Abstract
The adverse effects of invasive pests on ecosystems have gradually intensified, and the prevention of invasive pests is a long-standing research focus. Genetic control strategies are effective, sustainable and environmentally friendly methods for controlling pests and have received substantial attention worldwide. However, there is a lack of researches on the genetic control of Bactrocera dorsalis Hendel, a global invasive pest with strong flight ability. The wing of B. dorsalis as an important flight organ may be a main reason for its widespread occurrence. Here, we first analysed the wing structure of B. dorsalis and found that its wing has the typical characteristic of Diptera insect wing. The spatiotemporal expressions of the wing development genes were examined by quantitative real-time PCR, antibody staining and RNA in situ hybridization. The results indicated that the wing development genes were significantly upregulated in the pupal stage, and the regional expression of each gene was clarified. Wingless (wg), a key gene that significantly affects wing development, was selected from nine genes through RNA interference and used to simulate the field control of B. dorsalis. The offspring population and the fruit-borer rate decreased significantly after the simulated control. This study provides preliminary support for the application of genetic pest control by regulating the wing development gene and proposes a novel idea for solving the problem of the extensive spread of B. dorsalis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Key message
-
Bactrocera dorsalis is a global invasive pest with powerful flight ability. Interfering the flight ability by targeting wing development key genes is a potential way to control this pest.
-
Here, we proved the hypothesis by simulated control experiments using a key gene wingless related to wing development and proposed a new approach to control this economic important pest.
-
Our study lighted up the direction of practical application of RNA preparation for other pest control.
Introduction
In the process of invasion, the diversity of insect wings makes invasive insect pests more proficient than other terrestrial animals in terms of foraging, courtship, avoidance, dispersal and migration (Fraimout et al. 2018; Holway et al. 2002). Although they are slow flyers, insects have advantages in flight in comparison to birds and bats due to the speed and flexibility of their strong wings, which also help insects avoid predators (Grimaldi and Engel 2005). Wing-based diffusion is an important ecological strategy through which invasive pests adapt to environmental changes (Shen 2009).
As an important invasive pest, Bactrocera dorsalis Hendel, the oriental fruit fly, damages various fruits and vegetables (Clarke et al. 2005; Permpoon et al. 2011; White and Elson-Harris 1992). The first record of B. dorsalis was from India in 1794 (Clarke et al. 2019; Fabricius 1794); at present, B. dorsalis is widely distributed all over the world, including areas of Asia and Pacific regions, South Africa, Europe and, transitorily, Florida and California (Clarke et al. 2005; Nugnes et al. 2018; Stephens et al. 2007; Wan et al. 2011). Particularly, within the last 15 years, the fly has spread rapidly across sub-Saharan Africa and has begun a northward migration into central China (Clarke et al. 2019; Ekesi et al. 2016; Qin et al. 2018, 2019; Zeng et al. 2019). Compared with other similar species, B. dorsalis has a wider distribution (Liu et al. 2013; Stephens et al. 2007). Previous research showed that B. dorsalis has a stronger flight capacity than a similar species, Bactrocera correcta. The wing is an important organ that supports flight and may be an important factor for the widespread occurrence of B. dorsalis (Guo et al. 2018).
Chemical, physical and biological methods are commonly used to control B. dorsalis. However, chemicals tend to produce resistance and pesticide residues, while the cost of physical and biological control methods is relatively high. In recent years, the sterile insect technique (SIT), which is a genetic control strategy, has been widely used to control B. dorsalis because of its environmental friendliness (Aketarawong et al. 2011). Genetic pest control uses physical, chemical or genetic means to change the genetic material of pests to reduce their viability, reproductive potential or capacity to cause damage. According to genetic principles, pests with defective genes can mate with those from wild populations in nature, resulting in nonproduction or death of offspring (Schliekelman et al. 2005). As a result, the pest population will rapidly decrease after several generations (Wimmer 2005). It is also possible to use gene driving methods to modify insects so that strains with benign genes can gradually replace those with damaging effects (James 2005). Genetic pest control has the advantage of target specificity and long-lasting transmission of the genetic factors in the pest population without affecting other populations (Bruno Wilke and Marrelli 2012; Chen et al. 2007).
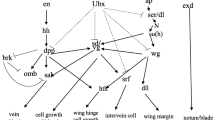
Insect wing types, which are mainly manifested in the shape, texture and covering of the wings, vary with insect species. Most insects have wings, but with lifestyle changes, the wings of some insect species degenerate or even disappear. For instance, differentiation between winged and wingless aphids is caused by the suppression or overexpression of key genes involved in wing development (Brisson et al. 2010). Researches on model insects have confirmed that the development of wings can be affected by controlling specific wing genes (Medved et al. 2015; Tomoyasu et al. 2005). Evidence has shown enhancement of flight performance by the genetic manipulation of wing shape in Drosophila melanogaster (Ray et al. 2016). Therefore, we considered whether the genes related to wing development could be regulated by a genetic control strategy to prevent the spread of B. dorsalis. In this paper, we first explored the wing developmental process and spatiotemporal expression patterns of wing development genes in B. dorsalis. We used RNA interference (RNAi) to screen for the key genes that seriously affect the wing development of this species and wingless (wg) was selected from nine genes. To preliminarily explore the genetic control of B. dorsalis in a closed greenhouse, we then performed a field control experiment in which wg was suppressed by nanocarrier-delivered RNAi. Our research provides candidate target genes for population substitution strategies and a novel approach for controlling B. dorsalis.
