering applications such as drainage management (Subba Rao and Wadhawan 1953) and land reclamation (Ivanov and Chu 2008). The process of bacterial clogging in porous media often leads to modifications to the soil permeability (Thullner 2010; Bloetscher et al. 2014; Lappan and Fogler 1996; Achal and Kawasaki 2016). Bio-clogging typically consist of two stages. The first stage involves the deposition of bacterial cells entrained in the fluid as it flows through the porous medium. Porosity of the medium and spatial distribution of deposited bacterial cells are dynamically related as the localized distribution of bacterial cells tends to reduce local porosity, leading to a re-direction of subsequent bacterial cells to adjacent locations. In general, bacterial deposition is a function of hydrodynamics and the flow field within the porous medium microstructure, and biochemical effects pertaining to the porous medium and cell surface properties. The former category may involve average grain size and heterogeneity (Tufenkji 2007), as well as porosity, tortuosity and the presence of hydrodynamically disconnected regions (Torkzaban et al. 2008). On the other hand, biochemical effects may be dependent on the specific porous medium material, bacterial species and phenotype, composition of surface-adhesion macromolecules (Tufenkji 2007), as well as hydrophobicity and ionic strength of the entraining fluid (Bai et al. 2016; Jacobs et al. 2007). Furthermore, appendages such as flagella and pili have commonly been observed to assist in reversible binding between cells and surfaces (Knights et al. 2021), although such phenomena has not extensively been studied in the context of deposition in porous media.
The second stage of bio-clogging occurs at larger timescales, where the deposited cells proliferate and secrete extracellular polymeric substances to form biofilms. In the case of Bacillus subtilis utilized in the present study, specific polysaccharides in the secreted EPS are linked to enhanced adhesivity relating to biofilm formation (Harimawan and Ting 2016). However, it is unclear if such EPS substances play a significant role in the earlier stage of bacterial deposition by mediating reversible binding between the cell and porous medium surfaces. As a whole, the resultant effect of the two stages of bio-clogging is the reduction of permeability of the porous media.
The effects of biofilm formation on the structure alteration of the porous media have been studied extensively thus far. However, relatively little attention has been given to the time period prior to biofilm formation, in which the clogging of microorganisms may impose transient effects on the hydraulic properties of porous media. Interactions between microorganisms and their surrounding environment are complex, being controlled by processes such as physical straining, as well as physical or biological properties such as the size and shape of the bacterial cells (Rinck-Pfeiffer et al. 2000; Bitton et al. 1974; Lawrence and Hendry 1996; Kinnari et al. 2009). To address this issue, many conventional experimental methods employing Darcy column or permeameter setups (Bai et al. 2016; Jeong et al. 2018; Vandevivere and Baveye 1992; Wadsworth et al. 2019; Zhou et al. 2009) have been implemented to measure the time-dependent adsorption rate of the influent (such as the breakthrough curve) (Deng et al. 2013; Yang et al. 2021; Bai et al. 2016), biomass accumulation and reduction of permeability (Lappan and Fogler 1996; Kim and Fogler 2000; Taylor and Jaffé 1990; Jeong et al. 2018). All these studies give good insights on the relationship between the microorganism deposition and permeability of the porous media. However, the lack of direct visualization is unable to reveal the mechanism that affects the deposition of bacteria in the porous media. It remains unclear if bacteria are preferentially deposited within certain regions of a porous environment based on physical conditions such as local pore size.
The recent development of microfluidic technologies, particularly soil-on-chip devices, opens doors toward better visualization of bacterial transport in terms of the real-time spatial distribution of bacterial cells. Past studies adopting microfluidic setups using evenly or randomly distributed arrays of circular micropillars (Mann et al. 2012; Lockery et al. 2008; Coyte et al. 2017; Gaol et al. 2021; Creppy et al. 2019; Lam et al. 2012), as well as tightly packed hydrogel (Bhattacharjee and Datta 2019) have revealed interactions amongst the flow, surrounding environment, and resultant behavioral response of bacterial cells (Creppy et al. 2019; Bhattacharjee and Datta 2019; Coyte et al. 2017; Yap et al. 2006). Nevertheless, few works have been performed to quantify the effect of pore size and distribution on the deposition of bacteria cells, and conversely how bacterial cell deposition reduces the permeability of the porous medium in turn. Furthermore, there are very limited reports that explore the use of experimental designs involving three-dimensional representation of porous media, in efforts to better mimic a typical soil structure.
