Abstract
A low-cost, convenient and precise drug combination screening microfluidic platform is developed, in which cell culture chambers designed with micropillars integrate with three laminar flow diffusion channels. This platform has several distinct features, including minimum shear stress on cells, biocompatibility, optimum concentration distribution and automatic combinatorial gradient generation, which can potentially speed up the discovery of an effective drug combination for cancer ablations. The presented device can generate two-drug combination gradients at the optimum flow rate of 90 μL/h and can be applied to identify the optimal combination of two clinically relevant chemotherapy drugs. For demonstration, paclitaxel at 0.77 × 10−3 mg/mL and cisplatin at 0.23 × 10−4 mg/mL were studied against lung cancer cells (A549). This microfluidic device has the potential to provide a precise and robust screening for anticancer combinational drugs practiced in clinics.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Drug combination, in the field of cancer treatment, is recently considered as one of the most effective therapies. Since the 1960s, it has generally been recognized that the use of two or more chemotherapy drugs together is more effective (Bonadonna et al. 1976). Besides reducing the dose of single drug, the combinatorial approach can improve the drug efficiency and minimize their toxicity (Al-Lazikani et al. 2012). Drug combination could simultaneously target different molecular mechanisms, thereby reducing the drug resistance while increasing tumor cell killing (Lam 1997). Combination drug therapy has been successfully applied for tuberculosis curative (Espinal et al. 2000), palliative care in prostate cancer (Beltran et al. 2011) and other microbial infections (Ding et al. 2012; Silva et al. 2016). Combinatorial drug chemistry has not only served as a great route for drug discovery, but also provided invaluable insights in basic research (Honda et al. 2013; Malo and Geuna 2000). However, traditional drug screening methods usually need high operating cost and are very time-consuming. Therefore, cell-based, reliable, rapid, high-throughput and large-scale screening assays are highly demanded for a successful combinatorial drug screening before conducting expensive in vivo experiments (Kim et al. 2010; Lam 1997).
During recent years, microfluidics-based devices have attracted great attention due to their contributions in cell-based drug screening (Bijl et al. 2002; Kalchman et al. 2013). Several drug screening devices have been reported for determining potential concentrations and combinations of multiple reagents (Lee et al. 2010; Shi et al. 2013; Wellhausen et al. 2012). Most of these studies have focused on the special properties of microfluidics in diffusion and mixing. In 2004, Neils et al. (2004) developed a microfluidic network to generate four titrations of two kinds of solutions to deliver 16 mixture combinations in the separate outlet. However, this device just reported dye-based experiment and has not been used for in vitro drug screening studies. Jang et al. (2011) demonstrated a tree-like generator to realize 100 combinatorial dilutions from two input solutions. This device was a high-throughput microfluidic screening system, but the potential effect of the medium flow to chamber was not considered, which could produce a shear stress and could impact the phenotype response of cells. Moreover, although the authors discussed its potential applications for drug screening, this device was not practically demonstrated for in vitro drug screening studies. Ahadian et al. (2014) used dielectrophoresis (DEP) to create 3D chemical gradients within hydrogels and nanoparticles for high-throughput drug screening applications. The particle distribution was controlled in the hydrogel by electrophoresis to form the desired concentration. However, only single drug screening was reported, and drug combination study was not considered.
Laminar flow-based linear chemical gradient studies have also been reported (El-Ali et al. 2006). This method can generate combinatorial concentration gradients in microfluidic devices using two laminar flows (Frank and Tay 2013). An “Y”-shaped microfluidic channel was designed to create a concentration gradient in 256 microfluidic cell cultures, while cell viability was used to determine the concentration gradient in chambers. Again only one drug screening was reported (Somaweera et al. 2015). In addition to using laminar flow diffusion methods, some scientists have designed microvalves to achieve combinational concentration and drug screening (Kim et al. 2012; Xu et al. 2012). Li et al. (2007) utilized structure-based approach to generate linear and nonlinear chemical gradients. Shamloo et al. (2008) described a fluid convection microfluidic device to minimize the effect of fluid flows on cell culture chamber. Atencia et al. (2009) designed a new microfluidic device called “microfluidic palette” with a 1.5-mm circular chamber for diffusion and three access ports for the delivery to study the chemotactic response of the bacteria P. aeruginosa to glucose. Some scientists have also utilized nanomechanical fluctuation or cell-substrate impedance to conduct rapid screening of anticancer drugs (Tran et al. 2013; Wu et al. 2016). Although these devices achieved cell-based drug screening on chip, screening of two or more drugs in combination remained rarely tackled.
