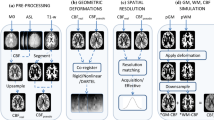
Abstract
Objective
In this perfusion magnetic resonance imaging study, the performances of different pseudo-continuous arterial spin labeling (PCASL) sequences were compared: two-dimensional (2D) single-shot readout with simultaneous multislice (SMS), 2D single-shot echo-planar imaging (EPI) and multishot three-dimensional (3D) gradient and spin echo (GRASE) sequences combined with a background-suppression (BS) module.
Materials and methods
Whole-brain PCASL images were acquired from seven healthy volunteers. The performance of each protocol was evaluated by extracting regional cerebral blood flow (rCBF) measures using an inline morphometric segmentation prototype. Image data postprocessing and subsequent statistical analyses enabled comparisons at the regional and sub-regional levels.
Results
The main findings were as follows: (i) Mean global CBF obtained across methods was were highly correlated, and these correlations were significantly higher among the same readout sequences. (ii) Temporal signal-to-noise ratio and gray-matter-to-white-matter CBF ratio were found to be equivalent for all 2D variants but lower than those of 3D-GRASE.
Discussion
Our study demonstrates that the accelerated SMS readout can provide increased acquisition efficiency and/or a higher temporal resolution than conventional 2D and 3D readout sequences. Among all of the methods, 3D-GRASE showed the lowest variability in CBF measurements and thus highest robustness against noise.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Stroke is a major global public health issue and the third leading cause of death in industrialized countries; early detection and treatment play important roles in limiting brain damage and improving patient outcomes. Thus, there is a need to improve both the timely identification of stroke patients eligible for accessible therapies and the evaluation of the benefit-risk ratio for patients. Longitudinal studies on cerebral blood flow (CBF) evaluation after stroke initially exhibited a reduced CBF response followed by an increased CBF after a month, which then progressively approximates the normal CBF response pattern over the next 3 months [1, 2]. In this clinical context, noninvasive, noninjected, fast and robust perfusion measurement techniques are essential to efficiently and sequentially monitor blood supply perturbations longitudinally in the brain and to broaden our understanding of how they relate to the resulting neurological deficits. The need for and the impact of a quick medical response in stroke cases have clearly been restated in a comment by von Kummer and colleagues [3].
Arterial spin-labeled (ASL) perfusion magnetic resonance imaging (MRI) uses magnetically labeled arterial blood water as an endogenous tracer to obtain quantitative regional Cerebral Blood Flow (rCBF) measures [4, 5]. ASL is a repeatable and completely noninvasive technique that has a wide range of clinical applications and advantages over exogenous contrast-based methods. This technique is particularly useful in pediatric and renal failure patients, in whom the use of external tracer is restricted [6, 7]. ASL preparations can be combined with both two-dimensional (2D) and three-dimensional (3D) readout approaches.
Currently, the main challenges of ASL techniques that limit its widespread use in routine evaluations of brain tissue viability are as follows: (1) long acquisition times due to the intrinsically low signal-to-noise ratio (SNR), which requires the accumulation of a sufficient number of control-label image pairs to obtain perfusion maps of diagnostic quality; (2) a decreased SNR for high-resolution perfusion images when 2D ASL images are acquired with increased in-plane and through-plane resolution (i.e., those with a greater number of slices covering the same brain volume); and (3) increased susceptibility to patient motion-induced artifacts due to the long scan time. These limitations are addressed and mitigated by continued innovations in the image 2D and 3D readout [4, 8,9,10,11] and labeling [7, 8, 12, 13] and background suppression [14,15,16,17,18]components of ASL sequences, which have recently enabled the technique to be employed in clinical routines.
Simultaneous multi-slice acceleration (SMS) uses multiband radiofrequency (RF) pulses to simultaneously excite multiple spatially distributed slices and to read out these images in a single echo train. The superimposed signals that are acquired from multiple slices are further unwrapped by unaliased image reconstruction [19]. This technique was further developed to reduce the acquisition time in diffusion or functional studies based on 2D single-shot EPI readouts [20, 21]. The simultaneous acquisition of multiple slices enables a coupled reduction of the total acquisition time and accelerated through-plane coverage, if desired, or is improved through-plane resolution without a time penalty for whole-brain applications (although the SNR/contrast-to-noise ratio (CNR) reduction in control/label pairs will be proportional to the slice thickness reduction). SMS imaging appears to be an attractive solution for whole-brain ASL to increase the spatial resolution in the slice direction using a 2D EPI readout. Indeed, SMS has recently been implemented to improve the coverage of data acquisition within a given repetition time in pulsed ASL (PASL). 2D SMS-PASL was compared to the standard single-band (SB) 2D EPI reference technique [22] and to 3D GRASE [23] for brain perfusion imaging using the flow-sensitive alternating inversion recovery (FAIR) [24] labeling technique with a standard low image resolution. These studies found that perfusion signal differences between the SB and SMS readouts were minimal. More recently, Li et al. [25] also investigated PCASL combined with an SMS factor of six for high-resolution whole-brain applications. In this study, by combining simulations and experimental validation, the authors demonstrated that the amplified noise (which was given by the coil geometry factor, i.e., the g-factor) and slice leakage (i.e., slice-to-slice contamination which was expected to be more pronounced from the high through-plane acceleration factor used) had a minimal impact on CBF quantification. They demonstrated that 2D EPI with SMS greatly benefited to high-resolution whole-brain PCASL studies in terms of improved spatial SNR and temporal SNR (tSNR) efficiencies. It also showed an increased compliance with the assumptions of the commonly used single-blood compartment model, which resulted in improved CBF estimates.
While constituting important initial findings, none of these studies evaluated the impact of accelerated techniques using the statistical analysis that is generally used in clinical practice to address clinical issues and, specifically, using PCASL preparation and comparing the results to those of 3D GRASE with the same preparation. It has been shown in the past that slightly different combinations of ASL parameters can ultimately produce regional differences between ASL sequences that are acquired on the same scanner and from the same subjects [26]. These results highlight the need for preclinical studies to verify and validate a chosen technology and protocol before large-scale clinical trials are designed.
