Abstract
Avian haemosporidians are vector-transmitted blood parasites that are distributed worldwide, abundant in many bird families and well studied across Europe and North America. Since avian haemosporidians have been poorly examined in the Palearctic migratory flyways of the Western Balkans, the goal of this study was to investigate which species of three haemosporidian genera, Plasmodium, Haemoproteus and Leucocytozoon, infect both resident and migratory passerines in Serbia. The prevalence, distribution and parasitaemia of avian haemosporidian infections were screened using both a nested PCR method and microscopy. Out of 202 birds sampled at seven localities, 66 were infected with haemosporidians, and the total prevalence was 32.7%. The great majority of infected birds (29 individuals) had moderate levels of parasitaemia. The most abundant haemosporidian genus was Haemoproteus at 26.1% prevalence. All infected birds were adults; none of the tested juveniles were infected. Mixed infection was only recorded in one bird. We identified 31 genetic lineages of haemosporidians. Two new cytochrome b lineages, of Plasmodium and Leucocytozoon, were identified and found in the hosts Common Chaffinch (Fringilla coelebs) and Golden Oriole (Oriolus oriolus), respectively. We identified three new host records for previously known lineages. The lineage GRW06 (Plasmodium elongatum) occurred in Common Chaffinch, while the lineages PARUS20 and PARUS25 (Leucocytozoon sp.) were recorded in Willow Tit (Poecile montanus) and Crested Tit (Lophophanes cristatus), respectively. We found statistically significant differences in the prevalence of three haemosporidian genera among residents and partial migrants. The difference in mean parasitaemia was significant only between residents and partial migrants.
Zusammenfassung
Diversität der Blutparasiten bei wildlebenden Sperlingsvögeln in Serbien unter besonderer Beachtung zweier neuer Abstammungslinien
Vogel-Hämosporidien, weltweit verbreitete Blutparasiten, die durch Vektoren übertragen werden, kommen bei vielen Vogelfamilien häufig vor und sind in Europa und Nordamerika gut untersucht. Da über Vogel-Hämosporidien im paläarktischen Zugkorridor auf dem westlichen Balkan jedoch nur wenig bekannt ist, war das Ziel dieser Studie, herauszufinden, welche Arten der drei Hämosporidien-Gattungen Plasmodium, Haemoproteus und Leucocytozoon nicht-ziehende und ziehende Sperlingsvögel in Serbien infizieren. Die Prävalenz, die Verbreitung und die Parasitämie von Vogel-Hämosporidien-Infektionen wurden mit Hilfe von verschachtelter PCR und Mikroskopie ermittelt. Von 202 Vögeln, die an sieben Standorten beprobt wurden, waren 66 mit Hämosporidien infiziert. Die Gesamtprävalenz betrug 32,7%. Der Großteil der infizierten Vögel (29 Individuen) wies moderate Parasitämiespiegel auf. Die häufigste Hämosporidiengattung war Haemoproteus mit einer Prävalenz von 26,1%. Alle infizierten Tiere waren Altvögel, wohingegen keine der untersuchten Jungvögel infiziert waren. Eine gemischte Infektion wurde lediglich bei einem Vogel festgestellt. Wir haben 31 genetische Abstammungslinien von Hämosporidien-Parasiten identifiziert. Zwei neue Cytochrom b-Abstammungslinien von Plasmodium und Leucocytozoon wurden identifiziert und beim Buchfinken (Fringilla coelebs) und Pirolen (Oriolus oriolus) nachgewiesen. Zudem haben wir drei neue Wirte für zuvor bekannte Abstammungslinien identifiziert. Die Linie GRW06 (Plasmodium elongatum) kam bei Buchfinken vor, während die Linien PARUS20 und PARUS25 (Leucocytozoon sp.) bei Weidenmeisen (Poecile montanus) bzw. Haubenmeisen (Lophophanes cristatus) nachgewiesen wurden. Wir haben statistisch signifikante Unterschiede in der Prävalenz der drei Hämosporidiengattungen bei Standvögeln und Teilziehern gefunden. Der Unterschied in der mittleren Parasitämie war nur zwischen Standvögeln und Teilziehern signifikant.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Haemosporidians (phylum Apicomplexa) are distributed worldwide, from tropical and temperate to subpolar climates, with the exception of Antarctica where cold weather prevents the development of insect vectors for these parasites (Bennett et al. 1993). Avian haemosporidians have been well studied across parts of Europe (Western and Northern Europe and Eastern Balkans) (Bensch et al. 2000; Waldenström et al. 2002; Bensch and Åkesson 2003; Scheuerlein and Ricklefs 2004; Ventim et al. 2012) and North America (Ricklefs et al. 2005; Fallon et al. 2006), but the distribution of avian haemosporidians in wild birds of the Balkan Peninsula has been examined only in Bulgaria (Valkiūnas et al. 1999; Shurulinkov and Golemansky 2003; Zehtindjiev et al. 2009; Dimitrov et al. 2010). The prevalence of haemosporidians and parasitaemia in birds used to be studied by light microscopy. Using this methodology, the prevalence of the genera Haemoproteus and Plasmodium in Bulgaria was estimated to be 6.3% and 9.5%, respectively (Valkiūnas et al. 1999; Shurulinkov and Golemansky 2003) and 1.3% for the genus Leucocytozoon (Shurulinkov and Golemansky 2003). However, the recent development of efficient protocols for the amplification of a specific part of the mitochondrial cytochrome b (Cytb) gene (Bensch et al. 2000; Hellgren et al. 2004) has allowed scientists to conduct more detailed studies on the diversity, distribution and evolution of avian haemosporidians (Fallon et al. 2003; Bensch et al. 2007; Zehtindjiev et al. 2009). Using a nested PCR protocol, Dimitrov et al. (2010) showed that, of all examined birds in Bulgaria, 43% were positive for Plasmodium and 48% for Haemoproteus.