Furthermore, Kim et al. (2012) demonstrated a fully automatic and programmable microfluidic cell culture platform, which had 64 individually cell culture chambers and 64 pairwise concentration combinations. This device has been used to screen combinatorial drug treatments against PC3 prostate cancer cells and does not require continuous perfusion to maintain gradient, which minimizes the amount of drug. In 2016, Khoo et al. (2016) designed an integrated microfluidic system with microwells to evaluate patient drug response using patient-derived circulating tumor cell (CTC) cultures. This device aims to rapidly evaluate the drug response of CTCs from blood of an individual patient throughout the course of treatment and can be a potential tool for guiding drug discovery development.
Another device utilized printing technology to print the target drug to the substrate. Ding et al. (2015) developed print-to-screen (P2S) platform, which utilized a microprinter to generate large-scale combinatorial anticancer drugs. In this P2S platform, 10 anticancer drugs were selected to generate a 23 × 23 array. This device achieved a high-throughput and relative large-scale screening approach for microfluidic screening, but this device required complex and time-consuming preparation methods.
Nevertheless, most devices realized a two-dimensional (2D) cell culture mode, which are less consistent compared to in vivo conditions. In order to better imitate physiological tissues and further improve consistency, three-dimensional (3D) cell culture methods have obtained massive attentions. Xu et al. (2013) designed a microfluidic three-dimensional (3D) co-culture drug sensitivity test platform. A mono-lung cancer cell line, a mixture of lung cancer and stromal cell lines, and cells from fresh lung cancer tissues were cultured in 3D under continuous cell culture media supplementation, imitating the actual tumor microenvironment in vivo and clinical reality.
The aim of this paper is to present a low-cost, convenient and precise on-drug cell culture and drug combination screening platform to acquire factorial design data eligible for statistical regression modeling (Ding et al. 2013) for any two drugs of interest. The platform integrates on-chip cell culture with laminar flow mixer to generate the dose/ratio combinations of drugs. This device has 42 independent carefully designed cell culture chambers, in which cells can be incubated with minimal shear stress. The serpentine structure in channel accelerates the diffusion of two laminar streams. With continuous infusion of media with drugs, cells in the chamber are consistently exposed to different drug combinations. The presented platform requires only the initial injection of the drug stock solution and no more intervention during the following drug combination study, which ousts the experimental errors induced by abundant and repeated drug dilution or administration handlings. As a whole, this microfluidic device offers a low-cost, convenient and automatic solution for drug combination screening compared to traditional methods for drug screening. The use of this microfluidic device is demonstrated by incubating lung cancer (A549) cells in the chamber to study the potential combinations of two clinically relevant drugs (paclitaxel and cisplatin).
2 Materials and methods
2.1 Materials
The following materials were used in this study:
SU-8 photoresist and its developer (MicroChem, USA), poly(dimethylsiloxane) (PDMS) and its curing agent (Dow Corning GmbH, USA), fluorescence dye (Fluka, GER), buffered saline (PBS, pH 7.4, VWR, Radnor, Pennsylvania), CCK-8 (Dojindo, Shanghai, China), paclitaxel and cisplatin (TopScience, USA).
2.1.1 Cells and cell culture
A549 cells were cultured in a humidified, 5% CO2 environment at 37 °C in DMEM (Dulbecco’s modified Eagle’s medium) with 10% fetal bovine serum and 15 mL/L antibiotic solution (penicillin–streptomycin solution). The media was changed every 2 days. Before the experiment, the cells were taken from the culture flasks and concentrated using a centrifuge, and then diluted with DMEM to the required concentrations.
2.2 Design and fabrication of device
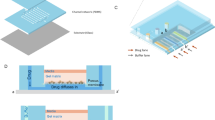
In this experiment, the device (Fig. 1a) was fabricated using standard soft photolithographic method (Unger et al. 2000) (Fig. S1†). As illustrated in Fig. 1b, the microfluidic device consisted of three main mixing channels, each channel had two inlets, and each side of the single main channel contained 7 square-shaped cell culture chambers. The single main mixing channel was 2 cm in length and 300 μm in width, while the square-shaped cell culture chambers had a side length of 900 μm. The channel depth for mixing channels and cell culture chambers were 100 μm.