The goal of this study was thus to evaluate the performance of a 2D SMS single-shot PCASL sequence by applying (i) 2D EPI without any slice acceleration, (ii) 2D EPI with three different levels of increasing SMS acceleration factors and correspondingly increasing through-plane resolution, and (iii) 3D GRASE with BS in healthy volunteers prior to decision making for a larger clinical trial. To this end, the analysis pipeline that would be used to statistically produce and mine CBF data in clinical or preclinical studies was tested for all PCASL variants.
Materials and methods
This study was conducted in seven healthy human subjects (mean age, 38 ± 7 years; one female). We obtained the approval from local ethical committee (French IRB, CPP Lyon Sud-Est II, IRB No: 2015-A01802-47) and performed in compliance with the Code of Ethics of the World Medical Association (Declaration of Helsinki) and the standards that were established by the Institutional Review Board and National Institutes of Health. All subjects provided informed written consent for this study. MRI was performed using a 3-Tesla MAGNETOM Prisma MRI scanner (Siemens Healthcare, Erlangen, Germany) equipped with a 64-channel head/neck coil. The body coil was used for RF transmission, and the 64-channel phased-array coil was used for signal reception. The total acquisition time for the protocol was ~ 40 min. Individual sequence durations and the main MRI parameters of interest are listed in Table 1. All ASL methods were performed during the same scanning session.
The multiband technique allows the acquisition of more slices within a given volume coverage at higher SMS factors, enabling each slice to be thinner than the slices acquired using the SB technique. This feature was utilized to increase the number of slices per volume when increasing the acceleration factor and reducing the slice thicknesses (4-6 mm) and partial volume in the slice direction. A prototype 2D EPI sequence was used with SMS factors of 1, 3, 4 and 6, leading to the optimized number of slices and hence the achievable slice thickness and brain coverage, as shown in Fig. 1. Detailed MRI parameters are listed in Table 1.
Summary of study design illustrating brain coverage and acquisition strategies using optimized protocols exploiting different simultaneous-multi slice acceleration factors (SMS). Number of colors in brain coverage indicates the SMS acceleration factor used and lines represents total number of slices acquired in each case: for every excitation one slice in each color group is acquired simultaneously in an ascending order
To achieve a fair comparison, the prototype 3D GRASE sequence was optimized to achieve a spatial resolution comparable to that of 2D EPI with the maximum acceleration factor used in this experiment (Table 1) while to being in line with the recommended implementation of 3D PCASL [4]. The labeling parameters for the 3D sequence were identical to those for the 2D EPI sequences, except for the insertion of two-pulse BS with the timing optimized to suppress static tissue signals [10]. This setup yielded a calibration factor of 70% for 3D GRASE and 85% for 2D EPI, with no BS [4, 10, 14]. For the 3D protocol, an in-plane parallel acceleration factor of 2 was used to minimize the ETL, as recommended [4]. Inline M0-weighted images were acquired without the ASL preparation before the control/label series and with TR = 4000 ms in all ASL protocols. The same acquisition order was used for every subject: the 2D techniques were sequentially scanned first, with an increasing acceleration factor, followed by the 3D sequence.
High-resolution sagittal T1 images were collected using a 3D magnetization-prepared 2 rapid acquisition gradient echo (MP2RAGE) sequence [29]. This sequence provides T1-weighted 3D brain images, as well as automated brain segmentation, quantitative T1 maps, quality control and morphometric reports generated directly on the scanner during image reconstruction. High-resolution T2 maps were obtained using the prototype GRAPPATINI multi-TE [30] sequence. Parameters details used for structural image acquisition are described in Annexes.
The evaluation of the respective performance of each method was conducted by extracting regional absolute CBF values using an inline morphometric segmentation prototype (MorphoBox) [27] and the commonly used Montreal Neurological Institute (MNI) structural atlas [28] implemented within the postprocessing platform FMRIB Software Library, www.fmrib.ox.ac.uk/fsl) (FSL), enabling comparisons at the regional and subregional levels. Details of brain segmentation, pre-processing steps motion correction and Partial Volume Correction (PVC) using the spatially regularized method [31] can be found in supplementary material (Annexe). PVC was particularly essential in this study, because the slice thicknesses across methods varied and was twice as large for some acquisitions compared to others.
Comparison of the tSNR
A voxelwise tSNR was classically calculated for each approach using the mean CBF divided by a standard deviation (SD) across time points according to [10]:
with
as the mean temporal CBF at each pixel coordinate (i,j), and
as the standard deviation at each pixel coordinate (i, j).
In order to offer a fair comparison of the tSNR between 2D and 3D sequences, a second tSNR calculation method was required for optimal matching of the temporal resolution of the two series. To do so, every 5 consequent CBF images of the 2D time series were averaged (subset-averaged CBF) prior to Eq. 1 was applied. The whole-brain mean classical tSNR and corrected tSNR were thus calculated for each ASL method. Spatial tSNR maps were produced to allow qualitative and visual comparisons.
Statistical analysis
The results from the statistical analysis were labeled according to the readout dimension (2D or 3D), followed by the use of BS, parallel acceleration and SMS factor. 3D-BS-P2 indicates the 3D PCASL sequence with BS acquired with iPAT 2 and no SMS. Likewise, 2D P1S1 refers to a 2D EPI sequence with no BS, no iPAT and no slice acceleration; 2D P1S3, SMS factor 3; 2D P1S4, SMS factor 4; and 2D P1S6, SMS factor 6. Indeed, for every subject, five CBF maps corresponding to each image acquisition approach were obtained, with CBF values extracted in 29 regions of each maps for comparison.