Since there is no information about the distribution of avian haemosporidians in Serbia, our aim was to obtain primary information on the occurrence of blood parasites in wild birds there. As part of the African-Eurasian Flyway, the Western Balkans and Serbia are important for breeding populations of several species of long-distance migrants that winter in tropical Africa. In this study, our objective was to investigate the distribution, prevalence and parasitaemia of the genera Haemoproteus, Plasmodium and Leucocytozoon among different migratory and resident passerines in Serbia. We consider that the results obtained are valuable for planning further experimental research on avian malaria.
Methods
Using both a nested PCR method and light microscopy, we screened a total of 202 individuals of 43 species of 21 families of passerines for the presence of Haemoproteus spp., Plasmodium spp. and Leucocytozoon spp. Birds were caught with mist nets during the breeding season (April–July). All birds were identified, ringed, aged as juveniles or adults and measured using a standard protocol (Svensson 1992). Sampled birds were divided into three groups based on their migratory status: residents (remaining in their territory all year round), migrants (annually migrating) and partial migrants (some individuals are migratory, some are not) (Berthold 1996). Bird nomenclature follows del Hoyo and Collar (2014). A small amount of blood (approximately 20 μl) was collected from each bird by puncturing the brachial vein, and a drop of this used for the preparation of blood smears (one smear per bird). The remaining blood was saved for the DNA analysis. We obtained blood smears for 199 individuals (one smear per bird) and 134 blood samples (one blood sample per bird). Both blood smears and blood samples were obtained for 134 birds.
Data were collected from seven localities in Serbia, of which four are wetlands [Ludaš Lake (46°06′N 19°49′E), Ponjavica (44°44′N 20°45′E), Mala Vrbica fishpond (44°36′N 22°40′E) and Gruža reservoir (43°57′N 20°41′E)]; one is Pannonic semidesert steppe [Deliblatska Sands (44°51′N 21°06′E)]; and two are mountains [Mt Tara (43°54′N 19°30′E) and Mt Rtanj (43°48′N 21°50′E; foothills)]. Samples were collected in 2007 only at Gruža reservoir, and from 2011 to 2016 from the other localities. The majority of blood samples (48.5%) were collected from birds at Mt Tara.
The blood slides were air dried, fixed in 96% ethanol for 3 min in the field and stained with Giemsa in the laboratory. Blood films were examined with a Leica DMLS light microscope for about 10 min at low magnification (× 400), and then at least 100 fields were studied at high magnification (× 1000), as described by Valkiūnas et al. (2008a). The intensity of infection was estimated as the percentage of parasite counts per 1000 or 10,000 erythrocytes, as recommended by Godfrey et al. (1987).