Materials and methods
Insect rearing
The laboratory population of B. dorsalis was reared on an artificial diet at 25 ± 1 °C under a relative humidity of 70% and a 14 h:10 h L:D photoperiod. Papaya was used as a bait to induce the adults to lay eggs. Eggs and larvae were cultured using an artificial diet in glass bottles (Yuan et al. 2006). The third-instar larvae were then transferred to wet soil for pupation. Adults were fed with a mixture of sucrose and soybean peptone in cages to complete their life cycle. B. dorsalis were collected from Guangzhou, Guangdong, China, and the population was reared in the laboratory for approximately 20 generations.
Immunohistochemistry
Dissected wing discs from the second- and third-instar larvae of B. dorsalis and D. melanogaster were fixed using fixative and then stained with antibodies according to standard procedures. The primary antibodies used were mouse anti-En (Engrailed), 1:200, mouse anti-Wg, 1:200, rat anti-Ci (Cubitus interruptus), 1:200, mouse anti-Ubx (Ultrabithorax), 1:200, mouse anti-Ptc (patched), 1:200 (Developmental Studies Hybridoma Bank, DSHB, IA, USA), rabbit anti-pMad (phosphorylated mothers against decapentaplegic), 1:200 (Cell Signalling Technology, MA, USA), rabbit anti-Vg (Vestigial), 1:200 (GeneTex, CA, USA), rabbit anti-Hh (Hedgehog), 1:200 (Abcam, Cambridge, UK) and rabbit anti-Omb (Optomotor-blind) (1:1000) (Clontech, Mountain View, CA, USA). Secondary antibodies were goat anti-mouse DyLight 488, 1:200 (Agrisera, Sweden) and goat anti-rabbit cy5, 1:200 (Abcam, Cambridge, UK). Other staining reagents include rhodamine-phalloidin (1:50, Invitrogen, Waltham, USA), DAPI (1:500, Sigma-Aldrich, Shanghai, China).
Wing disc cryosections
After secondary antibody staining, wing discs were refixed in 4% paraformaldehyde for 30 min and then washed and soaked in a 30% sucrose solution at 4 °C overnight. Wing discs were oriented in Tissue-Tek (Sakura Finetek, Torrance, CA, USA), frozen and cut into 25 μm sections on a freezing microtome (Jinhua YIDI Medical Appliance Co., Ltd, Zhejiang, China).
RNA in situ hybridization
Total RNA samples were isolated from B. dorsalis with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. First-strand cDNA was synthesized from 1 μg of RNA using the PrimeScript RT reagent kit with gDNA Eraser (Perfect Real Time) (TAKARA, Kyoto, Japan). The cDNA templates were employed for amplification using gene-specific primers (Table S1) and GO Taq® Hot Start DNA polymerase (Promega, Madison, WI, USA). The reverse transcription PCR (RT-PCR) product was subsequently gel-recovered, purified and ligated into the pGEM-T vector (Promega, Madison, WI, USA). The vector was then transformed into E. coli DH5α-competent cells (Takara, Kyoto, Japan) and sequenced. The recombinant plasmids were extracted using the QIAGEN® Plasmid Midi Kit (Qiagen, Hilden, Germany) followed by restriction analysis, single endonuclease digestion and linearization. The purified template DNA was used for the synthesis of RNA hybridization probes using a DIG RNA Labeling Kit (SP6/T7) (Roche, Basel, Switzerland) following the manufacturer’s protocol. The probes were dissolved in 25 μl of nuclease-free water (Promega, Madison, WI, USA) and 250 μl of hybridization buffer and then stored at − 80 °C until use.
Dissected wing discs were fixed using 4% paraformaldehyde at 4 °C for 30 min and then washed with 1 × PBS and hydrochloric acid buffer. Subsequently, the wing discs were washed with formamide solution at 4 °C for 10 min and then incubated with hybridization buffer containing the probes at 55 °C for 22 h. The wing discs were washed twice with 0.1 × SSC at 60 °C, followed by washing with a mixture of anti-digoxigenin alkaline phosphatase-conjugated antibody (DIG Nucleic Acid Detection Kit, Roche, Basel, Switzerland) and blocking buffer in TBS (Sigma-Aldrich, WI, USA). After a few rinses with 0.05% Tween (Amresco, ID, USA), the wing discs were washed with DAP buffer for 5–10 min. Finally, 20 μl of NBT/BCIP substrate solution was added to 980 μl of DAP buffer, and the mixture was used for the colour reaction, which was incubated overnight at 37 °C. The wing discs were photographed and observed the next day.