CBF obtained using each ASL method is provided as the mean ± standard deviation (M ± SD) after normality testing with the Shapiro–Wilk test. To compare the CBF values that were obtained by each ASL method while accounting for grouping factors (segmentation masks, GM/WM subregions), we used a linear mixed-effects model (Stata mixed package, Stata College Station, Texas). We applied this model with the grouping factors, and their interactions were set as fixed effects, while the subject variable was set as a random effect. A similar linear mixed-effects model was applied separately to the data with the MNI atlas mask but without the GM/WM grouping factor. Pairwise comparisons of the estimates were adjusted for multiple comparisons using the Bonferroni correction. Bland–Altman plots were generated for pairwise comparisons of the global CBF values that were obtained by the different methods, and the pairwise correlations were also computed and reported. To test the significance of differences in the correlation coefficient, we applied the method that was proposed by Lee and Preacher, 2013 [32], whereby each correlation coefficient is converted into a z-score using Fisher’s r-to-z transformation, and the asymptotic covariance of the estimates is computed. Calculation for a test of the difference between two dependent correlations with one variable in common was conducted using QuantPsy software (available from http://quantpsy.org.).
The mean CBF within GM and WM was extracted from the mean CBF maps in native space, and their ratio was computed. Contrast differences among the methods were compared with a mixed linear model, with the methods defined as fixed effects and the subjects defined as random effects. Pairwise comparisons of the estimates were adjusted for multiple comparisons using the Bonferroni correction.
The mean tSNR values were compared using a linear mixed model, with the methods defined as fixed effects and the subjects defined as random effects. Post hoc comparisons were carried out using Bonferroni correction. Data analysis was conducted using Stata 15.1 (StataCorp, College Station, TX, USA). For all analyses, significance was accepted at p < 0.05.
Results
Using SMS acceleration, the image acquisition window was reduced from 1131 ms for 2D P1S1 to 386 ms for 2D P1S3 and 2D P1S4 and 288 ms for 2D P1S6. Representative control images for each protocol variant and the corresponding absolute CBF color maps from one subject are shown in Fig. 1.
Comparison of the mean CBF values obtained using each ASL method
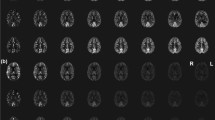
Individual analyses (subject-by-subject) were first conducted for quality control and to illustrate the variability between subjects (data not shown); then, using all seven subjects, the mean CBF maps per method were calculated. The MNI-registered triaxial color-rendered mean CBF maps (ml/min/100 g) that were calculated from all seven subjects are shown in Fig. 2. The absolute mean rCBF values that were calculated using subject-specific T1 and T2 pairs from all five methods are listed in Tables 2 and 3. The mean CBF values for the whole-brain global GM or global WM did not differ significantly across the 2D methods and were slightly higher for the 2D methods than the 3D BS-P2 method, marginally either due to a higher noise level or vascular signal in the absence of BS in 2D. The subject-specific mean regional T1 and T2 values (used for the T1 and T2 corrections) are also offered in Supplementary material in Table a and b.
Figure 3 shows the MNI-registered mean CBF maps across the subjects, separately for each acceleration factor, and the mean differences from the reference. One can appreciate the mean regional differences in the relative CBF between the acquisition protocols. 2D EPI showed a higher relative CBF in most regions. Slightly lower perfusion was observed for 2D P1S1 (SMS = 1) in the frontal lobes than for all other acceleration factor protocols. Lower perfusion was also noted in the frontal WM for 2D P1S6 than for the 2D reference protocol. Moreover, the variance distributions (Fig. 3, column SD) showed different spatial patterns between 2D and 3D acquisitions. 2D sequences displayed a higher SD in the structures with field inhomogeneity (orbitofrontal cortex, gyrus rectus, lower parts of the inferior and the middle frontal gyri), likely because of susceptibility-induced signal dropout in the 2D EPI images. On the other hand, the spatial variance of the 3D acquisition was more concentrated in deep brain structures. Consequently, the tSNR of 3D BS-P2 was reduced in the deep gray nuclei and surrounding structures compared to that of the 2D sequences, whereas the opposite behavior (3D BS-P2 superior to 2D) was observed in other brain regions.
The linear mixed-effects model analysis showed a systematic and significant effect of the ASL method on the estimated CBF value (p < 0.001), as shown in Fig. 4. There were no interactions between the effect of the ASL method on the CBF value for other terms of the model, including the method used to obtain segmentation masks (FSL vs. MorphoBox) and/or the ROI position (GM/WM, frontal, temporal, etc.).
a CBF values and their 95% confidence interval obtained from the linear mixed-effects model across ASL methods. The modelling has used absolute CBF values obtained using subject specific T1 & T2. b GW/WM ratio for all acceleration factors in 7 subjects derived based on mean CBF values.From these plots we can infer that there is no interaction between GM/WM regions and ASL methods
When considering the GM regions, all 2D ASL methods produced higher CBF values than the 3D BS-P2 method. The largest difference in CBF was observed with 2D P1S3, reaching 9.5 ml/100 g/min, 95% CI [7.8, 11.3] (p < 0.001). The difference in CBF with 2D P1S6 was 7.9 [6.2, 9.7] ml/100 g/min (p < 0.001), whereas that with 2D P1S4 was 5.2 [3.4, 5.9] ml/100 g/min (p < 0.001). The smallest difference in CBF was that of 2D P1S1, at 2.5 [0.7, 4.2] ml/100 g/min (p = 0.005), as compared to 3D BS-P2 method.
Figure c and d provides as supplementary material (see Annexes) are showing the linear regressions and the Bland–Altman analysis comparing the mean CBF values obtained in all brain regions for all subjects enrolled in the study. As a statistical indicator, the reproducibility coefficient for each acquisition scenario shows identical trends that can be observed for both CBF extraction methods. The mean CBF values for whole GM and WM regions in the brain that were obtained using all the methods were highly correlated, with r2 = 0.998/0.990/0.980/0.916 for the 2D P1S3/2D P1S4/2D P1S6/3D BS-P2 methods, respectively. The values and correlation coefficients, corroborate the findings of previous studies that compared 2D and 3D strategies [33].