The DNA was extracted by a standard ammonium acetate method (Richardson et al. 2001) and quantified by NanoDrop (Implen Nano photometer P330). The DNA was diluted to a standard concentration of approximately 25 ng/μl and used as a template for amplification of a 479-base pair fragment of the mitochondrial Cytb of parasites by nested polymerase chain reaction (PCR) assay (Hellgren et al. 2004). The initial PCR was carried out with the primers HaemNFI/HaemNR3, which amplified all three genera of the haemosporidians (Plasmodium, Haemoproteus and Leucocytozoon). The PCR reaction was performed in 25 μl total volume containing, per reaction, 1.5 μl MgCl2 (25 mM; Applied Biosystems), 2.5 μl GeneAmp 10X PCR Buffer II (Applied Biosystems), 2.5 μl dNTP (1.25 mM; Thermo Scientific), 0.1 μl AmpliTaq DNA polymerase (5 U/µl; Applied Biosystems), 1 μl of each primer (10 μM), 15.4 μl of double distilled H2O and 1 μl of diluted total genomic DNA template (25 ng/μl). One negative and two positive controls were used in every PCR run of 16 samples. Nested PCR was carried out with the primers HaemF/HaemR2 for Haemoproteus and Plasmodium, and HaemFL/HaemR2L only for Leucocytozoon. A nested PCR reaction was also performed in 25 μl total volume containing 2 μl of the first PCR product as the template and the same reagents. The presence of haemosporidian infection in the samples was evaluated by running 2.5 μl of the final PCR products on 2% agarose gel. All positive samples were sequenced by Sanger sequencing; reaction was with the BigDye Terminator version 1.1 Cycle Sequencing Kit (product no. 4336776; Applied Biosystems). Samples were loaded onto an ABI PRISM 3100 capillary sequencing robot (Applied Biosystems). The sequences were edited in BioEdit (Hall 1999) and aligned using ClustalW algorithm in MEGA version 6 (Tamura et al. 2013). After aligning, the sequences were identified by using the BLAST algorithm in the MalAvi database (Bensch et al. 2009) and GenBank sequences (Benson et al. 2014). Two unique lineages were recorded for the first time and deposited in GenBank (accession nos. MF543057 and MF374497).
A χ2-test with Bonferroni correction was used for pairwise comparisons to examine the association between the birds’ migratory status and the presence of parasites. The Kruskal–Wallis test, followed by post hoc Mann–Whitney U signed-rank test with Bonferroni adjustment for pairwise comparisons, was used to assess the differences in parasitaemia according to migratory status. All statistical analyses were carried out using R version 3.3.2. (R Core Team 2016).
Results
According to both independent microscopy and PCR analysis, out of the 202 passerines of 43 species examined, 66 individuals were found to be infected with haemosporidians (Table 1). All infected individual were adults. None of the juveniles were found to be infected.
Total prevalence for the three haemosporidian genera was 32.7%. This was scored by combining positive results from independent microscopy and screening samples with both blood smears and PCR. However, the prevalence differed between the methods, and was higher when PCR was used in combination with blood smears (41.8%) than when only blood smears were used (21.6%) (Pearson χ2\(\chi_{(1)}^{2}\) = 7.8724, p < 0.01). Considering that blood smears were only sufficient for the estimation of parasite prevalence and parasitaemia, for further analyses we only used data obtained by PCR. Of the 134 analysed birds, infected ones showed 26.1% prevalence for Haemoproteus spp., 9.7% for Plasmodium spp., and 6.7% for Leucocytozoon spp.. In addition, we identified 31 genetic lineages: 15 lineages of Haemoproteus and eight lineages each of Plasmodium and Leucocytozoon.
Two unique lineages, one of Plasmodium sp. and one of Leucocytozoon sp., were recorded for the first time and deposited in GenBank. Lineage CCF25 (MF543057) of the genus Plasmodium was recorded in the resident species Common Chaffinch (Fringilla coelebs), from Deliblatska Sands. Lineage ORIORI04 (MF374497) from the genus Leucocytozoon was found in the migratory species Golden Oriole (Oriolus oriolus), from Mala Vrbica fishpond.
We identified three new host records for previously known lineages. In one case, we found a new host record for the cosmopolitan parasite lineage cyt b GRW06 (Plasmodium elongatum) that occurred in Common Chaffinch at Deliblatska Sands. Lineages PARUS20 and PARUS25 (Leucocytozoon spp.) were recorded for the first time in Willow Tit (Poecile montanus) and Crested Tit (Lophophanes cristatus), respectively, at Mt Tara. Only one naturally infected Bullfinch (Pyrrhula pyrrhula) was positive for a mixed infection with Haemoproteus fringillae and Leucocytozoon majoris. The mixed infection was recorded by using a nested PCR approach.
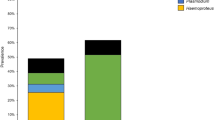
Among the birds examined, 78 were resident, 42 were migratory and 14 were partially migratory. Resident (28 individuals) and migratory birds (21 individuals) were infected with all three examined haemosporidian genera, while seven partial migrants had infections only with Haemoproteus spp. The majority of resident (15 birds) and migratory (12) hosts were infected with Haemoproteus spp., while Plasmodium spp. infection was present in seven residents and six migrants. Leucocytozoon spp. infection was established in seven residents and two migrants. Resident birds were the most infected group among the birds examined. We found a statistically significant difference between the prevalence of resident and partial migrants (\(\chi_{(1)}^{2}\) = 12.145, p = 0.00049), but there were no significant differences between the prevalence in residents and migrants (\(\chi_{(1)}^{2}\) = 1.239, p = 0.79), nor between the prevalence in migrants and partial migrants (\(\chi_{(1)}^{2}\) = 4.988, p = 0.07).