Synthesis of double-stranded RNA (dsRNA) and RNAi
The prepared cDNAs were used as templates for the amplification of gene products. The gene-specific primers were consistent with those used for RNA in situ hybridization, which are listed in Table S1. The pMD18-T vector (TAKARA, Kyoto, Japan) was used for the gene clone. The plasmids were extracted using a TIANprep Rapid Mini Plasmid Kit (TIANGEN, Beijing, China). The extracted plasmids were used as templates, and primers with the T7 promoter were designed for amplification and purification. The purified products were prepared for dsRNA synthesis using the T7 RiboMAX™ Express RNAi System (Promega, Madison, WI, USA) according to the manufacturer’s protocol. The dsRNAs were dissolved in nuclease-free water (Promega, Madison, WI, USA), and their quality and concentration were determined via agarose gel electrophoresis and with a Quawell UV–Vis Q5000 spectrophotometer (Quawell Technology Inc, San Jose, CA, USA) and were then stored at − 20 °C until use.
Each first-instar larva of B. dorsalis was treated with 1 µg of dsRNA mixed with 1 µl of the gene nanocarrier, a star polycation (SPc) constructed as a highly efficient gene vector (1 mg/ml), which was provided by Professor Yin Meizhen at Beijing University of Chemical Technology (Li et al. 2019). The technology of nanocarrier-dsRNA has been proven to effectively improve RNAi efficiency (Shen 2014). Three biological replicates were performed for each treatment, and every biological replicate included 20 individuals. Mature larvae were removed and placed in the soil for pupation, and phenotypic comparisons were conducted after eclosion. The growth and development of the individuals after RNAi were continuously observed. The corresponding biostatistical analyses were carried out, and the mortality and malformation rates of the individuals after RNAi were determined.
Preparation of artificial baits with nanocarrier-dsRNA
PCR amplification with the ExTaq enzyme was implemented using a plasmid with the target gene wg as the template, and the primers were the same as those in the previous step, which are listed in Table S1. After phenol–chloroform extraction and purification of the PCR products, 12 μg of product, 8 μl of T7 RNA polymerase, 10 μl of RNase inhibitor, 80 μl of transcription buffer and 8 μl of NTP (100 mM) were added, and ddH2O was used to bring the mixture to a volume of 400 μl. Then, samples were cooled at room temperature after incubation for 4 h at 37 °C and 5 min at 75 °C. Next, a large amount of dsRNA was extracted by phenolic chloroform. The concentration of dsRNA was measured, diluted to 500 ng/μl and then subpacked and stored at − 20 °C.
The SPc acts as a highly efficient but low-cost gene vector for pest management. A total of 500 μg of dsRNA was incubated with 500 μl of SPc for 0.5 h and added to 50 g of artificial diet. After mixing well, preservation film was used to wrap the mixture tightly, forming a spherical shape. Baits containing ddH2O (blank control), dsGFP (dsGFP control) and dswg were prepared.
Preliminary control of B. dorsalis by dsRNA preparation
A simulated control experiment was carried out in a closed greenhouse. Three groups of treatments (blank control, dsGFP control and treatment with dswg) were set up. Each group was treated with six replicates, which corresponded to six tomato seedlings with fruits. Each tomato seedling was hung with three baits and covered with nylon mesh to form an independent small ecological environment. The baits containing ddH2O, dsGFP and dswg were hung on the tomato plants of the blank control, the dsGFP control group and the dswg-treated group, respectively.
First, a total of 60 adults with 30 of each sex were released into each small ecological environment when all the tomato fruits were bagged. After approximately 10 days of mating and oviposition, all adults and bags were removed. The eggs hatched, and larvae were fed tomato fruits or baits. The larvae pupated in approximately 10 days, and the adults emerged in another 10 days. The statistical emergence rate and the attraction rate of artificial baits and tomato fruits (also considered as the fruit-borer rate) in the simulated control experiments served as the criteria for determining the effect of the dsRNA preparations. Because the emergence period among individuals was inconsistent, continuous observation and statistics were employed to ensure the accuracy of the simulated experiment.
Quantitative real-time PCR (qRT-PCR)
The expression levels of wing development genes in different periods and RNAi efficiency of dsRNA were tested via qRT-PCR (primers in Table S2). QRT-PCR was performed with an Applied Biosystems 7500 Real-time PCR system (Life Technologies, Grand Island, NY, USA). The reaction mixture (25 μl total volume) contained 12.5 μl of SYBR® Premix Ex Taq™ II (Tli RNaseH Plus) (TAKARA, Kyoto, Japan), 0.5 μl of ROX reference dye, 1 μl of each primer (10 μM), 1 μl of cDNA and 9 μl of RNase-free water. The following thermal cycling profile was used: 95 °C for 30 s; 40 cycles of 95 °C for 5 s and 60 °C for 34 s; 95 °C for 15 s; 60 °C for 1 min; and 95 °C for 15 s. Three independent biological replicates were performed, and 18S rRNA was used as a reference gene (Hu et al. 2014). Relative expression was calculated via the 2−ΔΔCt method (Livak and Schmittgen 2001).
Statistical analysis
The results of qRT-PCR are presented as the mean ± standard deviation (SD) of three independent biological replicates. Comparisons between the means of two independent samples were performed with Student’s t test, and multiple comparisons were performed with a one-way ANOVA followed by Tukey’s posthoc test in SPSS 17 (IBM Corporation, Armonk, NY, USA). A significant difference was considered to exist when p < 0.05. Graphs were generated using OriginPro 8 (OriginLab Corporation, Northampton, MA, USA).