Figure c (Supplementary material) shows that the global variability had the same order of magnitude for all methods. The Bland–Altman analysis also shows that the difference in CBF obtained using 2D P1S6 had a higher coefficient of variance (7.9%) than that obtained using lower acceleration factors, i.e., 4.8% for 2D P1S3 and 7.1% for 2D P1S4. Nevertheless, as shown in Fig. 3, the regional variability in CBF was higher for SMS methods across the cohort of subjects than for 3D BS-P2.
Individual Bland–Altman plots reveal some variability of the CBF per region between subjects for the same acceleration factor and confirm the consistency of the analysis, whether the brain mask was obtained using MorphoBox or the MNI atlas. Identical trends can be observed for the two extraction methods, with similar bias and correlations obtained per subject.
Comparison of the GM/WM CBF contrast ratio
The GM/WM CBF ratios calculated with all of the methods are reported in Table 2 and are also illustrated in Fig. 4b. A higher GW/WM CBF contrast ratio of 3.35 ± 0.83 was obtained for 3D BS-P2 than for all 2D EPI methods due to optimized BS. GW/WM CBF contrast ratios of 2.80 ± 0.58, 3.01 ± 0.57, 2.51 ± 0.52 and 2.76 ± 0.52 were obtained for 2D P1S1, 2D P1S3, 2D P1S4 and 2D P1S6, respectively. T test calculations showed that the GM/WM CBF contrast ratio was significantly higher for 3D BS-P2 than 2D P1S1 (p < 0.02), 2D P1S4 (p < 0.0007) and 2D P1S6 (p < 0.006) but not 2D P1S3 (p > 0.06). There were no significant differences (p < 0.05) in the GW/WM CBF ratio among the 2D EPI sequences.
Comparison of the tSNR
Classical and corrected tSNR values for all ASL methods are reported in Fig. 5. Detailed tSNR values calculated for each approaches are reported in Table c and d (Supplementary material). The mean classical tSNR for the 3D BS-P2 method was 3.32 ± 1.62, and that for 2D P1S1, 2D P1S3, 2D P1S4 and 2D P1S6 was 1.10 ± 0.65, 1.18 ± 0.68, 1.18 ± 0.68 and 1.06 ± 0.61, respectively. Post hoc comparisons of classical tSNR values by T-test also showed a significantly higher tSNR for 3D BS-P2 than for all of the 2D methods (p < 0.001). 3D BS-P2 showed an almost 3-fold greater tSNR than the 2D EPI methods, while no significant difference in the tSNR could be proven among all 2D EPI sequences (p > 0.7). Identical tSNR values for 2D P1S3 and 2D P1S4 were found, which could be due to similar image acquisition parameters, such as the number of slices per band, TE, TR and echo spacing. The 2D P1S6 method, with the highest acceleration factor among all 2D methods, showed the lowest tSNR, which is in line with previous studies [23, 25].
Alternatively, corrected tSNR values calculated from subset-averaged CBF images (the average of every 5 consequent CBF images of the 2D time series) are reported in Fig. 5b and Table d (Supplementary material). The mean tSNR for 2D P1S1, 2D P1S3, 2D P1S4 and 2D P1S6 was 2.48 ± 1.69, 2.84 ± 1.92, 2.70 ± 1.82 and 2.41 ± 1.63, respectively. T-test showed that only non-multiband 2D P1S1 and 2D P1S6 had a tSNR significantly lower than that of 3D BS-P2, with p < 0.003 and p < 0.008, respectively, whereas the other two 2D EPI methods did not show a tSNR significantly lower than that of 3D BS-P2, with p > 0.05. Among the 2D EPI methods, 2D P1S3 and 2D P1S4 showed a tSNR nonsignificantly higher than that of 2D P1S1 and 2D P1S6.
For every series the MCFLIRT calculate the mean rotation and translation of the brain along the time series enable to know the level of subject motion. The level of random motion was very similar along the different time series acquired and remained lower than 0.82 degree and 0.78 mm of rotation and translation motion, respectively. The outputted rotation and translation parameters of the MCFLIRT routine used for retrospective motion correction (MCFLIRT) are reported in Table e and f (supplementary material).
Discussion
The results from the statistical analysis demonstrate that using SMS excitation in conjunction with PCASL labeling offers an increased number of slices to acquire per excitation, without major perceived penalties, such as reduced spatial SNR and tSNR efficiencies of distal slices in caudocranial 2D acquisitions [25]. The global CBF (mean of all subjects) values as well as the CBF values obtained at individual subject levels for different ASL schemes were in agreement with those of previous studies [4, 20, 33], and similar trends were found among different ASL imaging approaches. Nevertheless, Fig. 4a shows that even if the difference in CBF between GM and WM remains equivalent among different strategies, the CBF values cannot be considered equal. Our results are consistent with those of the PASL study reported by Kim et al., who concluded that up to a fivefold SMS acceleration factor and a 3 cm SMS slab distance, the SMS technique provided whole-brain coverage with perfusion signals and tSNR values similar to those of non-accelerated 2D acquisition methods. Furthermore, the differences across 2D EPI methods did not reach significance, which was also demonstrated by Feinberg et al. [23].
Figure 4b shows that the GM/WM CBF ratio obtained for 3D BS-P2 was higher than that obtained for any of the 2D strategies. These differences in the GW/WM CBF ratio are likely to be related to differences in imaging, BS and magnetization transfer (MT) effects that cumulatively impact the degree of agreement between techniques. In 3D BS-P2, BS using spatially selective saturation and inversion pulses helps to minimize signal fluctuations that are induced by noise and subject motion, which, in turn, improves the ASL signal. 3D BS-P2 is a segmented k-space readout known to deliver higher SNR values, and well-timed BS attenuating static tissue signals would almost certainly be reasons for obtaining a high GM/WM CBF contrast ratio. The signal fluctuations and noise observed in the CBF images obtained by 2D EPI, especially in WM, might be likely due to MT effects associated with the application of the multiband RF pulse. Variations in CBF measurements were more evident as the acceleration factor increased, which can be observed in the SD images of 2D P1S6 shown in Fig. 3. However, future work should probably involve scrutinizing MT effects in detail, which is of special interest in the case of SMS, since rCBF values estimated using standard models that do not account for MT effects can be significantly altered.