In this work, we quantify the bacteria deposition in porous media in-situ and its effect of permeability alteration through a microfluidic device integrated with pressure differential measurements. Our study focuses on the primary influence of hydrodynamics on bacterial deposition and does not explicitly consider effects due to surface properties of the porous medium and bacterial cells or other biochemical mechanisms such as reversible binding. We show, first, how the pore size and distribution of the porous medium affects the initial spatial distribution of bacteria in the pre-clogging stage. Second, we investigate the mutualistic dynamics between spatial distribution of pores and deposition of bacterial cells and the resultant profile of reduction in permeability of the porous medium. Our developed approach may serve to probe fundamental questions pertaining to spatial distribution of pores that facilitate bacteria deposition, estimation of number of cells required to decrease a specific value of permeability and the correlation between spatial distribution of bacteria and structural configuration of the porous medium.
2 Material and methods
2.1 Design and fabrication
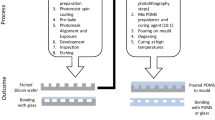
The schematic diagram of the permeability detection device is shown in Fig. 1a. The setup comprises of cascading microfluidic devices, the T-junction and the main device with two in-parallel channels, fabricated with soft-lithography method (Whitesides et al. 2001; Tran et al. 2018; Marcos and Stocker 2006; Kong et al. 2020). The photoresist, SU-8 3005 of thickness 20 μm is spin-coated onto a silicon master, the microfluidic devices are molded by pouring PDMS (Sylgard 184, Dow Chemical, Midland, MI, USA) over the wafer and curing at 60 °C for 2 h. Holes of diameter 1.5 mm (Harris Uni-Core puncher, USA) are punched through the PDMS, and the PDMS is then bonded to the microscope glass slide by plasma cleaning treatment. The main device comprises a main channel and a side channel in parallel Fig. 1b. The main channel of width 200 μm contains a square observation chamber located at the midpoint between inlet and outlet branches, with dimensions of 300 × 300 μm. Porous media (collagen or microbeads) is injected and packed into the chamber via inlet #1. A set of 3 × 4 arrays of micropillars are deployed at each of two regions, respectively located up- and down-stream from the chamber. The diameter of each micropillar is 35 μm and the separation distance between adjacent micropillars is 15 μm. The side channel of length 2630 μm and width 50 μm is introduced between inlets #4 and #5 to prevent excessive pressure buildup in the device. The pressure difference, denoted as ∆p, is measured via two manometers (HD 755–0.5 psi, Extech Instruments, MA, USA) connected to inlets #4 and #5. The measurement is used to evaluate the permeability of the porous media (Supplementary).
a Schematic diagram of cascading setup of microfluidic devices. Two syringe pump are connected to a T-junction and the outlet is connected to inlet #2 of the main device. Porous media are packed via inlet #1 to the square chamber. Microscope objective is focused at chamber to capture images of bacterial cells deposition. b Dimension of observation chamber and parallel side channel. c Image of microbeads in the observation chamber
2.2 Bacteria culture and preparation
The bacteria used is Bacillus subtilis OI1085 (Ullah and Ordal 1981). B. subtilis is a rod-shaped bacterium of length 5 μm, radius 1 μm and swimming speed 25 μm/s (Turner et al. 2016). B. subtilis is cultured by transferring the frozen stock into culture tube with 5 ml of Nutrient Broth (BD234000). This culture tube is incubated at 37 °C, 250 rpm for 24 h. The concentration of the stock bacteria culture is sampled to be in the order of 108 cells/ml. In this experiment, the infusion speed is kept constant while varying the bacterial concentration flowing into the device per unit time. Stock solution is centrifuged at 5000 rpm for 5 min (Eppendorf, Centrifuge 5804R) and sedimented bacteria is resuspended into 5 ml of autoclaved DI water which minimizes bacterial reproduction over the course of experiment. The experiment is conducted at flowrate of 2 μl/min with three levels of bacterial concentration, 4 × , 2 × and 1 × , respectively.
2.3 Experimental setup and operation
The device is operated in three steps. First, microbeads of diameter 14–20 μm (Cospheric PSMS-1.07) are infused into the observation chamber (Fig. 1c) via inlet #1, where they are packed in a random order and subsequently sealed. Next, DI water is infused for 90 min via syringe pump 1 into the T-junction which is connected to inlet #2 of the main chamber. Pressure readings are recorded at 1-s intervals between readings from the manometers connected to inlets #4 and #5. This step calibrates the steady-state permeability of the porous media formed from the randomly packed microbeads. The flow is stopped to allow the device to rest for 15 min. Last, bacteria suspension is infused for 90 min via syringe pump 2 and pressure readings are recorded at the same intervals as the previous step. Image sequences are captured by the microscope objective at 45-s intervals. In total, quintuplicate sets of experimental runs were conducted at a flowrate of 2 μl/min and varying three levels of bacterial concentration.