Lineages MW1 and ARW1 of Haemoproteus belopolskyi were recorded in two different host genera, while two lineages (ACAGR1 and SGS1) of the genus Plasmodium were found in more than one host family (Supplementary material, Table 2).
Most of the hosts from the different localities shared parasite lineages, but some of the parasites were recorded only at particular sites. Among 17 lineages recorded in total at Mt Tara, we found nine lineages of the genus Haemoproteus, six of Leucocytozoon and two of Plasmodium, which were specific to this area. Two different haplotypes (RBS4 and RBS2) of Haemoproteus lanii were recorded at two distant localities: Mt Tara and Mala Vrbica fishpond, respectively. Moreover, the Kruskal–Wallis χ2-test showed the absence of an overall statistical difference in the prevalence of haemosporidian genera among all examined localities (\(\chi_{(1)}^{2}\) = 6, p = 0.42). However, a Binominal test for proportions showed significant difference only between infected birds on Mts Tara and Rtanj (\(\chi_{(1)}^{2}\) = 11.829, p = 0.01).
The most infected host species was Eurasian Blackbird (Turdus merula) at 80% of the examined birds infected only with Haemoproteus spp. Common Chaffinch and Eurasian Blackcap (Sylvia atricapilla) also had very high infection levels, at 70% and 58.3% of examined individuals, respectively. Common Chaffinches were infected with Haemoproteus spp. and Plasmodium spp., while Eurasian Blackcaps were infected with Haemoproteus spp.
The level of calculated parasitaemia varied between 0.01% (low and chronic parasitaemia) and 4.8% (high parasitaemia). A low parasitaemia level, 0.01%, was found in 23 birds. The great majority of hosts (29 birds) had moderate parasitaemia levels of between 0.01 and 1%, while high parasitaemia levels above 1.1% were found in 14 birds. The highest level of parasitaemia, 4.8%, was found in one Red-backed Shrike (Lanius collurio) sampled at Mala Vrbica fishpond in 2016. Birds with chronic parasitaemia levels were infected with all three haemosporidian genera, as were hosts with moderate parasitaemia levels, while those with high parasitaemia levels were infected with only Haemoproteus spp. and one Plasmodium spp. Most resident birds had either a low or moderate level of parasitaemia, while a high level was predominantly found in partial migrants.
Mean parasitaemia levels differed significantly between the migratory groups (Supplementary material, Fig. 1). The highest mean level of parasitaemia, 1.9%, was found in partial migrants such as the European Robin (Erithacus rubecula) and Song Thrush (Turdus philomelos), while it was lower in residents and migrants, ranging from 0.3 to 0.6%. However, the Kruskal–Wallis test with a post hoc Mann–Whitney U-test and Bonferroni correction showed that a statistically significant difference in average parasitaemia existed only between residents and partial migrants (U = 269, p = 0.02); the differences were not statistically significant between residents and migrants (U = 426, p = 0.939), or between migrants and partial migrants (U = 72.5, p = 0.137).
We detected significant differences in average parasitaemia levels between the examined migratory groups. For resident birds, there was a significant difference in parasitaemia only between hosts infected with Haemoproteus and Leucocytozoon (U = 116.5, p = 0.0032), while there were no differences between birds infected with Haemoproteus and Plasmodium (U = 90.5, p = 0.168) nor between birds infected with Plasmodium and Leucocytozoon (U = 17, p = 0.262). Migrants showed a different pattern: a significant difference was observed among birds infected with Haemoproteus and Plasmodium (U = 82.5, p = 0.0034), while there were no differences among birds infected with Haemoproteus and Leucocytozoon (U = 18.5, p = 0.653) and Plasmodium and Leucocytozoon (U = 8.5, p = 0.382).
In our study, the females had higher average levels of parasitaemia (0.86%) than males (0.65%). However, there were no statistically significant differences in average parasitaemia between the sexes (U = 416.5, p = 0.0652).
Discussion
Many studies of avian haemosporidians using traditional microscopy (Valkiūnas et al. 1999; Shurulinkov and Golemansky 2003) or molecular approaches (Zehtindjiev et al. 2009; Dimitrov et al. 2010) have been published for Southeastern Europe, but not for the Western Balkans. We present the first results of a molecular and microscopic overview of avian malaria parasites in Serbia, in the Western Balkans.