The nominal PLD in this study was identical for all techniques (PLD = 1800 ms) and was set as recommended in the ASL white paper [4]; thus, it was not optimized individually using multi-TI acquisition. A fixed PLD was chosen to ensure the same decay of the spin label during the PLD, which directly controlled the loss of sensitivity associated with the decrease in signal strength. Nevertheless, in the case of 2D EPI acquisitions without BS, the choice of a given PLD could influence the CBF results due to the presence of vascular transit artifacts, resulting in a possible underestimation of CBF if the PLD is too short compared to the bolus arrival time (BAT) [34]. We did not observe any visible transit artifacts when inspecting the mean CBF maps from each subject, even though the spatial distribution of CBF was different across protocols. Although all the methods showed a similar range of CBF in the posterior circulation, 2D methods showed higher CBF values than did the 3D method in the orbitofrontal cortex, the gyrus rectus, and lower parts of the inferior and middle frontal gyri regions. Such differences can probably be attributed to the different BS, labeling and acquisition schemes [26]; differences in an effective PLD concomitant with a BAT mismatch also likely contribute to methods for rapid BAT quantification [11], allowing an improved experimental measure of CBF, which is likely to better reflect the physiopathology but at the cost of significant extra acquisition time.
Moreover, for 2D EPI acquisitions, the image readouts were performed as groups of slices from inferior to superior, with intrinsic simultaneous and interleaved coverage of the lower and upper brain regions. The BAT is naturally longer in watershed regions than in other regions of the brain, which are supplied by the most distal branches of their arteries [4]. The most simultaneous coverage offered by the SMS protocols is hence likely to result in lower sensitivity to BAT effects and better specificity, i.e., ability to detect differences in a population and grades of disease. Indeed, with SMS techniques, the higher the SMS factor, the shorter the relative delay time between slices: this property allows for a more comparable time for labeled spins to perfuse through the target tissue.
Indeed, SMS excitation has the advantage of reducing the long PLD time with multiple slices collected simultaneously at the same PLD time but in different spatial locations. A necessary condition is that all images should be acquired after all labeled blood spins pass through the imaging slices, i.e., when SMS excitation is applied. Therefore, it is possible that the labeled blood reaches slices of the inferior band, but not those of the most superior band, if the PLD time is shorter than the transit time. The aforementioned condition may not be fulfilled in some categories of patients, and further work on whether this variability could affect a group discrimination analysis is needed [35, 36].
The mean CBF and SD maps across subjects are shown in the left and central columns of Fig. 3; 3D BS-P2 showed less CBF variability between subjects than the 2D EPI variants. Indeed, the 2D P1S1, selected as the reference for comparison, exhibited a higher SD and comparable mean CBF compared to 2D P1S3 and 3D BS-P2; consequently, the reference 2D P1S1 had a lower tSNR than its counterparts, as shown in the right column of Fig. 3. These observed patterns seem to indicate rather regional differences resulting from different temporal labeling dynamics in SB vs. SMS or 3D methods, rather than typical g-factor-induced SNR leakage. The g-factor and induced SNR leakage was not investigated in this study, but has been reported to eventually result from simultaneously excited slices that are too close and an inherent incapacity to differentiate similarity in the received sensitivity profiles [37]. Additionally, as shown in column 3 of Fig. 3, the SD, i.e., arterial signal fluctuations combined with noise, is by far the highest in the cerebrospinal fluid and vascular regions, suggesting that the pulsating motion is a major factor, with a potential impact of this noise on measures in adjacent cortical regions. In pathologies affecting deep gray nuclei, care should be taken to investigate the impact of such nonuniform noise distributions.
Another factor that could influence the CBF values between techniques could be the differences in the recovery period between the control/label acquisitions due to differences in the marginal TE and readout duration. For example, 2D P1S1 images were acquired with a TE = 22 ms and a total readout during of 1131 ms, whereas 2D P1S3 images were acquired with a TE = 24 ms and a readout of 386 ms. In this case, the shorter readout time for 2D P1S3 limits the signal dropout, thereby increasing the perfusion signal, especially if CBF is quantified using a fixed value of T1blood. Indeed, the known variability of T1 could introduce regional differences [38,39,40]. But this shorter readout time could also potentially increase the vascular signal for upper slices of the brain. In our study, we carefully used subject-specific (T1, T2) pairs, as this procedure will be mandatory in patients, particularly to monitor CBF in longitudinal studies in which the status of the lesion and brain tissue can vary considerably. The SD of T1 and T2 values in our subjects was 18% and 6%, respectively. These biological differences show that interindividual differences in normal subjects are relatively small, as is their impact on CBF; however, for example, in stroke patients, the edema in the ischemic area-at-risk coupled with the presence of early/late hemorrhage would result in large T1 and T2 tissue changes over time. Similarly, in patients with tumors, in whom imaging will serve for treatment monitoring and follow-up, important tissue remodeling would most likely and importantly modify the local T1 and T2 values.
Overall, 2D EPI methods demonstrated lower classical tSNR values than the 3D BS-P2 method, and the sequence performance results that were obtained in our control population are generally consistent with those that have previously been reported in other control cohorts [10, 22, 23, 33]. The higher tSNR for 3D BS-P2 could be attributed to BS, which suppresses static brain tissue signals up to 85% and, therefore, improves the temporal stability of different images. However, the similarity of the different 2D results, also in terms of tSNR is not fully explainable. For example, the 2D P1S6 approach has a larger g-factor penalty and lower SNR from thinner slices, yet it was not proportionally reflected in the results compared to other variants and it may be indicative of a physiological noise dominated regime.