Imaging
Fluorescence images were obtained with a digital fluorescence microscope (Advanced Microscopy Group, Bothell, WA, USA) and a confocal microscope (Leica TCS-SP2-AOBS, Leica Microsystems Inc., Wetzlar, Germany). Images of the RNAi phenotypes and the protein expression of B. dorsalis were obtained using a UV-C optical confocal microscopic imaging system (Olympus, Tokyo, Japan).
Results
The structure of wing discs of B. dorsalis
Through dissection of the second- and third-instar larval wing discs of B. dorsalis, we found that the wing discs of B. dorsalis are saclike in shape and very similar to those of the model insect Drosophila melanogaster. With the development of the larvae, the wing discs gradually enlarged. At the beginning of the third instar, the morphological furrows were differentiated to prepare for drastic metamorphosis in the pupal stage (Fig. 1). To understand the three-dimensional structure of the wing discs of B. dorsalis, frozen sections of wing discs cut horizontally and vertically were used for morphological observations. Transverse sections showed that the wing discs consisted of two layers of cells, the pseudostratified epithelium (PE) and disc proper (DP), similar to those of D. melanogaster (Fig. 1E). The longitudinal section showed the location of the furrows on the wing discs (Fig. 1F). During metamorphosis, the peripodial membrane (PM) of the wings partially degraded and fused with the epidermis, which caused the wing discs to turn inside out and fold to form double layers. Afterwards, the cells stretched and flattened with development to become the adult wings.
Dynamic developmental process and stereo structure of the wing discs of Bactrocera dorsalis. The imaginal discs were oriented with the anterior left and the dorsal up, and a monolayer of epithelial cells was arranged in a saclike structure with the apical sides facing the lumen. A–D represent planar structures of wing discs in second, early third instar, middle third instar and late third instar larvae of Bactrocera dorsalis, respectively; × 20 magnification. E, F represent the transverse and longitudinal sections of wing discs in Bactrocera dorsalis, respectively; × 20 magnification. Arrows point to cell layers of the pseudostratified epithelium (PE) and disc proper (DP); the arrowheads represent the morphogenetic furrows. Phalloidin specifically stains the filamentous actin, while DAPI stains the nucleus. Scale bars are 50 µm
Spatiotemporal dynamic expression pattern of wing development genes in B. dorsalis
Nine wing development genes were identified by previous transcriptomic analysis (Fig. S1) (Guo et al. 2018). To explore the expression pattern of the wing development genes of B. dorsalis, the expression levels of nine genes were analysed in different developmental stages using qRT-PCR. Figure 2 shows the temporal dynamic expression analysis of nine wing development genes. The expression of wing development genes was significantly upregulated in the pupal stage, illustrating that the expression of wing genes was active during the pupal stage of drastic metamorphosis. The high expression of related genes in the pupal stage may play crucial roles in the development of wings.
Temporal expression patterns of nine genes of Bactrocera dorsalis at different developmental stages. A–I represent the expression levels of wg, vg, hh, en, ci, ap, dpp, omb and ubx in different developmental stages of Bactrocera dorsalis. Different letters above the bars indicate significant differences at p < 0.05 as determined by Student’s t test. E, Egg; L1, first-instar larvae; L2, second-instar larvae; L3, third-instar larvae; P2, 2-day pupae; P5, 5-day pupae; AM, male adults; AF, female adults
The structure and developmental process of the larval wing discs of B. dorsalis and D. melanogaster are very similar. First, we applied antibodies commonly used with D. melanogaster to perform immunohistochemical experiments on B. dorsalis. En and pMad antibodies were conservatively expressed in the two species (Fig. 3A, B, G, H). En was expressed in the posterior compartment, which clarified the boundary between the anterior and posterior (A/P) compartments. However, more antibodies involved in the study were not recognized effectively. As shown in Fig. S2, Ptc could not be recognized in the wing discs of B. dorsalis. Therefore, to understand the expression of wing development genes more intuitively in the wing discs of B. dorsalis, we used RNA in situ hybridization to reveal the spatial expression patterns of the related genes. Combined with the results of a previous study on D. melanogaster, we found that the expression domains of wing development genes in B. dorsalis were highly similar to those in D. melanogaster. Wg, the morphogen, was expressed along the dorsal/ventral (D/V) compartment boundary and the border of the wing pouch in B. dorsalis (Fig. 3C). The expression of Vg in the wing disc of B. dorsalis was mainly concentrated in the wing pouch region (Fig. 3D). Another morphogen, Hh, was expressed in the posterior compartment of the wing disc of B. dorsalis (Fig. 3E). The transcription factor En was expressed in the posterior compartment of the wing disc of B. dorsalis, as noted above (Fig. 3B), which directed the synthesis of Hh. In contrast, Ci was expressed in the anterior compartment of the wing disc of B. dorsalis, which was complementary to the region of En expression in the wing disc (Fig. 3F). The expression of Apterous (Ap) in the wing disc of B. dorsalis was concentrated in the dorsal compartment (Fig. 3I). Decapentaplegic (Dpp) was expressed in the A/P zone of the wing disc of B. dorsalis. As the downstream target of Dpp, the Omb expression pattern was similar to that of Dpp (Fig. 3J, K). In addition, Ubx was weakly expressed in the wing disc of B. dorsalis (Fig. 3L).