2.4 Bacteria counting
Samples of flow media were captured from the inlet and outlet to estimate the concentration and trapping efficiency. First, for the three levels of concentration, bacterial suspension is diluted in DI water by 4 × , 2 × and 1 × , respectively. Next, 20 μl of each solution is transferred onto a microscope glass slide and a glass cover slip is applied. The resultant height of the sandwiched liquid film is approximately 17 μm. The microscope viewfinder is approximately circular with diameter 618 μm. The objective used is 20 × , Optovar 1.6 × and depth of focus is approximately 28 μm (Meinhart et al. 2000; Marcos and Stocker 2006). Finally, the resultant images are used to count the bacterial cells and estimate the bacterial density and number of cells trapped by comparing inlet and outlet samples.
3 Results and discussion
A total of 15 experimental runs were conducted based on 5 replicates per dilution level of bacteria-containing solution (stock, 2 × and 4 ×). For each experimental run, the initial permeability of the bacteria-free porous medium configuration was determined from the steady-state value of measured pressure during DI-water calibration. After a brief relaxation period of 15 min, the flow of bacterial-containing solution was applied for 90 min and the gradual increase in pressure difference was recorded at 1-s intervals. One replicate per dilution level was selected at random (runs i to iii) as a representative for discussion throughout this chapter, whilst the other 12 experimental runs (runs iv to xv) are retained in the Supplementary text.
3.1 Estimation of number of bacteria at inlet, outlet and the percentage of bacteria trapped
We first sought to quantify the average number of bacterial cells trapped in the porous medium across the 5 replicates per dilution level (Fig. 2a), as well as to calculate the average ratio of cells trapped versus introduced (Fig. 2b). In Fig. 2a, we plot the average number of bacteria collected at each of the three concentration levels – stock solution, two times dilution in DI water, and four times dilution in DI water, denoted by “stock”, “2 × ” and “4 × ” in the horizontal labels, respectively. The grey (white) bar represents the concentration of bacteria at the inlet #2 (outlet #3), denoted by nin (nout). The average sampling of stock solution (4 × diluted solution) has the highest (lowest) number of bacteria collected, however, we note that the ratio between the number of bacteria in stock solution and 4 × diluted solution is not exactly 4 × . Such variations could arise from the initial size of the inoculum transferring from frozen stock to culture medium and also losses during the bacteria harvesting process via centrifuge.
In Fig. 2b, we plot the percentage of bacteria trapped, expressed as TE = (1 – nout/nin) × 100%, at each of the three bacterial dilutions, represented using a typical box plot. We observe a difference in the average percentage of bacteria trapped among the three levels. Based on experiments conducted, the average percentage of bacteria trapped at “stock”, “2 × ”, “4 × ” is 56.11%, 38.79% and 39.77%, respectively. The variation in the TE over 15 runs could be due to heterogeneity in the spatial distribution of voids occurred from randomly packed microbeads in the observation chamber. However, we note that total average of bacteria trapped is 44.89 ± 9.72%. Based on this finding, we discern that higher bacterial concentration sample does not lead to a significantly higher average percentage of bacteria trapped (leftmost and rightmost percentile plots in Fig. 2b).
3.2 Bacteria deposition causes pressure increase
Through microscopic observation, we investigated the change in pressure difference across the porous medium when bacteria are deposited. First, we infused DI water to calibrate the permeability of the porous media, κDI, via measurement of upstream and downstream pressure difference, denoted by ∆pDI. A higher ∆pDI signifies that the fluid faces high resistance to flow through the porous media, consequently resulting in lower κDI, and vice versa. Thereafter, we infused the flow containing bacterial cells to determine the effect of bacteria deposition on the reduction of permeability of the porous medium, recording the measured pressure difference, denoted by ∆pBact, as well as recording the spatial distribution of trapped bacterial cells.
The abovementioned steps were repeated for three levels of bacterial density and tabulated in Fig. 3 alongside the corresponding bacterial distribution at 30-min intervals in Fig. 4b-d. The black crosses in Fig. 3 represent the calibration pressure difference ∆pDI. First, we observe that the pressure gradually rises over approximately 2000 s, which is attributed to the high hydraulic resistance due to the small microchannel height of 20 μm. As Figs. 3 and 4 depict the pressure trends for only three individual experimental runs selected at random, the pressure rise due to the intrinsic hydraulic properties for the other twelve runs can be observed in the panels (a) of Figures S6-S9 as well. Second, ∆pDI is observed to fluctuate slightly within a nominal average range across each run (see the cross marks in Fig. 3a and the panels (a) of Figures S6-S9), possibly due to environmental disturbances. Therefore, for all experimental runs, we assume that ∆pDI reaches the steady state when the fluctuation is within ± 5% of the nominal average value in each trial. Last, the average steady-state ∆pDI varies among runs (black crosses in Fig. 3 (i-iii)) which arises from heterogeneity in the porous media configuration due to the random arrangement and spatial distribution of microbeads (Fig. 4a (i-iii) and the corresponding spatial distribution analysis in first row of Fig. 6).