Finally, the motion correction was performed using MCFLIRT (FSL) [41] for both 2D and 3D scans in our study with level of estimated motion reported to be less than 1 degree rotation and 1 mm translation motion, and very similar range of retrospectively evaluated random motion between the different acquired series. Indeed, since the sequences were always acquired in the same order, there was a potential for effects of possibly larger motion artifacts related to loss of volunteer compliance during the entire protocol, with the potential for better settling at the beginning and tiring at end. These offline motion corrections enable to compare the level of motion, and eliminate any possibility of causing CBF or tSNR variability due to control-label image subtraction error. Even though the scans with the highest SMS factor and the 3D GRASE scans were always performed last in this study, there is no evidence that they might have been more affected by increased subject fidgeting toward the end of the session. Note that between-shot motion in a segmented 3D GRASE acquisition will manifest not just as misregistration between imaging volumes but will also introduce ghosting due to k-space inconsistencies within in the multiple acquired sub-k-space shots.
Regarding the sensitivity to motion, it is worth noting that Murphy and Brunberg estimated that to obtain satisfactory images, approximately 14% of subjects require sedation or general anesthesia, which poses significant risks [42]. It is ethically impossible to perform such a preliminary study in the stroke patient population that we target and get a clear estimation of the expected level of motion. Nevertheless, it is likely that in such cases, which necessitate an imaging sequence with high motion robustness, 2D EPI sequences with a short acquisition window capable of acquiring more time points for a given scan time have the edge over their 3D counterparts and might be more suitable. However, the benefit for better motion management offered by SMS has not been directly studied or evaluated in this work and remains to be definitely shown in practice. One of the greatest advantages of the 3D sequence is the ability to achieve very good BS. 3D GRASE single shot readout with BS greatly reduces motion sensitivity and is especially valuable in patient studies, with benefits that tend to be greater in less cooperative populations [4]. However, 3D segmented readout having long scan time renders the sequence inherently more sensitive to motion though delivers higher SNR. For multisite and multiplatform studies using ASL as a biomarker, significant differences between ASL protocols may result from hardware differences that can severely affect the absolute CBF and should absolutely be considered in addition to previously reported findings [43].
One limitation but also the originality of the study is that the voxel sizes of the images across ASL protocols were different due to different slice thicknesses. This needs to be taken into account as it could affect the CBF comparisons of the methods, even though care was taken using the same spin labeling (PCASL) scheme in all the acquisitions. Additionally, the analyses was performed on ROIs that were sufficiently large to yield unchanged mean CBF values within the ROI, as long as the blood that supplies the microvasculature arrived within the ROI [44]. Since our analysis was based on large brain ROIs and not at the individual voxel level, the mean CBF should be largely independent of the voxel resolution; however, some residual effects of the voxel size due to differences in partial volume contamination and physiological noise cannot be excluded. However, it is worth noting that testing various approaches and protocol implementations remains the best way to make sure that the best implementation is indeed transferred for further clinical use.
A second limitation of the study is that of no in-plane parallel imaging acceleration was used in the 2D protocol: this study was designed to focus on the unique effect of the simultaneous multiband feature and through-plane accelerations. The main reason was motivated by the concern to warranty the exclusion of any additional spatial variation of the noise that could results also from iPAT implementation, further complexifying on the analysis. Note that in-plane parallel imaging could have allowed us to have more consistent slice thickness throughout the different protocols, which was explicitly not our goal here, since we wanted to explore the effect of different slice thicknesses. Another reason was also that factor-of-6 SMS acceleration would not have been achievable in combinations with in-plane acceleration 2. The reader can refer to the study from Ivanov et al. who investigated the effect of both acceleration factors in ASL (in-plane (PAT) and through-plane (SMS)) at 7T [45].
To summarize, SMS has some important advantages, such as its ability to increase the slice coverage without substantially increasing the readout time [23], which is of crucial importance in cases of whole-brain perfusion measurement with a high number of slices when using the single-slice 2D EPI acquisition method. In conventional 2D EPI techniques, the discrepancy in spin status across slices is high, because the PLD of the last acquired slice is a few hundred milliseconds greater than that of the first slice, which causes a difference in the longitudinal decay of labeled blood [46]. 2D EPI sequences with SMS overcome this problem through their capacity to shorten the acquisition window using higher acceleration factors; therefore, it would probably be the most suitable technique by which to implement a multi-TI protocol aimed at scouting the most accurate BAT.
In conclusion, 3D GRASE showed the lowest variability and thus the highest robustness for ASL perfusion imaging in the brain compared to the 2D EPI alternatives in healthy subject population. All 2D EPI protocols with SMS can probably be considered equivalent even though regional differences in the CBF values can be observed from one to the other, which in turn adds a factor of variability that should be considered in multicenter studies. Whether motion correction is more efficient over a 2D longitudinal series than a 3D longitudinal series with powerful BS still needs to be investigated and will determine which of the two approaches is the most suitable technique by which to perform follow-up examinations in a large cohort of patients who are more likely to exhibit involuntary head movements, such as those suffering from brain injury or patients who are affected by neurodegenerative disorders, such as Parkinson’s disease.