Spatial expression patterns of wing development genes in wing discs of Bactrocera dorsalis. A, B represent the expression of En antibody on the wing discs of Drosophila melanogaster and Bactrocera dorsalis, respectively. G, H represent the expression of the pMad antibody on the wing discs of Drosophila melanogaster and Bactrocera dorsalis, respectively. C–F show the expression domains of Wg, Vg, Hh and Ci. I–L show the expression domains of Ap, Dpp, Omb and Ubx. × 20 magnification of the wing disc of Drosophila melanogaster is shown, while × 10 magnification of the wing disc of Bactrocera dorsalis is shown. Blue or purple colour represents the expression domain of each gene
RNAi efficiency and biostatistics
Using qRT-PCR technology, the interference efficiencies of the corresponding genes were determined and are shown in Fig. 4. All genes were significantly downregulated after interference, indicating that RNAi was effectively implemented in B. dorsalis. After RNAi, the wing deformity rate of the individuals was determined, and the growth and development of individuals were continuously observed. Corresponding biostatistical analyses were carried out, and the mortality was calculated because of the rest adults only survive for several days and would eventually die. The data listed in Table S3 indicate that the highest wing malformation rate (73.33%) and the highest mortality (31.67%) were caused by gene interference with wg.
RNAi efficiency and biostatistics of simulated control for Bactrocera dorsalis. A–I represent the expression levels of wg, vg, hh, en, ci, ap, dpp, omb and ubx after RNAi in Bactrocera dorsalis. The symbols “*” and “**” above the bars represent significant differences at p < 0.05 and p < 0.01, respectively. CK, the control groups; dsGFP, dsGFP control groups
Functional verification of the wing development genes of B. dorsalis
Compared with the control group, there was no obvious abnormality in the wing development of the GFP control group (Fig. 5A, B), while the individuals in the RNAi treatment group had different degrees of wing dysplasia (Fig. 5C–K). The wings of the dpp-RNAi group were significantly shrunken and stunted (Fig. 5C), and the ap-RNAi group also showed a phenotype with insufficient wing development and obvious curl at the end of the wing (Fig. 5D). As the downstream target gene of the Dpp signalling pathway, omb may have a function similar to that of dpp. The wings of omb-RNAi individuals were shrunken and could not expand normally (Fig. 5E). After silencing of hh, distorted wings were observed (Fig. 5F). The wings of the en-RNAi group were significantly bent, and the individuals were unable to fly normally, while the ci-RNAi group showed a winged-crispatura phenotype (Fig. 5G, H). The key gene that makes the hind wing specialize into a haltere, ubx, is very important for wing development. After ubx was knocked down in B. dorsalis, the forewing was unable to stretch normally, and the haltere also disappeared (Fig. 5I).
Typical phenotypes after RNAi for seven genes in Bactrocera dorsalis. Compared with the blank control group (A) and the GFP-RNAi control group (B), the treated groups, including dpp-RNAi (C), ap-RNAi (D), omb-RNAi (E), hh-RNAi (F), en-RNAi (G), ci-RNAi (H), ubx-RNAi (I), wg-RNAi (J) and vg-RNAi (K), showed different wing phenotypes
Wg, as the morphogen, plays an important role as an organizer controlling the appearance of the D/V compartment. After gene interference with wg, the wing hinge of the wg-RNAi group was curved or even folded, and a partial nick was also formed at the edge of the wing (Fig. 5J, Fig. S3). The vg-RNAi group with phenotypes of nicked, folded, or bent wings was similar to the wg-RNAi group (Fig. 5K, Fig. S3). Overall, wg-RNAi and vg-RNAi produced the most severe wing deformity. More importantly, we observed not only the deformity of wings but also defects of other organs, such as the distortion of the body and the abnormal development of the legs (Fig. 5, Fig. S3).
Preliminary application of dsRNA preparation for controlling B. dorsalis
During the simulated control experiment, the adults could live and lay eggs normally in the small ecological environment (Fig. 6A). The eggs and the larvae of the next generation were found in the artificial bait. After pupation and emergence, the number of adults that emerged was counted. The results showed that the number of offspring in the blank control group was 201 ± 10.89, while the numbers of offspring in the nanocarrier/dsGFP- and nanocarrier/dswg-treated groups were 198 ± 8.11 and 62 ± 12.49, respectively. Compared with the control group, the population number in the treatment group fed the RNA preparation decreased significantly (Fig. 6B).
Simulated field control of Bactrocera dorsalis. A shows the schematic illustration of the simulated field control. The baits containing nanocarrier/dsRNA were hung on the tomato plants of the blank control (CK), dsGFP control group and the dswg-treated group. Each group was treated with six replicates, which corresponded to six tomato seedlings covered with nylon mesh to form an independent small ecological environment. Thirty females and thirty males were released into each nylon mesh. B, C show the number of offspring and the attraction rate of tomato fruits and artificial baits after simulated control of Bactrocera dorsalis. The symbols “**” above the bars (B) represent significant differences at p < 0.01. Different letters above the bars (C) indicate significant differences at p < 0.05
We also performed a statistical and comparative analysis of the attraction rate of tomato fruits and artificial bait of the three groups, for which the blank control group and the nanocarrier/dsGFP group showed the attraction rate of tomato fruits (also considered as the fruit-borer rate) of 58.28 ± 3.89% and 50.05 ± 3.87%, respectively. The fruit-borer rate of the nanocarrier/dswg treatment group of 29.34 ± 4.18% was significantly lower than that of the control group (Fig. 6C). The attraction rates of artificial bait were much higher than the fruit-borer rate in the three groups (Fig. 6C), which indicated that B. dorsalis had a better attraction to the artificial bait, and thus, the harm to the host was reduced to a certain extent.