Scheme of microfluidic device. a Optical image of the actual device. b Microfluidic device designed with three main mix channels. Various combinations of drug A (paclitaxel) and drug B (cisplatin) are generated in channel 1, various concentrations of drug A (paclitaxel) and buffer are generated in channel 2, and various concentrations of drug B (cisplatin) and buffer are generated in channel 3. c Detailed internal structure of one single channel
The channels and culture chambers were connected by capillaries with inside diameter of 90 μm and length of 270 μm, as shown in Fig. 1c. Four micropillars were evenly designed to reduce the shear stress on cells when the medium or anticancer drug fluid was flowing through the channel. The diameter of each micropillar was 90 μm and its height was 100 μm. The distance between two micropillars was 90 μm, so that the A549 cells could easily pass in between. Polyethylene tubing (PE-20, OUPLI, Shanghai, China) were used to connect the microfluidic device to syringe pump (LSP04-1A, LongerPump, Baoding, China) with three 1-mL syringes containing cell culture medium and drug solutions. The inside diameter of polyethylene tubing is 0.6 mm.
2.3 Microfluidic device simulation
To simulate fluid flow and reagent diffusion in the device and to find optimal parameters for system operation, a finite element three-dimensional model was constructed and analyzed using COMSOL Multiphysics 5.0 (Stockholm, Sweden). The steady-state Navier–Stokes equations were solved to obtain the flow field. For demonstration purposes, simulation result for a single main mixing channel was displayed. To predict the optimum convection flow rate for drug, a finite element 3D simulation was performed with injection flow rates from 10 to 150 μL/h. The simulation parameters are shown in Table S1†.
2.4 Characterization of concentration gradients in the device
Fluorescein dye at 0.5, 1, 1.5, 2, 2.5, 3 and 3.5 µM was used to calibrate the chip. The dye was diluted with 1X PBS. To calibrate fluorescence detection, fluorescein dyes were injected into a single channel successively to generate a non-gradient concentration. The dye was injected into the channel at 50 μL/h with a syringe pump. Before each injection, all chambers in the chip were filled with PBS buffer. After 30 min, fluorescent images of each chamber with different dye concentrations were taken using a CCD camera and analyzed using ImageJ (version 1.45, National Institutes of Health).
To examine the relationship between flow rates and drug concentration distribution at equilibrium state in each chamber, fluorescein dye at 3 μM was injected into the channel at various flow rates of 10, 30, 60, 90, 120 and 150 μL/h in a single main mix channel. After 20 min to create an equilibrium concentration gradient across the channel, the fluorescent images of each chamber with different flow rates were acquired and analyzed using the same setup as introduced above.
2.5 Cell viability study in the microfluidic device
Before A549 cell suspension in DMEM loaded into the chip, the PDMS device was sterilized in autoclave (150 °C, Panasonic, Japan) for 30 min, followed by UV light irradiation overnight. To seed A549 cells into each culture chamber, cells at a density of 2 million/mL were injected into the chip at 2.5 mL/min with a syringe pump. Cell culture medium was then continuously injected into the chip at 50 μL/h, and the chip was kept in an incubator at 37 °C. Cell images in each chamber were recorded every 12 h until the cells became confluent for all the chambers. To examine the effect of micropillars structures on cell growth rate, cells incubated in the chambers either with micropillars or without micropillars were compared after 12-h incubation.
2.6 Combinatorial screening of anticancer drugs
Two anticancer drugs, namely paclitaxel and cisplatin, were selected based on their unique anticancer mechanisms (Liebmann et al. 1993; Theodossiou et al. 1998). These drugs were received in powder forms and dissolved in DMSO to prepare 10 mg/mL solutions. Then, they were diluted in DMEM to the target concentration.
A549 cell suspension prepared in cell culture medium was continuously injected into the chip at 2.5 mL/min until the cells were uniformly distributed in the chamber. Cell culture medium was then continuously injected into the chip at 50 μL/h for 24 h to seed the cells, and the chip was kept in an incubator at 37 °C. Then, paclitaxel (10−3 mg/mL), cisplatin (10−4 mg/mL) and cell culture medium were continuously injected into the chip from the inlets at 90 μL/h. After another 24-h incubation in the incubator, cell image from each chamber was taken and live cell number was calculated using ImageJ.
In order to compare cell density acquired from different chambers on different channels, cell density readout from each chamber was normalized to the blank control (no drug treatment) on the same channel. In this chip, chamber No. 14 in channel 2 and channel 3 was blank control. Therefore, the standardized cell growth rate was defined as averaged percentage of live cell number after and before drug injection in these two chambers,
Then, the normalized cell viability in each chamber was calculated using the following equation:
2.7 Cell experiments in 96-well plates
A549 cells were plated at a density of 10,000 cells per well in 96-well plates and incubated at 37 °C and 5% CO2 for 24 h. The supernatant was removed and replaced by combinatorial treatment of paclitaxel and cisplatin. Drug concentration is shown in Table S2† based on the results acquired from session (2.4). After 24-h incubation, the drug containing medium was removed, and the cells in each well were then incubated in 100 μL culture medium containing 10 μL CCK-8 for 30 min. The reported results were obtained from three independent experiments.