The change in pressure due to infusion of bacteria, ∆pBact, is positively correlated with the estimated number of deposited bacteria in the observation chamber. The time-dependent ∆pBact is represented by gray dots in Fig. 3. We observe a similar trend of gradual increase in ∆pBact in the first 2000s as reported for ∆pDI (black crosses and gray dots in Figs. 3 and S6(a) to S9(a). While ∆pDI reaches steady state at about 4000 s, ∆pBact continues to rise across the entire experiment duration (gray dots in Figs. 3 and S6(a) to S9(a)). We plot the differential images to visualize the bacterial deposition at 30-min intervals in Fig. 4b–d (i-iii) to explain the difference in slope of gray dots in Fig. 3(i-iii). Each snapshot is converted to grayscale and the differences in the numerical pixel values between the snapshot at time t and 0 are plotted. Black pixels indicate no bacterial deposition while illuminated pixels indicate bacterial presence and the pixel intensity is representative of the number of deposited bacteria at a given location. We find that, first, the differential images get brighter over time (Fig. 4b, d), indicating the number of bacteria deposited increases over time, resulting in the increment of ∆pBact. Second, the differential image is brightest (dimmest) for Fig. 4d (i) (Fig. 4d (iii)) due to the three levels of inlet bacterial concentrations. Hence, we expect the first (third) run traps most (least) bacteria, which leads to the largest (smallest) slope in ∆pBact of the corresponding experimental data in Fig. 3(i) and (iii). Similar findings have also been obtained from the other experimental runs as shown in Figures S6(a) to S9(a).
3.3 Effects of bacterial deposition on the permeability of the porous medium
Here we develop the relationship between the numbers of deposited bacteria with the pressure increase, hence the reduction of permeability of the porous medium. The total hydraulic resistance, Rtot, of the porous medium can be expressed as
where Q = 2 μL/min is infused into the device, ∆p is the differential pressure measured at the outlets #4 and #5 shown in Fig. 1b.
Since our device setup involves a parallel configuration of main and side channels, we express the total resistance, Rtot, as
where Rm and Rs is the resistance of the main and side channel, respectively. The side channel resistance is calibrated in prior by blocking the main channel with PDMS and measuring the pressure drop across the side channel (Supplementary Figure S2(a)). We find that Rs = (4.86 ± 0.49) × 1013 Nm/s, which is assumed to be constant throughout this study (Tran et al. 2018). We further relate Rm to the fluidic viscosity μ, permeability, κ, and geometry of observation chamber using Darcy Law (Larson 1981),
where the observation chamber of length, L, = 0.3 mm and cross sectional area, A = 0.006 mm2. By substituting Eqs. (1) and (3) into Eq. (2), we therefore obtained the permeability of the porous media:
In our experiment, we use Eq. (4) to evaluate κ for DI water and bacterial runs, denoted by κDI and κBact, respectively, and obtain the FINAL reduction of permeability of the porous media, ∆κ = κBact – κDI. In Fig. 5a, we plot ∆κ for 15 experiments as a function of the number of bacteria trapped in the observation chamber, n. Each set of crosses in Fig. 5 represents an experimental run. We find that, first, the number of deposited bacteria has a weak relationship with bacteria concentration introduced. The number of deposited bacterial cells ranges from 7 × 103 to 1.5 × 105 cells in the observation chamber packed with microbeads. The stock solution contains the highest bacteria concentration, and hence the number of deposited bacteria is expected to be the largest. At 2 × and 4 × dilution, we are not able to conclusively determine the relationship between the bacterial concentration and the resultant estimated number of deposited bacteria. This could arise from significant variation of the stock bacterial concentration before dilution, and the heterogeneous spatial distribution of voids amongst runs. (based on Fig. 2).