References
Salinet ASM, Panerai RB, Robinson TG (2014) The longitudinal evolution of cerebral blood flow regulation after acute ischaemic stroke. Cerebrovasc Dis Extra 4:186–197
Saa JP, Tse T, Baum C, Cumming T, Josman N, Rose M, Carey L (2019) Longitudinal evaluation of cognition after stroke—a systematic scoping review. PLoS ONE 14:e0221735
von Kummer R (2019) Treatment of ischemic stroke beyond 3 hours: is time really brain? Neuroradiology 61:115–117
Alsop DC, Detre JA, Golay X, Gunther M, Hendrikse J, Hernandez-Garcia L, Lu H, MacIntosh BJ, Parkes LM, Smits M, van Osch MJ, Wang DJ, Wong EC (2015) “Recommended implementation of arterial spin-labeled Perfusion mri for clinical applications: A consensus of the ISMRM Perfusion Study group and the European consortium for ASL in dementia. Magn Reson Med 73(1):102–116
Detre JA, Leigh JS, Williams DS, Koretsky AP (1992) Perfusion imaging. Magn Reson Med 23:37–45
Pollock JM, Tan H, Kraft R, Whitlow CT, Burdette JH, Maldjian JA (2010) Arterial spin labeled MRI Perfusion. Magn Reson Imaging Clin N Am. https://doi.org/10.1016/j.mric.2009.01.008.Arterial
Sadowski EA, Bennett LK, Chan MR, Wentland AL, Garrett AL, Garrett RW, Djamali A (2007) Nephrogenic systemic fibrosis: risk factors and incidence estimation. Radiology 243:148–157
Günther M, Oshio K, Feinberg DA (2005) Single-shot 3D imaging techniques improve arterial spin labeling perfusion measurements. Magn Reson Med 54:491–498
Fernández-Seara MA, Edlow BL, Hoang A, Wang J, Feinberg DA, Detre JA (2008) Minimizing acquisition time of arterial spin labeling at 3T. Magn Reson Med 59:1467–1471
Vidorreta M, Wang Z, Rodriguez I, Pastor MA, Detre JA, Fernández-Seara MA (2013) Comparison of 2D and 3D single-shot ASL perfusion fMRI sequences. Neuroimage 66:662–671
Dai W, Shankaranarayanan A, Alsop DC (2013) Volumetric measurement of perfusion and arterial transit delay using hadamard encoded continuous arterial spin labeling. Magn Reson Med 69:1014–1022
Wang J, Zhang Y, Wolf RL, Roc AC, Alsop DC, Detre JA (2005) Amplitude-modulated continuous arterial spin-labeling 3.0-T perfusion MR imaging with a single coil: feasibility study. Radiology 235:218–228
Dai W, Garcia DM, de Bazelaire C, Alsop DC (2008) Continuous flow driven inversion for arterial spin labelling using pulsed radiofrequency and gradient fields. Magn Reson Med 60:1488–1497
Garcia DM, Duhamel G, Alsop DC (2005) Efficiency of inversion pulses for background suppressed arterial spin labeling. Magn Reson Med 54:366–372
Maleki N, Dai W, Alsop DC (2012) Optimization of background suppression for arterial spin labeling perfusion imaging. Magn Reson Mater Phy 25:127–133
Ye FQ, Frank JA, Weinberger DR, McLaughlin AC (2000) Noise reduction in 3D perfusion imaging by attenuating the static signal in arterial spin tagging (ASSIST). Magn Reson Med 44:92–100
Kilroy E, Apostolova L, Liu C, Yan L, Ringman J, Wang D (2014) Reliability of 2D and 3D pseudo-continuous arterial spin labeling perfusion mri in elderly populations—comparison with 15O-water PET. J Magn Reson Imaging 39:931–939
Wu WC, Edlow BL, Elliot MA, Wang J, Detre JA (2009) Physiological modulations in arterial spin labeling perfusion magnetic resonance imaging. IEEE Trans Med Imaging 28:703–709
Young IR, Larkman DJ, Hajnal JV, Ehnholm G, Herlihy AH, Coutts GA (2002) Use of multicoil arrays for separation of signal from multiple slices simultaneously excited. J Magn Reson Imaging 13:313–317
Moeller S, Yacoub E, Olman CA, Auerbach E, Strupp J, Harel N, Uǧurbil K (2010) Multiband multislice GE-EPI at 7 tesla, with 16-fold acceleration using partial parallel imaging with application to high spatial and temporal whole-brain FMRI. Magn Reson Med 63:1144–1153
Setsompop K, Cohen-Adad J, Gagoski BA, Raij T, Yendiki A, Keil B, Wedeen VJ, Wald LL (2012) Improving diffusion MRI using simultaneous multi-slice echo planar imaging. Neuroimage 63:569–580
Kim T, Shin W, Zhao T, Beall EB, Lowe MJ, Bae KT (2013) Whole brain perfusion measurements using arterial spin labeling with multiband acquisition. Magn Reson Med 70:1653–1661
Feinberg DA, Beckett A, Chen L (2013) Arterial spin labeling with simultaneous multi-slice echo planar imaging. Magn Reson Med 70:1500–1506
Kim SG, Tsekos NV (1997) Perfusion imaging by a flow–sensitive alternating inversion recovery (FAIR) technique: application to functional brain imaging. Magn Reson Med 37:425–435
Li X, Wang D, Auerbach EJ, Moeller S, Ugurbil K, Metzger GJ (2015) Theoretical and experimental evaluation of multi-band EPI for high-resolution whole brain pCASL Imaging. Neuroimage 106:170–181
Lövblad KO, Montandon ML, Viallon M, Rodriguez C, Toma S, Golay X, Giannakopoulos P (2015) Arterial spin-labeling parameters influence signal variability and estimated regional relative cerebral blood flow in normal aging and mild cognitive impairment: fAIR versus PICORE techniques. AJNR Am J Neuroradiol 36(7):1231–1236
Schmitter D, Roche A, Maréchal B, Ribes D, Abdulkadir A, Bach-Cuadra M, Daducci A, Granziera C, Klöppel S, Maeder P, Meuli R, Krueger G (2015) An evaluation of volume-based morphometry for prediction of mild cognitive impairment and Alzheimer’s disease. NeuroImage Clin 7:7–17
Collins DLL, Holmes CJ, Peters TM (1995) Automatic 3-D model-based neuroanatomical segmentation. Hum Brain 3:190–208