3 Results and discussion
3.1 Design and simulation results of microfluidic device
The goal of this microfluidic device is to identify the optimal combination of two anticancer drugs. Drug containing solutions were injected into the channels to generated three concentration gradient profiles in the three mixing channels by diffusion. The serpentine structure of the channel was designed to facilitate liquid mixing.
Previous studies have shown that PDMS has gas adsorption permeation and diffusion properties (Merkel et al. 2000; Singh et al. 1998). The dissolved gas concentration in PDMS is proportional to the gas pressure inside the PDMS channel (Luo et al. 2008). As the gas pressure inside the chambers increases, the PDMS sidewall will absorb the gas and the gas pressure inside the chamber will reach a new equilibrium. When the pressure in the chamber is higher than the pressure outside the device, the gas in the chambers will be gradually removed. Therefore, when the drug solution flowed into the chambers, due to the higher internal pressure, the gas inside the chamber would be repelled, and the liquid would smoothly enter. Furthermore, when the flow rate was increased, the filling time would be reduced due to elevated pressure in the chambers, as shown in Fig. S2†.
A finite element simulation of the microfluidic device was created to check various designing parameters, including diffusion constant, inlet flow rate, inlet concentration, dynamic viscosity and density. The geometry of the device is shown in Fig. 2a. The flow rate at the inlet was 90 μL/h. Because the Reynolds number (8 × 10−4) was significantly <1, the creeping flow interface could be used. The problematic convective term in the Navier–Stokes equations was therefore negligible, leaving the incompressible Stokes equations: (Bijl et al. 2002)
Simulation results for the concentration gradient distribution in one channel on the microfluidic device. a Geometry of a single main mixing channel used in this simulation. b Concentration gradient profile along the entire x-axis of the channel at the flow rate of 90 μL/h. c Velocity profile of the channel; the right side plot is an enlarged view of the S–S cross section. Velocity inside chambers was close to zero, while velocity in the main channel was obviously exacerbated. This result implied the cells cultured in the chamber would receive minimal impacts from the flow rate shear stress in the main channel
where u is the local velocity and p is the pressure.
According to the simulation of Navier–Stokes equations with a concentration dependent viscosity, at flow rate of 90 μL/h, the concentration gradient profile was stable along the whole x-axis of the channel (Fig. 2b). Figure 2c shows the velocity field for the case where viscosity is independent of concentration. The flow was symmetric and was not affected by the concentration field. Meanwhile, velocity vector in chambers was close to zero and the highest velocity was shown in main channel.
We also obtained the simulation results of the concentration distribution along different culture chamber numbers at different flow rates, including 10, 30, 60, 90, 120 and 150 μL/h. The concentration in each chamber was measured and was normalized using the injected solution as the reference. As shown in Fig. 3a, when the flow rate decreased, the diffusion in channel increased. The concentration and velocity field at the flow rates of 10 and 150 μL/h are calculated in Fig. S3†. At 10 μL/h, the liquids in the middle of the channel had a high diffusion. But at 150 μL/h, low diffusion profile was observed in the main channel.
Comparison of simulation results and experimental results of dye concentration gradient profiles at various flow rates. a Results obtained from simulation of the dye concentration distribution along chamber numbers for various flow rates: 10, 30, 60, 90, 120 and 150 μL/h. b Experimental results of dye concentration gradient profiles of the fluorescein dye at various flow rates, 10, 30, 60, 90, 120 and 150 μL/h. The normalized dye concentration of chambers was decreased with the distance further from the inlet. Experimental results for each flow rate agreed with the predicted simulation results. The flow rate of 90 μL/h was fixed for all subsequent experiments
3.2 Quantification of the concentration gradient inside the chip
To quantitatively analyze the concentration in each culture chamber, the average fluorescent intensity values of 14 chambers resulting from a series concentration of fluorescein dyes were used to obtain a calibration curve. Figure S4† shows the relationship between averaged intensity values and fluorescein dye concentrations at non-gradient condition. This curve allowed calculation of unknown dye concentration in each culture chamber based on the examination of the corresponding fluorescent intensity.
The gradient profile in this device depends on several factors, such as the concentration of solutions, design of the channel and the flow rate (Zhou and Lin 2014). By varying the flow rates of two streams, different gradient profiles were obtained. At various flow rates, the fluorescence intensity of each chamber was measured and was normalized using its intensity at the inlet as the reference. Figure 3b shows quantification of the normalized dye concentration for each chamber at various flow rates. The normalized dye concentration of chambers was decreased along with the distance from the inlet. The formation of the concentration gradient was due to the diffusion of laminar flow. Because of the diffusion, the dye was diluted; thus, the concentration was continuously decreased in the chambers further from the inlets. At the same time, when the flow rate decreased, the diffusion in channel increased. The experimental results at various flow rates agreed with the predicted simulation results.