Second, we observe that the reduction of permeability of the porous media in Fig. 5 varies in a non-linear trend to the number of deposited bacteria that can be expressed as
where C and D are experimentally determined constant, -2.51 × 10–13 m2 and −5.40 × 10–5, respectively, in 15 runs. The R2 is 0.102. We noted that the outlier points deviating from the fitting curve (Fig. 5a) may suggest that there are some effects on reduction of permeability due to spatial distribution of microbeads, initial permeability, and its resultant spatial distribution of bacterial cells. We plot ∆κ normalized with κDI as a function of n in Fig. 5b and obtain the normalized non-linear relation, which is expressed as
where the normalized coefficient \(\tilde{C}\) and D -0.26 and -3.96 × 10–5, respectively. The R2 for the normalized plot is 0.295. The normalized plot eliminates the effect of the initial permeability on ∆κ, and demonstrates that the number of deposited bacteria is the primary contributing factor in the reduction of permeability. More bacterial cells deposited, the less permeable the porous media is and vice versa. However, taking into further account the noticeable variation in ∆κ/κDI, we hypothesize that the spatial distribution of the deposited bacteria has a secondary effect on ∆κ/κDI and this effect is analyzed in the following section.
We observed that the onset of significant reduction of permeability does not occur immediately as it requires an initial minimum number of bacterial cells to first be deposited. This deposition leads to the increase in flow resistance through the porous media. However, when the number of bacterial cells reaches a critical number (specific to the characteristics of the porous medium used in this experiment), the rate of the reduction of permeability decreases. Based on our experiment results and the curve fitting, the critical number of bacterial cells is approximately 5000. The first 5000 bacterial cells deposited in the porous media significantly reduce the permeability, after reaching this number of cells, the reduction of permeability approaches a pseudo-steady state, where any further increase in the number of cells deposited results in minimal further reduction to the measured permeability.
3.4 Correlation between the bacterial distribution and pore size
We mapped the spatial distributions of voids and bacteria, pij (Figs. 6 and S6(d-e) to S9(d-e)). We evaluated the average light intensity, Iij, in the chamber and discovered that it is inversely correlated to the amount of microbeads accumulated and positively correlated to the amount of space occupied by void regions (Supplementary Figure S4). The microbeads in chamber are depicted in gray, with void regions being of a lighter shade while densely packed microbeads are darker, corresponding to Fig. 4a. We obtain the contour plots of the microbeads and bacterial spatial distribution in Figs. 6 and S6(d-e) to S9(d-e) to analyze the effect of void size on bacterial distribution. We find that void distribution is non-uniform in the observation chamber. The plots in Figs. 6 and S6(d-e) to S9(d-e) show that pij are larger at the edges of the chamber (see lighter colored region), indicating that the gaps between microbeads and edge are large. The microbeads progressively build up near the entrance where they are introduced (inlet #1); however, regions with densely packed microbeads are random in location, shape and size. For example, Fig. 6(i) shows “lung” patterns, while Fig. 6(iii) exhibits a densely packed region in the bottom right corner. The findings from these contour plots are consistent with the microscopic observations of the microbeads distributions in Fig. 4a (i-iii).
We also find that bacteria are likely to clog in the regions with loosely packed microbeads. From the bacterial distribution plots in Fig. 6, we observe that the spatial distribution patterns of bacteria are similar to those of the microbeads. It appears that the majority of bacterial cells settle at the edges, where the voids are large. A minority of bacteria in Fig. 6(i) are deposited at the lung-shaped region and few bacteria in Fig. 6(ii) and Fig. 6(iii) clog at the region where microbeads are densely packed.
This finding runs contrary to the more intuitive expectation that bacterial deposition is likely to occur preferentially at voids with narrow gaps due to physical confinement (Liu and Ford 2009), as our experimental results indicate instead that bacteria are more likely to be trapped in region with larger voids. One possibility is that bacterial accumulation in the wider void is related to the velocity distribution in the porous medium, with flow velocity being inversely related to the cross-sectional area available. This is supported by the findings of (Coyte et al. 2017) where flow is diverted to regions with lesser hydraulic resistance. The flow through a porous medium can be routed through either active or inactive paths, where the former have lesser hydraulic resistance and the latter have higher resistance. The fact that flow primarily passes through regions with lower resistance (larger voids), coupled with reduced flow velocity, suggests that enhanced bacterial deposition can be attributed to large quantities of cells that are experiencing favorable conditions for wall adhesion. Conversely, the evidence does not point to bacterial deposition occurring as a result of narrow voids becoming physically plugged by bacterial bodies. From this work, we thus raise the possibility that the mechanism of bacteria deposition is more dependent on flow velocity, pore size and bacterial cell adhesion than on direct trapping due to constriction in narrow passageways.
Aside from preferential flow paths through the porous medium space, another possibility is that there may be a degree of inertial trapping effects occurring in the large void spaces. As the narrow ways through the gaps between closely packed microbeads open up into a void space, the microstructure here resembles a series of channels and cavities (Haddadi and Di Carlo 2017), suggesting the presence of vortices in the void space that increase the probability of free-swimming cells becoming trapped by the circulating flow as a precursor to wall attachment. In general, the bacterial cells experience a shear-gradient lift force and a wall effect lift force in opposing directions, the balance of which determines the cell’s position along the channel cross-section (Hur et al. 2011). During the abrupt transition to the much larger void (cavity) space, the loss of wall effect lift force results in a net movement toward the center of the void space. However, more work is required to verify the presence of micro-vortices, the extent to which cells experience circulation or trajectory alteration within the large voids, and ascertain if enhanced cell deposition can be positively correlated to said experience.