Discussion
Population inhibition and population substitution are two main genetic pest control strategies. The SIT employs population inhibition, and males need to be released annually. Population substitution was first proposed to control the spread of mosquito-borne malaria by using strains with benign genes replacing strains with toxic genes in nature, which is conducive to the protection of species diversity compared with population inhibition (Bruno Wilke and Marrelli 2012). However, there are few studies on phytophagous pest control by means of population substitution. Our research on wing development genes provides candidate target genes for implementing population substitution strategies in the future.
Our study first revealed the morphology of wing discs and the dynamic process of wing development in the larval stage of B. dorsalis, which is a very important invasive pest. In the early stage of wing development, the related genes were normally expressed in wing discs of B. dorsalis, and then, furrows were formed, which provided conditions for the normal folding and extension of wings. Based on a previous analysis of the transcriptome, nine wing development genes were screened and identified. The wing genes of B. dorsalis were highly conserved compared with those of D. melanogaster, a representative Diptera insect.
At the temporal level, nine wing development genes were expressed more actively in the pupal stage, indicating that wing development genes were generally activated and highly expressed in pupae. From this result, we determined that the best time for performing RNAi in B. dorsalis is before the pupal stage as well as the larval stage. This section lays a foundation for follow-up studies of gene functions. Spatially, immunohistochemical experiments and RNA in situ hybridization revealed the expression domains of related wing development genes in the wing tissue of B. dorsalis. In addition, the coding proteins of different genes were limited to their respective expression domains to perform corresponding functions, consistent with D. melanogaster. Vg mediated the appearance of cell morphology by regulating the accumulation of actin, which was induced by the Wg signalling pathway (Zecca and Struhl 2007). The function of Ci in Drosophila is to coordinate the development of the A/P axis with En and Omb (Grimm and Pflugfelder 1996; Rodriguez et al. 2004). Ap can determine the fate of dorsal compartment cells and provide location information for the D/V boundary in Drosophila (Blair et al. 1994). The target of Hh, another morphogen, Dpp, whose signal molecule could be transported from organizer cells to both sides, formed a continuous concentration gradient on the wing disc, guiding the regionalized expression of target genes and regionally regulating the wing disc (Shen et al. 2010; Zhang et al. 2013). The specific expression of Ubx, a member of the Hox family, determines the morphological difference between wings and halters (Tomoyasu et al. 2005; Weatherbee et al. 1998). Because morphogen guides the development of most organs, we also found that not only wing deformity, but also other organs were defective after RNAi in the course of phenotypic observation. Therefore, it is reasonable to use wing development genes as candidate genes for controlling B. dorsalis.
In recent years, scholars mainly have used microinjection to carry out RNAi in B. dorsalis (Liu et al. 2015; Shi et al. 2019; Zhang et al. 2019). However, this injection method is not suitable for field applications for the genetic control of B. dorsalis. This study established mature methods and systems for dsRNA synthesis, which could successfully synthesize high-quality dsRNA for the implementation of RNAi. In this study, a double-control method was used to verify the possibility of high-efficiency RNAi in B. dorsalis by offering dsRNA as food with a nanocarrier. The nanocarrier could rapidly deliver dsRNA into the insect cells and efficiently inhibit the expression of selected genes. We then observed the phenotypes of wing defects or deformities. At the same time, the determination of interference efficiency using qRT-PCR also showed the effectiveness of the RNAi (Fig. 4). The success rate of gene interference has been greatly improved because of the advantages of nanocarriers, which not only provide a new method for studies of insect gene function but also promote the application of the RNAi feeding method in future pest control. Our study also found that with the help of nanocarriers, an average of 1 µg of dsRNA per fruit fly could be sufficient to achieve high RNAi efficiency, which provides a reference for future research on gene functions in B. dorsalis. After RNAi of wing development genes, we found that inhibiting the expression of corresponding genes could lead to individual death; therefore, we counted mortality as a biological indicator. In addition to the direct lethal effect, the loss of flight ability also contributes to the control of this pest. Based on the results of phenotypic and biological tests, the key gene wg was selected to synthesize dsRNA for preparing an RNA preparation for simulated field control because the highest wing malformation and mortality rates were observed after wg-RNAi.
We preliminarily carried out a simulated field control experiment in a greenhouse to test the efficacy of nanocarrier/dsRNA preparations, providing technical support for future applications in orchards or fields. B. dorsalis had a high attraction to the artificial bait containing the nanocarrier/dsRNA preparation (Fig. 6), which could attract females to lay eggs and reduce the harm inflicted on host fruits. The larvae fed on the dsRNA in the bait, RNAi was completed in the larvae, and the number of emerging adults was reduced. As the morphogen, Wg could influence the growth and development of insects, which may affect the hatching of eggs, result in the death of larvae, or failure of eclosion. Due to the high frequency of wing malformation, adults failed to fly and would eventually die, which also contribute to the control of this pest. In this study, a cost-saving farmland-type nanocarrier, SPc, was used to carry dsRNA into insect cells efficiently and rapidly, which could not only facilitate RNAi control of B. dorsalis but also contribute to the development of technology used in synergistic RNA preparation with nanomaterials.