From both the simulation and the experimental results, 90 μL/h was chosen as the optimum flow rate. At this flow rate, the chip required 20 min to develop an equilibrium gradient and then remained unchanged with a continuous injection until the pump was stopped. At this flow rate, 2.16 mL of each drug solution is required.
3.3 Viability of A549 on chip
A549 cells were monitored in the microfluidic chip for 3 days while fresh cell culture medium was continuously supplied through the channels. When the cells were infused into the culture chambers, they were randomly distributed in the chambers. In this case, cell culture medium without drug was used. The cell number increased significantly with continuous medium flow (Fig. 4a). After 96 h, the cells have completely filled the chambers and the growth conditions of the cells in chip were consistent with the normal condition commonly observed on standard cell culture plates. Cell images with micropillars and without micropillars are shown in Fig. 4b. With the presence of micropillars, the cells were uniformly distributed in the chamber. However, inside the chambers without micropillars, the cells could hardly reach complete confluence and would flow away during gradient drug solution producing. This result indicated that the cell culture chambers with micropillars could reduce the shear stress on cells and offer higher density of cultured cells.
a Time-based cell number curve and corresponding microscope photographs in chamber every 12 h for 3 days. Cell number increased significantly with continuous medium flow and the growth conditions of the cells in chip were consistent with the normal conditions observed from regular cell culture plates. b Comparison of cell distribution with the presence of micropillars and without micropillars; the cells were uniformly distributed in the chambers with micropillar structures
3.4 Combinatorial screening of anticancer drugs
The presented device was designed to generate three types of concentration gradients, namely single paclitaxel gradient, single cisplatin gradient and combination of paclitaxel and cisplatin gradient. We designed the microfluidic chip for investigating the effect of the combinatorial anticancer drugs on the viability of A549 cells. A549 cells were incubated on chip and exposed sequentially to different concentrations of paclitaxel and cisplatin. Cells images were taken at each culture chamber before and after drug under gradient conditions. The cell viability of A549 after 24 h was calculated as previously described in Sect. 2.5. Figure S5† shows the cell images under a combination of paclitaxel and cisplatin gradient in channel 1. Figure S6† shows the cell images at single paclitaxel gradient in channel 2. Figure S7† shows the cell images at single cisplatin gradient in channel 3. Figure S8† shows the cell number in each chamber before and after drug injections. In channel 2 and channel 3 (single drug gradient), the viability of the cells was decreased due to the increasing concentrations of drugs along the chamber. But in channel 1 (combination drug gradient), cell viability was not monotonically decreased or increased along the channel. Actually, several combination chambers at middle (neither at the beginning nor the end of the channel) showed much lower cell viability compared to single drug counterparts, indicating the combination of the two drugs when optimized had potential synergistic effects in reducing cancer cell viability. Figure 5a shows cell images in chamber 1 (only paclitaxel), chamber 14 (only cisplatin) and chamber 4 (combination of paclitaxel and cisplatin) before and after drug injection in channel 1. To make better visualization, the pseudo-color operation was used by computer-assisted program to distinguish the live cells and dead cells. The lowest cell viability was obtained in chamber 4, indicating synergistic cancer cell reduction synergy between the two drugs existed at the particular dose/ratio combination in that specific chamber. Cell viability in chamber with two-drug combination gradient in channel 1, paclitaxel gradient in channel 2 and cisplatin gradient in channel 3 is shown in Fig. 5b. The actual data of the cell viability in different chambers are shown in Table S4.