We adopted the two-dimensional (2D) correlation analysis for the light intensity matrix pij to determine the correlation between the spatial distribution of voids and bacteria. (Supplementary Figure S4). The correlation coefficient, R = 0, -1, + 1 represents none, perfectly negative, and perfectly positive correlation respectively between the light intensity matrices compared. We plot the 2D correlation coefficient, R, for 15 runs, with an average R value of 0.526 ± 0.22, and thus identify a mild positive correlation between the spatial distribution of voids and bacterial deposition. (Supplementary Figure S5).
3.5 Future work
The present work has focused on the macroscale property of permeability and how it is reduced, as a whole, by the deposition of organic matter in the form of bacterial cell bodies within the porous medium matrix. In this exploratory study, all parameters were kept constant except for the inherent heterogeneity in the actual arrangement of microbeads within the porous medium chamber and the subsequent randomness in bacterial distribution. With the promising results obtained thus far, it is a reasonable next step to extend the study by conducting a parametric analysis of the problem. From the perspective of porous medium configuration, it would be valuable to find out how bacterial deposition is affected by microbead size, which is a question of comparing length-scales (Dehkharghani et al. 2022), as well as to relate bacterial deposition and permeability reduction to other key measurements such as porosity and tortuosity.
One limitation of the porous medium based on microbeads is that it may not closely resemble a naturally occurring porous medium sample such as real soil. Firstly, there is an issue of biocompatibility as the microbead material may not promote or sustain long-term bacterial growth and subsequent formation of biofilm as the subsequent phase of bio-clogging. Secondly, the spherical profile of microbeads generates a relatively simple porous medium microstructure with a well-defined minimum gap size, reasonably consistent packing density, and lack of dead-end passages. Conversely, a real soil sample consists of particles of widely varying sizes and shapes, often angular and close-fitting, and hence introducing much more heterogeneity in tortuosity and interconnectivity of paths through the porous medium, while introducing significant complexity in terms of characterization and direct visualization. One possibility is to conduct subsequent studies using porous media based on transparent soil such as Nafion (Downie et al. 2012). Additionally, fluorescent tracer methods (Bhattacharjee and Datta 2019) can be employed for probing the path network of the transparent porous medium, allowing quantification of tortuosity and mean path length for detailed characterization of each randomly packed configuration of transparent soil grains, as well as characterization of bacterial transport through the porous medium.
One may also consider next steps from the perspective of bio-engineering. From the knowledge that bacterial deposition can be quantified and actively controlled, the question of optimizing permeability reduction or enhancement arises. This could be achieved by varying the surface characteristics the particles making up the porous medium, such as hydrophobicity (Bai et al. 2016) or adhesive surface coatings, as well as by extending the scope to consider deposition of more species of bacteria and microorganisms (Tufenkji 2007).
Lastly, it would be valuable to examine the early-stage dynamics of bacterial deposition to a deeper level of detail. For example, while the present study utilized dilutions to produce a range of deposited cell counts, it does not consider possible transient effects of the rate of bacterial deposition, or bacterial flux, on the internal flow field. It may be interesting to examine the significance of elasticity of the combined structure of porous medium and attached cells, where the cumulative effect of gradual blockage of the porous medium network may be different from that of abrupt and large-scale blocking events when a high bacterial density load is applied.
4 Conclusion
In conclusion, we have developed a microfluidic device and experimental technique that is able to characterize the permeability of a porous medium sample. By integrating the pressure differential measurement apparatus within the microfluidic channel configuration of the main experiment, our device allows simultaneous in-situ visualization of bacteria deposition and permeability quantification. First, from experimental results, we discerned that the average cell trapping efficiency of the porous medium is 44.89 ± 9.72%, and appears to be independent of bacterial concentration within the tested range of 2.05 × 107 to 2.85 × 108 cells/ml. Second, we observed that the number of bacterial cells deposited correlated to a linear reduction of permeability of the porous medium. Third, contrary to expectations, we observed that the majority of bacterial cells were preferentially deposited at regions corresponding to larger voids rather than smaller ones. We raised the possibility that this could be attributed primarily to fluid dynamics, where regions with smaller voids presented higher flow resistance and thus actively diverted the main flow through channels of larger cross-sectional area, and additionally to inertial trapping effects. From mass conservation, we expect that there is significant reduction in flow velocity through such regions, which may potentially further contribute to the likelihood of cell deposition due to surface adhesion. These findings suggest that bacterial deposition may actively be directed to occur selectively by strategically designing void regions and controlling parameters such as the shape, size and distribution of voids within porous media.