Overall prevalence obtained by: (1) independently scoring blood slides (without blood samples) with results from (2) molecular methods (PCR), in combination with microscopy where both samples were available, was 32.7%. However, the prevalence obtained by PCR (combined with microscopy) was significantly higher. Our results did not agree with those of Valkiūnas et al. (2008a), who showed a similar overall prevalence of infection for PCR and microscopy, probably due to the high quality of their slides.
Several studies on blood parasites in wild passerines across Europe that amplified Cytb showed Haemoproteus prevalence of 5.1%, 48% and 17.7% and Plasmodium prevalence of 15.4%, 43% and 82.3% in Germany, Bulgaria and Spain, respectively (Wiersch et al. 2007; Dimitrov et al. 2010; Ventim et al. 2012). As Valkiūnas et al. (2008a) noted, the genus Leucocytozoon was rarely detected due to unskilled microscopy and low prevalence. However, using PCR-based methods for Leucocytozoon, studies in European passerines recorded prevalences for Leucocytozoon of 30%, 2.4% and 85.3% (Valkiūnas et al. 2008a; Rönn et al. 2015; Schmid et al. 2017). Our results are consistent with the literature (Zehtindjiev et al. 2009; Dimitrov et al. 2010), as the most common haemosporidian genus that we found was Haemoproteus.
We recorded two new lineages from the genera Plasmodium and Leucocytozoon. Lineage CCF25 of the genus Plasmodium was found in Common Chaffinch and lineage ORIORI04 of the genus Leucocytozoon was isolated from Golden Oriole. To date, only Haemoproteus lineages (ORIORI01, ORIORI02 and ORIORI03) (Dimitrov et al. 2010) and Plasmodium rouxi (Valkiūnas 2005) have been isolated from Golden Oriole, but no Leucocytozoon.
The parasite lineage GRW06, of the morphological species Plasmodium elongatum, has so far been recorded in 57 bird hosts (MalAvi database), including Great Reed Warbler (Acrocephalus arundinaceus) and House Sparrow (Passer domesticus) from Bulgaria (Valkiūnas et al. 2008b). For the first time, we isolated the lineage GRW6 from Common Chaffinch at Deliblatska Sands in Serbia.
According to the occurrence of separate bands of both infections in the sequence electropherogram, only one Bullfinch had a mixed infection with Haemoproteus fringillae and Leucocytozoon majoris. The detection efficiency might vary due to low-quality sequences due to the combination of parasite lineages or the intensity of infection (Pérez-Tris and Bensch 2005; Zehtindjiev et al. 2012). We could not identify with certainty other mixed infections in our samples, probably due to weak peaks of the lineage where parasitaemia was low, as explained by Pérez-Tris and Bensch (2005).
In an examination of 460 wild passerines (both migrants and residents) in Bulgaria, Dimitrov et al. (2010) found 267 birds infected with haemosporidians and identified 52 lineages of these. Of these 52 lineages, 38 belonged to the genus Haemoproteus and 14 to the genus Plasmodium. Hellgren et al. (2009) obtained similar results in Western Europe, identifying 63 lineages of the genus Haemoproteus and 35 of Plasmodium. In our study of 202 wild birds, we found 66 individuals that tested positive for haemosporidians and we identified 31 genetic lineages, 15 of which belonged to the genus Haemoproteus and eight each to Plasmodium and Leucocytozoon, i.e. the most common haemosporidians in wild passerines in Serbia belong to the genus Haemoproteus.
Latta and Ricklefs (2010) showed that haemosporidian prevalence can differ significantly depending on the migratory status of birds. The most frequently infected bird species in their study were residents, mostly infected by Haemoproteus spp., while the endemic residents had the highest rates of infection; the parasite assemblage was dominated by Plasmodium lineages. In our study, among 78 residents, 42 migrants and seven partial migrants, residents were the most infected group of birds, with all three haemosporidian genera present in 28 individuals. Migrants (21 infected birds) were also infected by all three haemosporidian genera, whereas partial migrants (seven birds) were infected only with the genus Haemoproteus. Unlike Ventim et al. (2012), who only found the genus Haemoproteus in migratory birds, we found that residents were mostly infected by species of this genus. Moreover, infection with the genus Plasmodium was established both in resident birds (young and adults) and young migrants (Ventim et al. 2012). We found Plasmodium spp. and Leucocytozoon spp. infections in both residents and migrants. None of the juvenile birds in our sample were infected, which lead us to conclude that adults might obtain the haemosporidian infection in the non-breeding part of their range or that young birds were insufficiently exposed to vectors to become infected at the nest sites. However, we did not investigate the diversity of vectors at the breeding sites, which is the next step necessary to obtain sufficient information for a better understanding of haemosporidian infection in Serbian birds.