a A549 cell images in chamber 1 (only paclitaxel), chamber 14 (only cisplatin) and chamber 4 (combination of paclitaxel and cisplatin) before and after drug injection in channel 1. The lowest cell viability was obtained in chamber 4. The scale bar is 100 μm. b Cell viability in chambers with two-drug combination gradient in channel 1, paclitaxel gradient in channel 2 and cisplatin gradient in channel 3. The viability of the cells in channel 2 and channel 3 (single drug gradient) was decreased due to increasing concentrations of drug along the chamber. But in channel 1 (combination drug gradient), optimal drug treatment with minimal cell viability occurred neither at the beginning nor at the end of the channel but somewhere in between, indicating a synergistic drug interactions existed at specific dose/ratio condition. Comparison of A549 viability in microfluidic device and 96-well plates was investigated, including A549 cell viability response to c channel 1 with two-drug combination gradient, d channel 2 with paclitaxel gradient and e channel 3 with cisplatin gradient. The combination treatments in the chip lead to a lower viability of A549 cells due to the density of cells and the continuous flow of culture medium
3.5 Comparison of microfluidic device and cell culture plate
In order to support the results of concentration gradient in the chip, a similar experiment was carried out using the conventional well plate cultures. The results for exposing A549 cells to the different anticancer drugs with selected concentrations in microfluidic chip were compared with those in 96-well plates. Figure 5c shows the results of these comparisons in the combination gradient channel. Figure 5d shows the results of these comparisons in paclitaxel gradient channel. Figure 5e shows the results of these comparisons in cisplatin gradient channel. In general, the combination treatment in the chip leads to a lower viability of A549 than that of the well plates. There were several factors that can lead to differences in drug efficiency between these two approaches. Firstly, the density of cells in the chambers and plates was different, which may lead to the higher growth rate observed in chip. Secondly, the chip was working under the continuous flow of culture medium and continuous removal of the cell culture wastes, while the plate culture was performed at an immutable condition. When the concentration ratio of paclitaxel and cisplatin was 0.7–0.3 (chamber 4 in the chip), the minimal cell viability was obtained. This confirmed that the designed chip enables the identification of the optimal dose/ratio drug combination.
3.6 Quantitative analysis on the drug interactions
The goal of this study is to develop a lab-on-chip platform that could automatically create two-drug factorial design data with minimal experimental or operational variance. Factorial design is a commonly used statistical experimental design scheme in engineering and pharmacy to quantitatively investigate multifactor interactions and to identify the optimal combinations of multiple factors through a statistical modeling (Ding et al. 2013). The presented platform requires only the initial injection of the drug stock solution and no more intervention during the following drug combination study, which ousts the experimental errors induced by abundant and repeated drug dilution and administration handlings. To verify whether the data acquired from the presented chip could be applied to generate a robust statistical model similar to that from a conventional factorial design which has to be collected one piece at a time laboriously on a cell culture plate, we established a regression model with the drug combination data acquired from chip in (3.5) to give a quantitative analysis of drug interactions using MATLAB (R2013a, MathWorks, USA). Table S4 provides coefficients of the regression modeling. The mathematical model could be written as follows:
Figure 6 verifies the statistical accuracy of this regression model. In Fig. 6a, the residuals of the regression model follow a mean equal to zero at all fitted values, indicating the model was not biased at any particular fitted value. Normal Q–Q plot implies the residuals that follow a normal distribution, which satisfies the prerequisite for data acquired from the presented chip to be applied for regression modeling (Fig. 6b). In Fig. 6c, Cook’s distance indicates no obvious outliers in the data acquired from the chip, suggesting no data point should be ignored due to experimental operation error or equipment error, which highlights the merit of the presented chip in terms of minimizing data errors and maintaining high data quality. Figure 6d examines whether a regression model for Y or \(Y^{\lambda }\) should be fitted. Since \(\lambda\) shows an optimal value close to 1, no further transformation for Y is needed. All together, Fig. 6 indicates the quality of data generated through the presented chip could be faithfully applied for statistical analysis compatible to traditional factorial design.
Statistical analysis of the regression model generated with data from the presented chip. a Model residuals attain a mean equal to zero, indicating the statistical model is not biased for any particular fitted value. b Normal Q–Q plot supports the residuals that follow a normal distribution, which satisfies the prerequisite to apply the data for regression modeling analysis. c Cook’s distance implies no obvious outlier in the data, suggesting the data set acquired from the presented chip has minimal operation errors or equipment errors. d An investigation on whether Y or \(Y^{\lambda }\) should be applied for regression model indicates no \(Y^{\lambda }\) transformation is needed for the data set acquired from the presented chip. All together, the statistical analysis suggests the data from chip could be applied for statistical modeling in a similar way as conventional factorial design performed on cell culture plate
This mathematical model was used to determine the optimal combination of two drugs against A549 cells. When A = 0.77 × 10−3 mg/mL, B = 0.23 × 10−4 mg/mL, this combination leads to the lowest cell viability of A549 cells (Y = 0.34). The statistical analysis together with the cell phenotype indicates the present platform is highly reliable and can be used as a factorial design chip to screening combination drugs.
4 Conclusion
In summary, a low-cost, convenient and precise drug combination screening platform is developed by generating concentration gradient and drug combination on chip. At first, we simulated fluid flow and reagent diffusion in the device to determine the optimal parameters for system operation. Then we used fluorescein dye to calibrate the chip and to examine the relationship between flow rates and drug concentration distribution at equilibrium state in each chamber. In this section, the ideal flow rate was identified. Finally, the ideal flow rate was applied to screen optimal combinatorial anticancer drugs in the chip. Using this device, two anticancer drugs (paclitaxel and cisplatin) at various concentration gradients have been generated at optimum flow rate of 90 μL/h. Through a cell-based anticancer assay on chip and a mathematical model, the optimal combination drug treatments (paclitaxel: 0.77 × 10−3 mg/mL, cisplatin: 0.23 × 10−4 mg/mL) against A549 cells were determined.