References
Achal V, Kawasaki S (2016) Biogrout: a novel binding material for soil improvement and concrete repair. Front Microbiol 7:314–414
Arora KR (1987) Permeability of soil, in soil mechanics and foundation engineering. Soil Mechanics and Foundation Engineering, Nak Sarak, Delhi
Bai H, Cochet N, Drelich A, Pauss A, Lamy E (2016) Comparison of transport between two bacteria in saturated porous media with distinct pore size distribution. RSC Adv 6:14602–14614
Bhattacharjee T, Datta SS (2019) Bacterial hopping and trapping in porous media. Nat Commun 10:2075
Bitton G, Lahav N, Henis Y (1974) Movement and retention of Klebsiella aerogenes in soil columns. Plant Soil 40:373–380
Bloetscher F, Sham CH, Danko III JJ, Ratick S (2014) Lessons learned from aquifer storage and recovery (ASR) systems in the United States. J Water Resour Prot 06:1603–1629
Coyte KZ, Tabuteau H, Gaffney EA, Foster KR, Durham WM (2017) Microbial competition in porous environments can select against rapid biofilm growth. Proc Natl Acad Sci USA 114:E161–E170
Creppy A, Clément E, Douarche C, D’Angelo MV, Auradou H (2019) Effect of motility on the transport of bacteria populations through a porous medium. Phys Rev Fluids 4:013102
Dehkharghani A, Waisbord N, Guasto JS (2022) Self-transport of swimming bacteria is impaired by porous microstructure. arXiv preprint arXiv:2201.03059
Deng W, Cardenas MB, Kirk MF, Altman SJ, Bennett PC (2013) Effect of permeable biofilm on micro- and macro-scale flow and transport in bioclogged pores. Environ Sci Technol 47:11092–11098
Downie H, Holden N, Otten W, Spiers AJ, Valentine TA, Dupuy LX (2012) Transparent soil for imaging the rhizosphere. PLoS ONE 7:e44276
Gaol CL, Ganzer L, Mukherjee S, Alkan H (2021) Investigation of clogging in porous media induced by microorganisms using a microfluidic application. Environ Sci: Water Res Technol 7:441–454
Haddadi H, Di Carlo D (2017) Inertial flow of a dilute suspension over cavities in a microchannel. J Fluid Mech 811:436–467
Harimawan A, Ting YP (2016) Investigation of extracellular polymeric substances (EPS) properties of P. aeruginosa and B. subtilis and their role in bacterial adhesion. Colloids Surf, B 146:459–467
Hur SC, Mach AJ, Di Carlo D (2011) High-throughput size-based rare cell enrichment using microscale vortices. Biomicrofluidics 5:22206
Ivanov V, Chu J (2008) Applications of microorganisms to geotechnical engineering for bioclogging and biocementation of soil in situ. Rev Environ Sci Bio/technol 7:139–153
Jacobs A, Lafolie F, Herry JM, Debroux M (2007) Kinetic adhesion of bacterial cells to sand: cell surface properties and adhesion rate. Colloids Surf, B 59:35–45
Jeong HY, Jun S-C, Cheon J-Y, Park M (2018) A review on clogging mechanisms and managements in aquifer storage and recovery (ASR) applications. Geosci J 22:667–679
Kim DS, Fogler HS (2000) Biomass evolution in porous media and its effects on permeability under starvation conditions. Biotechnol Bioeng 69:47–56
Kinnari TJ, Esteban J, Martin-de-Hijas NZ, Sánchez-Muñoz O, Sánchez-Salcedo S, Colilla M, Vallet-Regí M, Gomez-Barrena E (2009) Influence of surface porosity and pH on bacterial adherence to hydroxyapatite and biphasic calcium phosphate bioceramics. J Med Microbiol 58:132–137
Knights HE, Jorrin B, Haskett TL, Poole PS (2021) Deciphering bacterial mechanisms of root colonization. Environ Microbiol Rep 13:428–444
Kong TF, Shen X, Marcos, Yang C, Ibrahim IH (2020) Dielectrophoretic trapping and impedance detection of Escherichia coli, Vibrio cholera, and Enterococci bacteria. Biomicrofluidics 14:054105
Lam RHW, Weng S, Lu W, Fu J (2012) Live-cell subcellular measurement of cell stiffness using a microengineered stretchable micropost array membrane. Integr Biol 4:1289–1298
Lappan RE, Fogler HS (1996) Reduction of porous media permeability from in situ Leuconostoc mesenteroides growth and dextran production. Biotechnol Bioeng 50:6–15
