Abstract
A macaque population produced by the hybridization of native Japanese macaques (Macaca fuscata) and introduced Taiwanese macaques (M. cyclopis) in Wakayama Prefecture was shown to possess three DNA haplotypes of the natural resistance-associated macrophage protein 1 (NRAMP1). Based on genotyping and comparison with M. fuscata populations, it was revealed that the introduced M. cyclopis population was polymorphic for the NRAMP1 locus. Extensive crossbreeding of the introduced species with the native species was confirmed using this genetic marker and the proportion of M. cyclopis genes was 57.4%. Results of statistical tests suggested non-random mating in the hybrid population.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Threats to native biological diversity by human activities are of concern worldwide (WRI 1992); the impact of introduced species on the extinction of native species or damage to ecosystems represents one of these major threats (IUCN 2000). In Japan, more than 29 introduced mammalian species were reported in 2002 (Ecological Society of Japan 2002). One of these, an artificially introduced population of Taiwanese macaques (Macaca cyclopis), was found to have hybridized with native Japanese macaques (M. fuscata) in Wakayama Prefecture on the western part of the Kii peninsula (Kawamoto et al. 1999, 2001). The Taiwanese macaques have been introduced since the 1950s (Kawamoto et al. 2001). The prefectural government started efforts in 2001 to eliminate the hybrid population to prevent the spread of exotic genes. Diagnostic genetic markers are important tools in biological monitoring to assess hybridization between different species and to confirm the transfer of genes through adult male migration in macaque populations. Here, I report a nuclear DNA variation of natural resistance-associated macrophage protein 1 (NRAMP1) in the hybrid population in Wakayama Prefecture with special reference to interspecific differences as well as its DNA polymorphism in Taiwanese macaques.
Methods
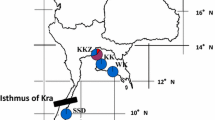
DNA sequences including intron 5 of the NRAMP1 (503 bp) were examined from 81 individuals of the hybrid population at Ooike, Wakayama Prefecture (Fig. 1). The samples from the Ooike population were distinguished according to natal origin by mitochondrial DNA (mtDNA) type. Three adult male samples from the same population had a type of M. fuscata mtDNA. Morphological features of tail length and pelage color also suggested M. fuscata origin and they were judged to be immigrants from native Japanese macaque populations. Including those M. fuscata samples, a total of 28 Japanese macaques from eight localities in Wakayama and Mie Prefectures (Fig. 1) and three captive Taiwanese macaques from KUPRI (Primate Research Institute, Kyoto University) were also examined to assess interspecific sequence differences (Table 1).
Genomic DNA was prepared from whole blood. PCR was performed according to Deinard and Smith (2001), and the amplification product was directly sequenced with an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems). Two additional internal primers (forward primer: 5′-TTTATCCTGCTCTCCCCTCT-3′ and reverse primer: 5′-AGTGGGAGTGTGGCATAGAA-3′) were used to determine the heterozygotes with confidence. The DNA sequences obtained in this study have been deposited in DDBJ (Accession Nos. AB162926–AB162928).
Results and discussion
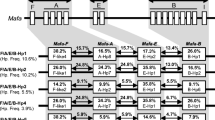
In the hybrid population, intron 5 of the NRAMP1 gene was polymorphic at three nucleotide sites of the 503-bp fragment. The first two positions from the 5′ end (sites A and B) showed C/T polymorphisms and the third (site C) showed A/C polymorphism (Fig. 2). Of the 27 possible genotypic combinations of the three variable sites, only six were observed in the hybrid population (Table 1). For native Japanese macaques, all samples showed a single homozygous genotype with the T-C-C haplotype. Individuals in the hybrid population had genotypes that were either identical to the homozygous native Japanese macaque haplotypes (T-C-C/T-C-C) or one of the heterozygous M. fuscata-specific haplotypes (C-C-A/T-C-C or T-T-A/T-C-C). The heterozygous hybrids had one of two distinct haplotypes that originated from Taiwanese macaques: haplotype C-C-A or haplotype T-T-A. Taiwanese macaques at KUPRI showed a single homozygous genotype with the T-T-A haplotype.
The present result suggests the presence of DNA polymorphism of the NRAMP1 gene in Taiwanese macaques. While haplotypes T-T-A in Taiwanese macaques and T-C-C in Japanese macaques were reported previously by Deinard and Smith (2001), this is the first report of the haplotype C-C-A in Taiwanese macaques. However, this newly observed NRAMP1 haplotype (C-C-A) is also known to occur in rhesus (M. mulatta), bonnet (M. radiata) and bear (M. arctoides) macaques (Deinard and Smith 2001).