This chip has several distinct advantages. Firstly, the microfluidic chip provides rapid combination gradient generation. Stable concentration gradient can be generated within 20 min and the optimum combination of any two drugs can be identified more conveniently. Secondly, the cells can robustly grow in the chip, which is the basis of subsequent experiments. Thirdly, the design of micropillars reduces the shear stress on cells because the fluid flow into the culture chambers is insignificant. Finally, the serpentine structure in channel accelerates the diffusion of two laminar streams. As a whole, this microfluidic device offers a low-cost, convenient and automatic solution for drug combination screening and can be applied in rapid clinical examination.
It should be noted that this study has examined only two drugs in combination. In practical drug combination study, the number of drugs could be larger. Therefore, a larger-scale combination screening platform to adapt the various emerging applications in complex combination drug screening shall be developed in the future.
References
Ahadian S, Ramón-Azcón J, Estili M, Obregón R, Shiku H, Matsue T (2014) Facile and rapid generation of 3D chemical gradients within hydrogels for high-throughput drug screening applications. Biosens Bioelectron 59:166–173
Al-Lazikani B, Banerji U, Workman P (2012) Combinatorial drug therapy for cancer in the post-genomic era. Nat Biotechnol 30:679–692
Atencia J, Morrow J, Locascio LE (2009) The microfluidic palette: a diffusive gradient generator with spatio-temporal control. Lab Chip 9:2707–2714. doi:10.1039/b902113b
Beltran H et al (2011) New therapies for castration-resistant prostate cancer: efficacy and safety. Eur Urol 60:279–290
Bijl H, Carpenter MH, Vatsa VN, Kennedy CA (2002) Implicit time integration schemes for the unsteady compressible Navier–Stokes equations: laminar flow. J Comput Phys 179:313–329
Bonadonna G et al (1976) Combination chemotherapy as an adjuvant treatment in operable breast cancer. N Engl J Med 294:405–410
Ding X, Sanchez DJ, Shahangian A, Al-Shyoukh I, Cheng G, Ho C-M (2012) Cascade search for HSV-1 combinatorial drugs with high antiviral efficacy and low toxicity. Int J 7:2281–2292
Ding X, Xu H, Hopper C, Yang J, Ho CM (2013) Use of fractional factorial designs in antiviral drug studies. Qual Reliab Eng Int 29:299–304
Ding Y et al (2015) Microfluidic-enabled print-to-screen platform for high-throughput screening of combinatorial chemotherapy. Anal Chem 87:10166–10171
El-Ali J, Sorger PK, Jensen KF (2006) Cells on chips. Nature 442:403–411
Espinal MA et al (2000) Standard short-course chemotherapy for drug-resistant tuberculosis: treatment outcomes in 6 countries. JAMA 283:2537–2545
Frank T, Tay S (2013) Flow-switching allows independently programmable, extremely stable, high-throughput diffusion-based gradients. Lab Chip 13:1273–1281
Honda Y, Ding X, Mussano F, Wiberg A, C-m Ho, Nishimura I (2013) Guiding the osteogenic fate of mouse and human mesenchymal stem cells through feedback system control. Sci Rep 3:3420
Jang Y-H, Hancock MJ, Kim SB, Selimović Š, Sim WY, Bae H, Khademhosseini A (2011) An integrated microfluidic device for two-dimensional combinatorial dilution. Lab Chip 11:3277–3286
Kalchman J et al (2013) A three-dimensional microfluidic tumor cell migration assay to screen the effect of anti-migratory drugs and interstitial flow. Microfluid Nanofluid 14:969–981. doi:10.1007/s10404-012-1104-6
Khoo BL et al (2016) Liquid biopsy and therapeutic response: circulating tumor cell cultures for evaluation of anticancer treatment. Sci Adv. doi:10.1126/sciadv.1600274
Kim J, Hegde M, Jayaraman A (2010) Co-culture of epithelial cells and bacteria for investigating host–pathogen interactions. Lab Chip 10:43–50
Kim J et al (2012) A programmable microfluidic cell array for combinatorial drug screening. Lab Chip 12:1813–1822
Lam KS (1997) Mini-review. Application of combinatorial library methods in cancer research and drug discovery. Anti-cancer Drug Des 12:145–167