As shown by Waldenström et al. (2002), Haemoproteus spp., which infect fewer host species, are considered host specialists, while Plasmodium spp. are predominantly host generalists. Accordingly, we detected two Haemoproteus and one Plasmodium lineage infecting different host species and genera. Using the same bird nomenclature as Waldenström et al. (2002), we observed the same pattern; host species infected with Plasmodium lineages were from different families, whereas host species infected with Haemoproteus lineages were from the same family.
According to Valkiūnas (2005), who followed the bird nomenclature after Sibley and Monroe (1990), the most infected bird family was Sylviidae. Applying the same systematics to this study, we observed the same pattern as Valkiūnas (2005) and Palinauskas et al. (2005): birds from the family Sylviidae were infected with all three examined haemosporidian genera, while species from the family Turdidae were infected only with Haemoproteus spp. This difference is probably due to the ecological demands of the family Turdidae during the breeding season; species from this family place their nests on the ground or 1.5–2 m above the ground (Cramp 1988) where there is higher activity of biting midges, which are potential vectors of Haemoproteus spp. (Diarra et al. 2014).
The most infected host species was Eurasian Blackbird, of which 80% of birds were infected; this in accordance with studies conducted in the UK and the Azores (Hatchwell et al. 2000; Hellgren et al. 2011), where 80% and 57% of the examined Eurasian Blackbirds were infected, respectively. Moreover, Bentz et al. (2006) found only one Haemoproteus lineage and two Plasmodium lineages in Eurasian Blackbird, while Hatchwell et al. (2000) found all three haemosporidian genera. Unlike these studies, we found only one lineage from the genus Haemoproteus (TURDUS2); this was most likely due to the lack of suitable insect vectors for other haemosporidian genera at the sampling habitats and other locations in Serbia. Eurasian Blackcaps were also highly infected, with a prevalence of 58.3%, but only with Haemoproteus spp., which is in line with the results of Arizaga et al. (2010) in Spain, where 34.1% of Eurasian Blackcaps were infected with both Haemoproteus and Plasmodium genera. Similarly to Eurasian Blackbird, the absence of other haemosporidian genera in Eurasian Blackcaps could be due to the lack of insect vectors for the Plasmodium and Leucocytozoon lineages at the sampling sites in Serbia.
Valkiūnas et al. (2008a) found that more than 80% of infected birds examined from Europe, North America and Africa had parasitaemia of less than 0.01%, which is considered to indicate chronic infection. However, in our study, a large number of infected birds (14.3%) had a moderate infection (mean 0.4%, σ = 32.79%). Since birds with a moderate infection were residents, one could assume that the breeding grounds in Serbia had suitable vectors of haemosporidia, and that birds were exposed to these vectors for a sufficient period of time to become infected.
Contrary to the theory that female birds are usually less infected than males due to their better immunity (Zuk 1990), many studies have reported that females had higher levels of parasitaemia than males (Hatchwell et al. 2000; Bentz et al. 2006; Asghar et al. 2011; Sorensen et al. 2016). In our study, although females had higher medium parasitaemia levels than males, we did not find any statistically significant difference between them.
This paper presents the first results on the distribution of haemosporidian genera in wild birds in Serbia. We found that resident birds, in contrast with migratory and partially migratory birds, were the most infected group; however, partial migrants had the highest levels of parasitaemia. Haemoproteus appears to be the most common haemosporidian genus in wild passerines in Serbia. Future research on avian parasites and their vectors in Serbia will give better insights into their relationships and distribution there.