Practical experiments can be carried out to further develop this theoretical control into a reality, allowing RNA preparations to be used in orchards to control fruit flies. We can enlarge the small-scale ecosystems used in the simulation experiment to a field scale, hang baits on each fruit tree, make continuous observations for multiple generations to ensure the control effect of the RNA preparations and then popularize the genetic pest control method for actual production. In addition, new techniques such as gene editing and gene driving can be used to produce strains with defective wing development genes, which can be released into wild populations to stabilize the defective genes, thus replacing natural insects with normal wings to gradually control the spread of invasive pests.
Conclusions
In this study, we develop a mature dsRNA synthesis system that can synthesize dsRNA with high interference efficiency and propose a new approach to control the invasive pest B. dorsalis by inhibiting wing developmental gene wingless for the first time. Combined with the latest nanomaterial technology, the dsRNA could be carried into cells efficiently, which may provide a solution for the field application of RNA preparations to control other pests. With the rapid development of new techniques such as gene transfection and gene drive systems, the artificial production of genetically modified insects for population substitution will be realized in the future.
Authors contribution
ZL, JS and SG conceived and designed the experiments. SG, XG and LZ performed the experiments. SG, ZZ, LL, JS and ZL analysed the data. SG, ZL and JS wrote the paper. All authors read and approved the manuscript.
References
Aketarawong N, Chinvinijkul S, Orankanok W, Guglielmino CR, Franz G, Malacrida AR, Thanaphum S (2011) The utility of microsatellite DNA markers for the evaluation of area-wide integrated pest management using SIT for the fruit fly, Bactrocera dorsalis (Hendel), control programs in Thailand. Genetica 139:129–140
Blair SS, Brower DL, Thomas JB, Zavortink M (1994) The role of apterous in the control of dorsoventral compartmentalization and PS integrin gene expression in the developing wing of Drosophila. Development 120:1805
Brisson JA, Ishikawa A, Miura T (2010) Wing development genes of the pea aphid and differential gene expression between winged and unwinged morphs. Insect Mol Biol 192:63–73
Bruno Wilke AB, Marrelli MT (2012) Genetic control of mosquitoes: population suppression strategies. Rev Inst Med Trop Sao Paulo 54:287–292
Chen C, Huang H, Ward CM, Su JT, Schaeffer LV, Guo M, Hay BA (2007) A synthetic maternal-effect selfish genetic element drives population replacement in Drosophila. Science 316:597–600
Clarke AR, Armstrong KF, Carmichael AE, Milne JR, Raghu S, Roderick GK, Yeates DK (2005) Invasive phytophagous pests arising through a recent tropical evolutionary radiation: the Bactrocera dorsalis complex of fruit flies. Annu Rev Entomol 50:293–319
Clarke AR, Li ZH, Qin YJ, Zhao ZH, Liu LJ, Schutze MK (2019) Bactrocera dorsalis (Hendel) (Diptera: Tephritidae) is not invasive through Asia: it’s been there all along. J Appl Entomol 143(8):797–801
Ekesi S, De Meyer M, Mohamed SA, Virgilio M, Borgemeister C (2016) Taxonomy, ecology, and management of native and exotic fruit fly species in Africa. Annu Rev Entomol 61:219–238
Fabricius JC (1794) Entomologia systematica emendata et aucta, vol 4. Impensis Christ. Gottl. Proft., Copenhagen
Fraimout A et al (2018) Phenotypic plasticity of Drosophila suzukii wing to developmental temperature: implications for flight. J Exp Biol 221:jeb166868
Grimaldi DA, Engel MS (2005) Evolution of the insects. Cambridge University Press, Cambridge
Grimm S, Pflugfelder GO (1996) Control of the gene optomotor-blind in Drosophila wing development by decapentaplegic and wingless. Science 271:1601–1604
Guo S, Zhao Z, Liu L, Li Z, Shen J (2018) Comparative transcriptome analyses uncover key candidate genes mediating flight capacity in Bactrocera dorsalis (Hendel) and Bactrocera correcta (Bezzi) (Diptera: Tephritidae). Int J Mol Sci 19:396
Holway DA, Lach L, Suarez AV, Tsutsui ND, Case TJ (2002) The causes and consequences of ant invasions. Annu Rev Ecol Syst 33:181–233
Hu J, Chen B, Li Z (2014) Thermal plasticity is related to the hardening response of heat shock protein expression in two Bactrocera fruit flies. J Insect Physiol 67:105–113
James AA (2005) Gene drive systems in mosquitoes: rules of the road. Trends Parasitol 21:64–67
Li J, Qian J, Xu Y, Yan S, Shen J, Yin M (2019) A facile-synthesized star polycation constructed as a highly efficient gene vector in pest management. ACS Sustain Chem Eng 7:6316–6322
Liu X, Jin Y, Ye H (2013) Recent spread and climatic ecological niche of the invasive guava fruit fly, Bactrocera correcta, in mainland China. J Pest Sci 86:449–458