Larson RG (1981) Derivation of generalized Darcy equations for creeping flow in porous media. Ind Eng Chem Fundam 20:132–137
Lawrence JR, Hendry MJ (1996) Transport of bacteria through geologic media. Can J Microbiol 42:410–422
Liu J, Ford RM (2009) Idling time of swimming bacteria near particulate surfaces contributes to apparent adsorption coefficients at the macroscopic scale under static conditions. Environ Sci Technol 43:8874–8880
Lockery SR, Lawton KJ, Doll JC, Faumont S, Coulthard SM, Thiele TR, Chronis N, McCormick KE, Goodman MB, Pruitt BL (2008) Artificial dirt: microfluidic substrates for nematode neurobiology and behavior. J Neurophysiol 99:3136–3143
Mann JM, Lam RHW, Weng S, Sun Y, Fu J (2012) A silicone-based stretchable micropost array membrane for monitoring live-cell subcellular cytoskeletal response. Lab Chip 12:731–740
Marcos, Stocker R (2006) Microorganisms in vortices: a microfluidic setup. Limnol Oceanogr Methods 4:392–398
Meinhart CD, Wereley ST, Gray MHB (2000) Volume illumination for two-dimensional particle image velocimetry. Meas Sci Technol 11:809–814
Rinck-Pfeiffer S, Ragusa S, Sztajnbok P, Vandevelde T (2000) Interrelationships between biological, chemical, and physical processes as an analog to clogging in aquifer storage and recovery (ASR) wells. Water Res 34:2110–2118
Subba Rao K, Wadhawan SK (1953) Studies in soil permeability. Proc Indian Acad Sci- Sect A 37:68–80
Taylor SW, Jaffé PR (1990) ’Biofilm growth and the related changes in the physical properties of a porous medium: 1. Experimental investigation. Water Resour Res 26:2153–2159
Thullner M (2010) Comparison of bioclogging effects in saturated porous media within one- and two-dimensional flow systems. Ecol Eng 36:176–196
Torkzaban S, Tazehkand SS, Walker SL, Bradford SA (2008) Transport and fate of bacteria in porous media: coupled effects of chemical conditions and pore space geometry. Water Resour Res 44
Tran QD, Marcos, Gonzalez-Rodriguez D (2018) Permeability and viscoelastic fracture of a model tumor under interstitial flow. Soft Matter 14:6386–6392
Tufenkji N (2007) Modeling microbial transport in porous media: traditional approaches and recent developments. Adv Water Resour 30:1455–1469
Turner L, Ping L, Neubauer M, Berg HC (2016) Visualizing flagella while tracking bacteria. Biophys J 111:630–639
Ullah AH, Ordal GW (1981) In vivo and in vitro chemotactic methylation in Bacillus subtilis. J Bacteriol 145:958–965
Vandevivere P, Baveye P (1992) Saturated hydraulic conductivity reduction caused by aerobic bacteria in sand columns. Soil Sci Soc Am J 56:1–13
Wadsworth FB, Vossen CEJ, Schmid D, Colombier M, Heap MJ, Scheu B, Dingwell DB (2020) Determination of permeability using a classic Darcy water column. Am J Phys 88:20–24
Whitesides GM, Ostuni E, Takayama S, Jiang X, Ingber DE (2001) Soft lithography in biology and biochemistry. Annu Rev Biomed Eng 3:335–373
Yang Y, Liu T, Li Y, Li Y, You Z, Zuo M, Diwu P, Wang R, Zhang X, Liang J (2021) Effects of velocity and permeability on tracer dispersion in porous media. Appl Sci 11:4411
Yap YF, Li D, Chai JC (2006) Flow of cells in microchannels. J Phys: Conf Ser 34:448–453
Zhou J, Zheng X, Flury M, Lin G (2009) Permeability changes during remediation of an aquifer affected by sea-water intrusion: a laboratory column study. J Hydrol 376:557–566
Acknowledgements
We thank Tian Fook Kong for many fruitful discussions. This research is supported by the Ministry of Education, Singapore, under its Academic Research Funds Tier 2 MOE2018-T2-2-052 and Academic Research Funds Tier 1 RT04/19.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Teo, T.W., Shen, X., Tan, P.Y. et al. A microfluidic approach to investigate the effects of bacteria deposition in porous media containing randomly packed microbeads via real-time pressure measurement. Microfluid Nanofluid 26, 58 (2022). https://doi.org/10.1007/s10404-022-02561-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10404-022-02561-z