References
Arizaga J, Esparza X, Barba E (2010) Haemosporidians in migratory Blackcaps (Sylvia atricapilla): a comparison between autumn and spring periods of passage. An de Biol 32:87–93
Asghar M, Hasselquist D, Bensch S (2011) Are chronic avian haemosporidian infections costly in wild birds? J Avian Biol 42:530–537
Bennett GF, Peirce MA, Ashford RW (1993) Avian Haematozoa: mortality and pathogenicity. J Nat Hist 27(5):993–1001 (Taylor & Francis Online)
Bensch S, Åkesson S (2003) Temporal and spatial variation of hematozoans in Scandinavian Willow Warblers. J Parasitol 89:388–391
Bensch S, Stjernman M, Hasselquist D, Östman Ö, Hansson B, Westerdahl H, Pinheiro RT (2000) Host specificity in avian blood parasites: a study of Plasmodium and Haemoproteus mitochondrial DNA amplified from birds. Proc R Soc Lond B 267:1583–1589
Bensch S, Waldenström Ј, Jonzén N, Westerdahl H, Hansson B, Sejberg D, Hasselquist D (2007) Temporal dynamic and diversity of avian malaria parasites in a single host species. J Anim Ecol 76:112–122
Bensch S, Hellgren O, Pérez-Tris J (2009) MalAvi: a public database of malaria parasites and related haemosporidians in avian hosts based on mitochondrial cytochrome b lineages. Mol Ecol Resour 9:1353–1358
Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW (2014) GenBank. Nucleic Acids Res 41:36–42
Bentz S, Rigaud T, Barroca M, Martin-Laurent F, Bru D, Moreau J, Faivre B (2006) Sensitive measure of prevalence and parasitaemia of haemosporidia from European Blackbird (Turdus merula) populations: value of PCR-RFLP and quantitative PCR. Parasitology 133:685–692
Berthold P (1996) Control of bird migration. Chapman and Hall, London, pp 25–26
Cramp S (ed) (1988) The birds of the Western Palearctic, vol V. Oxford University Press
del Hoyo J, Collar NJ (2014) HBW and BirdLife International illustrated checklist of the birds of the world: passerines, vol 2. Lynx, Barcelona
Diarra M, Fall M, Fall GA, Diop A, Seck MT, Garros C, Balenghien T, Allène X, Rakotoarivony I, Lancelot R, Mall I, Bakhoum M, Dosum AM, Ndao M, Bouyer J, Guis H (2014) Seasonal dynamics of Culicoides (Diptera: Ceratopogonidae) biting midges, potential vectors of African horse sickness and bluetongue viruses in the Niayes area of Senegal. Parasites Vectors 7:147
Dimitrov D, Zehtindjiev P, Bensch S (2010) Genetic diversity of avian blood parasites in SE Europe: cytochrome b lineages of the genera Plasmodium and Haemoproteus (Haemosporida) from Bulgaria. Acta Parasitol 55:201–209
Fallon SM, Ricklefs RE, Swanson BL, Bermingham E (2003) Detecting avian malaria: an improved polymerase chain reaction diagnostic. J Parasitol 89(5):1044–1047
Fallon SM, Fleischer RC, Graves GR (2006) Malarial parasites as geographical markers in migratory birds? Biol Let 2:213–216
Godfrey RD, Fedynich AM, Pence DB (1987) Quantification of hematozoa in blood smears. J Wildl Dis 23:558–565
Hall TA (1999) BIOEDIT: a user friendly biological sequence alignment editor and analysis program for Windows 95/98 NT. Nucleic Acid Symp Ser 41:95–98
Hatchwell BJ, Wood MJ, Anwar M, Perrins CM (2000) The prevalence and ecology of the haematozoan parasites of European Blackbirds, Turdus merula. Can J Zool 78(4):684–687
Hellgren O, Waldenström J, Bensch S (2004) A new PCR assay for simultaneous studies of Leucocytozoon, Plasmodium, and Haemoproteus from avian blood. J Parasitol 90(4):797–802
Hellgren O, Pérez-Tris J, Bensch S (2009) A jack-of-all-trades and still a master of some: prevalence and host range in avian malaria and related blood parasites. Ecology 90(10):2840–2849
Hellgren O, Križanauskiené A, Hasselquist D, Bensch S (2011) Low haemosporidian diversity and one key-host species in a bird malaria community on a mid-Atlantic island (São Miguel, Azores). J Wildl Dis 47(4):849–859
Latta SC, Ricklefs RE (2010) Prevalence patterns of avian haemosporida on Hispaniola. J Avian Biol 41:25–33
MalAvi. A database for avian haemosporidian parasites. http://www.mbio-serv.mbioekol.lu.se/Malavi. Accessed 23 Dec 2017
Palinauskas V, Markovets MY, Kosarev VV, Efremov VD, Sokolov LV, Valkiûnas G (2005) Occurrence of avian haematozoa in Ekaterinburg and Irkutsk districts of Russia. Ekologija (Lithuania) 4:8–12
Pérez-Tris J, Bensch S (2005) Diagnosing genetically diverse avian malarial infections using mixed-sequence analysis and TA-cloning. Parasitology 131(1):15–23
R Core Team (2016) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. http://www.R-project.org. Accessed 14 Oct 2017