Liu G, Wu Q, Li J, Zhang G, Wan F (2015) RNAi-mediated knock-down of transformer and transformer 2 to generate male-only progeny in the oriental fruit fly, Bactrocera dorsalis (Hendel). PLoS ONE 10:e0128892
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25:402–408
Medved V, Marden JH, Fescemyer HW, Der JP, Liu J, Mahfooz N, Popadic A (2015) Origin and diversification of wings: insights from a neopteran insect. Proc Natl Acad Sci USA 112:15946–15951
Nugnes F, Russo E, Viggiani G, Bernardo U (2018) First record of an invasive fruit fly belonging to Bactrocera dorsalis complex (Diptera: Tephritidae) in Europe. Insects 9:182
Permpoon R, Aketarawong N, Thanaphum S (2011) Isolation and characterization of Doublesex homologues in the Bactrocera species: B. dorsalis (Hendel) and B. correcta (Bezzi) and their putative promoter regulatory regions. Genetica 139:113–127
Qin Y et al (2018) Population structure of a global agricultural invasive pest, Bactrocera dorsalis (Diptera: Tephritidae). Evol Appl 11:1990–2003
Qin Y, Wang C, Zhao Z, Pan X, Li Z (2019) Climate change impacts on the global potential geographical distribution of the agricultural invasive pest, Bactrocera dorsalis (Hendel) (Diptera: Tephritidae). Clim Change 155:145–156
Ray RP, Nakata T, Henningsson P, Bomphrey RJ (2016) Enhanced flight performance by genetic manipulation of wing shape in Drosophila. Nat Commun 7:10851
Rodriguez DD, Felix JT, Diaz-Benjumea FJ (2004) The role of the T-box gene optomotor-blind in patterning the Drosophila wing. Dev Biol 268:481–492
Schliekelman P, Ellner S, Gould F (2005) Pest control by genetic manipulation of sex ratio. J Econ Entomol 98:18–34
Shen Z (2009) Insect ecology and ecological principles of IPM. China Agricultural University Press, Beijing
Shen D (2014) Systemically interfering with immune response by a fluorescent cationic dendrimer delivered gene suppression. J Mater Chem B 2:4653–4659
Shen J, Dahmann C, Pflugfelder GO (2010) Spatial discontinuity of Optomotor-blind expression in the Drosophila wing imaginal disc disrupts epithelial architecture and promotes cell sorting. BMC Dev Biol 10:23
Shi Y et al (2019) The ecdysis triggering hormone system, via ETH/ETHR-B, is essential for successful reproduction of a major pest insect, Bactrocera dorsalis (Hendel). Front Physiol 10:151
Stephens AEA, Kriticos DJ, Leriche A (2007) The current and future potential geographical distribution of the oriental fruit fly, Bactrocera dorsalis (Diptera: Tephritidae). Bull Entomol Res 97:369–378
Tomoyasu Y, Wheeler SR, Denell RE (2005) Ultrabithorax is required for membranous wing identity in the beetle Tribolium castaneum. Nature 433:643–647
Wan X, Nardi F, Zhang B, Liu Y (2011) The oriental fruit fly, Bactrocera dorsalis, in China: origin and gradual inland range expansion associated with population growth. PLoS ONE 6:e2523810
Weatherbee SD, Halder G, Kim J, Hudson A, Carroll S (1998) Ultrabithorax regulates genes at several levels of the wing-patterning hierarchy to shape the development of the Drosophila haltere. Genes Dev 12:1474–1482
White IM, Elson-Harris MM (1992) Fruit flies of economic significance: their identification and bionomics. Environ Entomol 22:1408
Wimmer EA (2005) Eco-friendly insect management. Nat Biotechnol 23:432–433
Yuan S, Kong Q, Xiao C, Yang S, Sun W, Zhang J, Li Z (2006) Introduction to two kinds of artificial diets for mass rearing of adult Bactrocera dorsalis (Hendel). J Huazhong Agric Univ 25:371–374
Zecca M, Struhl G (2007) Recruitment of cells into the Drosophila wing primordium by a feed-forward circuit of vestigial autoregulation. Development 134:3001–3010
Zeng Y et al (2019) Global distribution and invasion pattern of oriental fruit fly, Bactrocera dorsalis (Diptera: Tephritidae). J Appl Entomol 143:165–176
Zhang X, Luo D, Pflugfelder GO, Shen J (2013) Dpp signaling inhibits proliferation in the Drosophila wing by Omb-dependent regional control of bantam. Development 140:2917–2922
Zhang J et al (2019) Identification of COP9 signalosome subunit genes in Bactrocera dorsalis and functional analysis of csn3 in female fecundity. Front Physiol 10:162
Acknowledgements
The authors would like to thank Professor Yin Meizhen from Beijing University of Chemical Technology for providing the nanocarriers. This research was financially supported by Natural Science Foundation of Beijing Municipality [6172021, 6182020] and the National Natural Science Foundation of China [31872295].
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Communicated by G. Smagghe.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Guo, S., Guo, X., Zheng, L. et al. A potential genetic control by suppression of the wing developmental gene wingless in a global invasive pest Bactrocera dorsalis. J Pest Sci 94, 517–529 (2021). https://doi.org/10.1007/s10340-020-01263-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10340-020-01263-1