The NRAMP1 polymorphism reported here is a useful molecular tool for evaluating the genetic features of the hybrid population in Wakayama. The haplotypes C-C-A, T-T-A and T-C-C occur at an estimated frequency (with SE) 0.198±0.031, 0.377±0.038 and 0.426±0.038, respectively. Finally, based on this nuclear marker, the proportion of M. cyclopis genes in the population was estimated to be 57.4±3.9%, suggesting extensive crossbreeding of the M. cyclopis group with native M. fuscata. Reproductive features of the hybrid population can be further analyzed by testing for random association of gametes. The null hypothesis of random mating was not rejected when testing M. cyclopis haplotypes (C-C-A and T-T-A) together (χ2=1.10, 0.2<P<0.3, df=1), but was rejected when these three haplotypes were treated separately (χ2=8.29, 0.02<P<0.05, df=3). This was due mainly to individuals possessing the T-T-A haplotype (M. cyclopis type) where there was an excess of homozygotes and a deficiency of heterozygotes with the haplotype T-C-C (M. fuscata type). This detailed observation suggests the possibility of assortative mating in the hybrid population.
Although the possibility that the M. cyclopis founders had a haplotype identical to M. fuscata cannot be fully rejected, it is rather difficult to support this possibility for two reasons. First, the size of the founding population was small; the individuals had escaped from an open enclosure of a zoological park (Kawamoto et al. 2001). Second, the available data, including those on the M. cyclopis control in this study, do not show such evidence (Deinard and Smith 2001). Significant species difference was seen only in the A/C polymorphism at the third variable site, and thus, in practice, it is sufficient to type this nucleotide polymorphism to quantify the rate of hybridization in populations.
This is the first report to apply a DNA polymorphism at a single autosomal locus to the study of a hybrid population of primates. Other kinds of variable genetic characteristics, such as mtDNA, microsatellite DNA or amplified fragment length polymorphism (AFLP), have been commonly used in population genetics studies on hybrid populations (e.g., Evans et al. 2001; Nijman et al. 2003). However, most of these methods are weak in analyzing gametic association in hybrid populations due to aspects such as sex-biased mode of inheritance, high degree of polymorphism within species or difficulty in discrimination of specific alleles. The application of multiple nucleotide polymorphisms in DNA molecules could give us the insight to inquire into the cause and process of hybridization between species.
References
Deinard A, Smith DG (2001) Phylogenetic relationships among the macaques: evidence from the nuclear locus NRAMP1. J Hum Evol 41:45–59
Ecological Society of Japan (2002) Handbook of alien species in Japan (in Japanese). Chigin Shokan, Tokyo
Evans BJ, Supriatna J, Melnick D (2001) Hybridization and population genetics of two macaque species in Sulawesi, Indonesia. Evolution 55:1686–1702
IUCN (2000) IUCN guidelines for the prevention of biodiversity loss caused by alien invasive species. IUCN SSC Invasive Species Specialist Group, Gland, Switzerland
Kawamoto Y, Shirai K, Araki S, Maeno K (1999) A case of hybridization between the Japanese and Taiwanese macaques found in Wakayama Prefecture (in Japanese with English summary). Primate Res 15:53–60
Kawamoto Y, Ohsawa H, Nigi H, Maruhashi T, Maekawa S, Shirai K, Araki S (2001) Genetic assessment of a hybrid population between Japanese and Taiwanese macaques in Wakayama Prefecture (in Japanese with English summary). Primate Res 17:13–24
Nijman IJ, Otsen M, Verkaar ELC, de Ruijter C, Hanekamp E, Ochieng JW, Shamshad S, Rege JEO, Hanotte O, Barwegen MW, Sulawati T, Lenstra JA (2003) Hybridization of banteng (Bos javanicus) and zebu (Bos indicus) revealed by mitochondrial DNA, satellite DNA, AFLP and microsatellites. Heredity 90:10–16
WRI (1992) Global biodiversity strategy: guidelines for action to save, study and use earth’s biotic wealth sustainably and equitable. World Resources Institute, World Conservation Union (IUCN), and the United Nations Environment Programme, Washington D.C.
Acknowledgements
I wish to thank prefectural authority of Wakayama Prefecture for allowing me the opportunity to undertake this study. I am indebted to Mr. Kei Shirai and other members of Wildlife Management Office, Kawasaki, Japan, for sample collection and information. I am grateful to Mr. Shingo Maekawa, Drs. Hideo Nigi, Hideyuki Ohsawa, Shunji Gotoh, Yasuyuki Muroyama, Tamaki Maruhashi and Harumi Torii for their help and cooperation. I also thank Dr. Jan R. de Ruiter for helpful suggestions on the manuscript. This study was supported in part by Grants-in-Aid for General Scientific Research (13440256 and 16310156) and the MEXT Grant-in-Aid for the 21st Century COE Program (A2 to Kyoto University) from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Author information
Authors and Affiliations
Corresponding author
About this article
Cite this article
Kawamoto, Y. NRAMP1 polymorphism in a hybrid population between Japanese and Taiwanese macaques in Wakayama, Japan. Primates 46, 203–206 (2005). https://doi.org/10.1007/s10329-004-0119-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10329-004-0119-3