Richardson DS, Jury FL, Blaakmeer J, Komdeur J, Burke T (2001) Parentage assignment and extra—group paternity in a cooperative breeder: the Seychelles Warbler (Acrocephalus sechellensis). Mol Ecol 10:2263–2273
Ricklefs RE, Swanson BL, Fallon SM, Martínez-Abraín L, Cheuerlein A, Gray J, Latta SC (2005) Community relationships of avian malaria parasites in southern Missouri. Ecol Monogr 75(4):543–559
Rönn JAC, Harrod C, Bensch S, Wolf JBW (2015) Transcontinental migratory connectivity predicts parasite prevalence in breeding populations of the European Barn Swallow. J Evol Biol 28:535–546
Scheuerlein A, Ricklefs R (2004) Prevalence of blood parasites in European passeriform birds. Proc R Soc Lond B 271:1363–1370
Schmid S, Fachet K, Dinkel A, Mackenstedt U, Woog F (2017) Carrion Crows (Corvus corone) of southwest Germany: important hosts for haemosporidian parasites. Malar J 16(1):369
Shurulinkov P, Golemansky V (2003) Plasmodium and Leucocytozoon (Sporozoa: Haemosporida) of wild birds in Bulgaria. Acta Protozool 42(3):205–214
Sibley CG, Monroe BL (1990) Distribution and taxonomy of birds of the world. Yale University Press, New Haven, London
Sorensen MC, Asghar M, Bensch S, Fairhurst GD, Jenni-Eiermann S, Spottiswoode CN (2016) A rare study from the wintering ground provides insight into the cost of malaria infection for migratory birds. J Avian Biol 47:575–582
Svensson L (1992) Identification guide to European passerines, 4th edn. British Trust for Ornithology, Stockholm
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Valkiūnas G (2005) Avian malaria parasites and other haemosporidia. CRC, Boca Raton
Valkiūnas G, Iezhova T, Golemansky G, Pilarska D, Zehtindjiev P (1999) Blood protozoan parasites (Protozoa: Kinetoplastida and Haemosporida) in wild birds from Bulgaria. Acta Zool Bulg 51:127–129
Valkiūnas G, Iezhova TA, Križanauskienė A, Palinauskas V, Sehgal RNM (2008a) A comparative analysis of microscopy and PCR-based detection methods for blood parasites. J Parasitol 94(6):1395–1401
Valkiūnas G, Zehtindjiev P, Dimitrov D, Križanauskienė A, Iezhova TA, Bensch S (2008b) Polymerase chain reaction-based identification of Plasmodium (Huffia) elongatum, with remarks on species identity of haemosporidian lineages deposited in GenBank. Parasitol Res 102:1185–1193
Ventim R, Tenreiro P, Grade N, Encarnação P, Araújo M, Mendes L, Pérez-Tris J, Ramos JA (2012) Characterization of haemosporidian infections in warblers and sparrows at south-western European reed beds. J Ornithol 153:505–512
Waldenström J, Bensch S, Kiboi S, Hasselquist D, Ottosson U (2002) Cross-species infection of blood parasites between resident and migratory songbirds in Africa. Mol Ecol 11:1545–1554
Wiersch SC, Lubjuhn T, Maier WA, Kampen H (2007) Haemosporidian infection in passerine birds from Lower Saxony. J Ornithol 148:17–24
Zehtindjiev P, Ilieva M, Križanauskienė A, Oparina O, Oparin M, Bensch S (2009) Occurrence of haemosporidian parasites in the Paddyfield Warbler, Acrocephalus agricola (Passeriformes, Sylviidae). Acta Parasitol 54(4):295–300
Zehtindjiev P, Križanauskienė A, Bensch S, Palinauskas V, Asghar M, Dimitrov D, Scebba S, Valkiūnas G (2012) A new morphologically distinct avian malaria parasite that fails detection by established polymerase chain reaction-based protocols for amplification of the cytochrome b gene. J Parasitol 98(3):657–665
Zuk M (1990) Reproductive strategies and diseases susceptibility, an evolutionary viewpoint. Parasitol Today 6(7):231–233
Acknowledgements
The laboratory work was partially supported by the Natural History Museum in Belgrade, Serbia and the Department of Biology, Lund University, Sweden. Aleksandra Urošević is acknowledged for her help in the laboratory at the Institute for Medical Research, University of Belgrade, Serbia. We are thankful to Dr Staffan Bensch and two anonymous reviewers for valuable comments on earlier versions of the manuscript. We thank Dr Bojana Stanić for proofreading the manuscript and Nicola Crockford for correcting the English. The sampling in this study complies with the current legislation of the Ministry of Environmental Protection of the Republic of Serbia.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by K. C. Klasing.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Stanković, D., Jönsson, J. & Raković, M. Diversity of avian blood parasites in wild passerines in Serbia with special reference to two new lineages. J Ornithol 160, 545–555 (2019). https://doi.org/10.1007/s10336-019-01628-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10336-019-01628-z