Marques JP, Kober T, Krueger G, van der Zwaag W, Van de Moortele PF, Gruetter R (2010) MP2RAGE, a self bias-field corrected sequence for improved segmentation and T1-mapping at high field. Neuroimage 49:1271–1281
Hilbert T, Sumpf TJ, Weiland E, Frahm J, Thiran JP, Meuli R, Kober T, Krueger G (2018) Accelerated T 2 mapping combining parallel MRI and model-based reconstruction: GRAPPATINI. J Magn Reson Imaging 48:359–368
Chappell MA, Groves AR, MacIntosh BJ, Donahue MJ, Jezzard P, Woolrich MW (2011) Partial volume correction of multiple inversion time arterial spin labeling MRI data. Magn Reson Med 65:1173–1183
Lee IA, Preacher KJ (2013) Calculation for the test of the difference between two dependent correlations with one variable in common [computer soft-ware]. http://quantpsy.org
Dolui S, Vidorreta M, Wang Z, Nasrallah IM, Alavi A, Wolk DA, Detre JA (2017) Comparison of PASL, PCASL, and background-suppressed 3D PCASL in mild cognitive impairment. Hum Brain Mapp 38:5260–5273
Ferré JC, Bannier E, Raoult H, Mineur G, Carsin-Nicol B, Gauvrit JY (2013) Arterial spin labeling (ASL) perfusion: techniques and clinical use. Diagn Interv Imaging 94:1211–1223
Zhu J, Zhuo C, Qin W, Xu Y, Xu L, Liu X (2015) Altered resting-state cerebral blood flow and its connectivity in schizophrenia. J Psychiatr Res 63:28–35
Wolf Marc, Okazaki Shuhei, Eisele Philipp, Roßmanith Christina, Gregori Johannes, Griebe Martin, Günther Matthias, Gass Achim, Hennerici Michael, Szabo Kristina, Kern R (2018) Arterial spin labeling cerebral perfusion magnetic resonance imaging in migraine aura: an observational study. J Stroke Cerebrovasc Dis 27(10):101
Setsompop K, Gagoski BA, Polimeni JR, Witzel T, Wedeen VJ, Wald LL (2012) Blipped-controlled aliasing in parallel imaging for simultaneous multislice echo planar imaging with reduced g-factor penalty. Magn Reson Med 67:1210–1224
Dolui S, Wang Z, Wang DJJ, Mattay R, Finkel M, Elliott M, Desiderio L, Inglis B, Mueller B, Stafford RB, Launer LJ, Jacobs DR, Bryan RN, Detre JA (2016) Comparison of non-invasive MRI measurements of cerebral blood flow in a large multisite cohort. J Cereb Blood Flow Metab 36:1244–1256
Hales PW, Kirkham FJ, Clark CA (2016) A general model to calculate the spin-lattice (T1) relaxation time of blood, accounting for haematocrit, oxygen saturation and magnetic field strength. J Cereb Blood Flow Metab 36:370–374
Wu W-C, Jain V, Li C, Giannetta M, Hurt H, Wehrli FW, Wang DJJ (2010) In vivo venous blood T1 measurement using inversion recovery true-FISP in children and adults. Magn Reson Med 64:1140–1147
Jenkinson M, Bannister P, Brady M, Smith S (2002) Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage 17:825–841
Kieran KJ, Brunbergz JA (1997) Adult claustrophobia, anxiety and sedation in MRI. Magn Reson Imaging 15:51–54
Mutsaerts HJMM, Steketee RME, Heijtel DFR, Kuijer JPA, Van Osch MJP, Majoie CBLM, Smits M, Nederveen AJ (2014) Inter-vendor reproducibility of pseudo-continuous arterial spin labeling at 3 Tesla. PLoS One. https://doi.org/10.1371/journal.pone.0104108
Qiu D, Straka M, Zun Z, Bammer R, Moseley ME, Zaharchuk G (2012) CBF measurements using multidelay pseudocontinuous and velocity-selective arterial spin labeling in patients with long arterial transit delays: comparison with xenon CT CBF. J Magn Reson Imaging 36:110–119
Ivanov D, Poser BA, Huber L, Pfeuffer J, Uludağ K (2017) Optimization of simultaneous multislice EPI for concurrent functional perfusion and BOLD signal measurements at 7T. Magn Reson Med 78:121–129
Campbell AM, Beaulieu C (2006) Comparison of multislice and single-slice acquisitions for pulsed arterial spin labeling measurements of cerebral perfusion. Magn Reson Imaging 24:869–876
Acknowledgements
This work was performed within the framework of the LABEX PRIMES (ANR-11-LABX-0063) of Université de Lyon, within the program “Investissements d’Avenir” (ANR-11-IDEX-0007) operated by the French National Research Agency (ANR).
Funding
No funding was received for this study.
Author information
Authors and Affiliations
Contributions
MN: Study conception and design; acquisition of data; analysis and interpretation of data; drafting of manuscript. TT: Study conception and design; acquisition of data; drafting of manuscript; critical revision. JP: Critical revision, provided the prototype 2D and 3D ASL sequences and protocol parameter optimization. BM: Acquisition of data, provided the prototype MorphoBox for inline segmentation. TH: Acquisition of data, provided the prototype GRAPPATINI for fast T2 mapping. TK: Acquisition of data and MP2RAGE protocol parameter optimization; critical revision. FS: Analysis and interpretation of data; drafting of manuscript; critical revision. PC: Analysis and interpretation of data. MV: Study conception and design; acquisition of data; analysis and interpretation of data; drafting of manuscript; critical revision.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Nanjappa, M., Troalen, T., Pfeuffer, J. et al. Comparison of 2D simultaneous multi-slice and 3D GRASE readout schemes for pseudo-continuous arterial spin labeling of cerebral perfusion at 3 T. Magn Reson Mater Phy 34, 437–450 (2021). https://doi.org/10.1007/s10334-020-00888-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10334-020-00888-8