Lee K, Kim C, Jung G, Kim TS, Kang JY, Oh KW (2010) Microfluidic network-based combinatorial dilution device for high throughput screening and optimization. Microfluid Nanofluid 8:677–685. doi:10.1007/s10404-009-0500-z
Li C-W, Chen R, Yang M (2007) Generation of linear and non-linear concentration gradients along microfluidic channel by microtunnel controlled stepwise addition of sample solution. Lab Chip 7:1371–1373
Liebmann JE, Fisher J, Teague D, Cook JA (1993) Sequence dependence of paclitaxel (Taxol) combined with cisplatin or alkylators in human cancer cells. Oncol Res 6:25–31
Luo C, Zhu X, Yu T, Luo X, Ouyang Q, Ji H, Chen Y (2008) A fast cell loading and high-throughput microfluidic system for long-term cell culture in zero-flow environments. Biotechnol Bioeng 101:190–195
Malo S, Geuna A (2000) Science–technology linkages in an emerging research platform: the case of combinatorial chemistry and biology. Scientometrics 47:303–321
Merkel T, Bondar V, Nagai K, Freeman B, Pinnau I (2000) Gas sorption, diffusion, and permeation in poly (dimethylsiloxane). J Polym Sci Part B Polym Phys 38:415–434
Neils C, Tyree Z, Finlayson B, Folch A (2004) Combinatorial mixing of microfluidic streams. Lab Chip 4:342–350
Shamloo A, Ma N, Poo M-M, Sohn LL, Heilshorn SC (2008) Endothelial cell polarization and chemotaxis in a microfluidic device. Lab Chip 8:1292–1299
Shi Y, Gao XH, Chen LQ, Zhang M, Ma JY, Zhang XX, Qin JH (2013) High throughput generation and trapping of individual agarose microgel using microfluidic approach. Microfluid Nanofluid 15:467–474. doi:10.1007/s10404-013-1160-6
Silva A, Lee B-Y, Clemens DL, Kee T, Ding X, Ho C-M, Horwitz MA (2016) Output-driven feedback system control platform optimizes combinatorial therapy of tuberculosis using a macrophage cell culture model. In: Proceedings of the national academy of sciences, 201600812
Singh A, Freeman B, Pinnau I (1998) Pure and mixed gas acetone/nitrogen permeation properties of polydimethylsiloxane [PDMS]. J Polym Sci Part B Polym Phys 36:289–301
Somaweera H, Haputhanthri SO, Ibraguimov A, Pappas D (2015) On-chip gradient generation in 256 microfluidic cell cultures: simulation and experimental validation. Analyst 140:5029–5038
Theodossiou C, Cook J, Fisher J, Teague D, Liebmann J, Russo A, Mitchell J (1998) Interaction of gemcitabine with paclitaxel and cisplatin in human tumor cell lines. Int J Oncol 12:825–832
Tran TB, Cho S, Min J (2013) Hydrogel-based diffusion chip with Electric Cell-substrate Impedance Sensing (ECIS) integration for cell viability assay and drug toxicity screening. Biosens Bioelectron 50:453–459
Unger MA, Chou H-P, Thorsen T, Scherer A, Quake SR (2000) Monolithic microfabricated valves and pumps by multilayer soft lithography. Science 288:113–116
Wellhausen M, Rinke G, Wackerbarth H (2012) Combined measurement of concentration distribution and velocity field of two components in a micromixing process. Microfluid Nanofluid 12:917–926. doi:10.1007/s10404-011-0926-y
Wu S et al (2016) Quantification of cell viability and rapid screening anti-cancer drug utilizing nanomechanical fluctuation. Biosens Bioelectron 77:164–173
Xu Y, Lv Y, Wang L, Xing W, Cheng J (2012) A microfluidic device with passive air-bubble valves for real-time measurement of dose-dependent drug cytotoxicity through impedance sensing. Biosens Bioelectron 32:300–304
Xu Z et al (2013) Application of a microfluidic chip-based 3D co-culture to test drug sensitivity for individualized treatment of lung cancer. Biomaterials 34:4109–4117. doi:10.1016/j.biomaterials.2013.02.045
Zhou Y, Lin Q (2014) Microfluidic flow-free generation of chemical concentration gradients. Sens Actuators B Chem 190:334–341
Acknowledgements
This work was supported by National Natural Science Foundation of China (81301293) and National Science and Technology Major Projects for “Major New Drugs Innovation and Development” (2014ZX09507008).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sun, J., Liu, W., Li, Y. et al. An on-chip cell culturing and combinatorial drug screening system. Microfluid Nanofluid 21, 125 (2017). https://doi.org/10.1007/s10404-017-1959-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10404-017-1959-7