Abstract
The two cultivated Luffa species can be severely infected by Tomato leaf curl New Delhi virus (ToLCNDV) with up to 100% yield loss. Here, 52 Luffa genotypes were screened for ToLCNDV resistance after natural field infection. Mean vulnerability index (VI) ranged from 0.00 to 75.33; genotypes IIHR-137 and IIHR-138 had no symptoms (VI 0), 16 genotypes were resistant (VI 0–25), 15 were moderately resistant (VI 26–50), and 19 were moderate to susceptible (VI > 50). Ten of the most resistant genotypes and five susceptible checks were then challenge-inoculated using whiteflies or sap in an insect-proof net house; only IIHR-137 [L. cylindrica (L.) Roem.] was symptomless (VI 0.00), and 3–5% of plants of IIHR-138 [L. cylindrica (L.) Roem.] and IIHR-Sel-1 [L. acutangula (L.) Roxb.] had only mild symptoms; genotype Arka Prasan was most susceptible (VI 80.96). Asymptomatic plants were confirmed as infected using polymerase chain reaction. Susceptible genotypes rapidly developed leaf curling, then a severe mosaic 10 days post-inoculation. The resistant inbred lines identified are good candidates for a breeding program for ToLCNDV-resistant cultivars.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ridge gourd [Luffa acutangula (L.) Roxb.] and sponge gourd [L. cylindrica (L.) Roem.] (Cucurbitaceae) are the two main cultivated Luffa species and grown during the spring–summer and rainy seasons in subtropical and tropical regions. China, India, Korea, Japan, and Central America are the major regions for commercial cultivation of Luffa spp. (Dhillon et al. 2016). In India, Andhra Pradesh, Tamil Nadu, Karnataka, Gujarat, Maharashtra, Assam, and West Bengal account for a significant share. Tender, immature fruits of both species are cooked and eaten. The high fibre content in fruits aids digestion and excretory system functioning (Swetha and Muthukumar 2016). Fibre from dried fruits of ridge gourd has potential application in sound insulation and textile industry, like other commercial lignocellulosic fibres (Karthik and Ganesan 2015).
However, many biotic and abiotic constraints affect ridge gourd commercial production, including a severe threat from begomoviruses transmitted by whiteflies (Bemisia tabaci). For example, Tomato leaf curl New Delhi Virus (ToLCNDV) can cause 100% yield loss of ridge gourd in epiphytotic conditions (Patil et al. 2017). Leaf curl disease, first reported in India on tomato (Solanum lycopersicum L.) (Padidam et al. 1995), has now been found in many countries such as Pakistan, Bangladesh, Iran, Sri Lanka, Malaysia, Taiwan, Thailand, Indonesia, Tunisia, Spain, Italy, and Greece (Zaidi et al. 2017), causing damage to 44 eudicot plant species including vegetables, ornamentals, weed species and fibre crops, but tomato and cucurbits have been identified as some of the most susceptible (Ito et al. 2008; López et al. 2015; Moriones et al. 2017; Sáez et al. 2016; Zaidi et al. 2017). The virus is bipartite, and both of the two circular single-stranded DNA molecules (DNA-A and DNA-B) encode transcripts that are necessary for infectivity (Sangeetha et al. 2018; Zaidi et al. 2017).
In nature, ToLCNDV transmission by the whitefly species complex Bemisia tabaci is circulative and persistent (Zaidi et al. 2017). Studies have also suggested that ToLCNDV can be effectively transmitted mechanically and through seed in select cucurbit hosts (López et al. 2015; Sáez et al. 2016; Sangeetha et al. 2018; Sohrab et al. 2014). Symptoms on the various hosts include yellow spots, yellow mosaic, reduced leaf size, short internodes, leaf curling, thickened leaf margins, and darkening, puckering, and severe stunting of the entire plant. Virus management generally focuses on vector control with pesticides and biological and cultural practices (Legg et al. 2014). However, factors such as seed transmission, whitefly migration kinetics, and expansion of the host range of the virus, complicate the development of comprehensive virus management strategies (Zaidi et al. 2017). Planting genetically resistant varieties provides a simple, effective strategy to control ToLCNDV in affected crops. Because cucurbits are economically important hosts, the search for sources of resistance is a priority, especially because the use of only one or two resistance lines leads to adverse levels of genetic vulnerability. Considering the extent of losses and the epidemiology of the virus, germplasm must be screened in hotspot areas.
So far worldwide, ToLCNDV-resistant accessions have been selected for Indian melon (Cucumis melo subsp. agrestis var. momordica and wild agrestis accessions), sponge gourd [L. cylindrica (L.) Roem.], and Cucurbita moschata accessions (Islam et al. 2010; López et al. 2015; Sáez et al. 2016). Therefore, here we screened all available Luffa germplasm to find ToLCNDV-resistant sources to incorporate into a commercial ridge gourd cultivar with high productivity.
Materials and methods
Forty-four genotypes of Luffa acutangula and eight of L. cylindrica were screened at the experimental farm of the Division of Vegetable Crops, ICAR-IIHR, Bengaluru, Karnataka, India, during 2017–2018. Commercially cultivated varieties and advanced breeding lines maintained as inbreds following selfing at the experimental farm were included in the present experiment.
Screening in the field after natural infection
Fifty-two genotypes were initially screened in the field during March–June 2017 when the whitefly population was high to favor disease development. Fifteen plants of each genotype (five plants per replication) were observed for disease symptoms. Seedlings of each genotype were planted in a randomized block design 14 days after sowing (DAS) in raised beds covered with plastic mulch with 150 cm between beds and 50 cm between plants. All other recommended practices were followed, except that no insecticides were applied to avoid reducing whitefly proliferation and thus virus transmission and disease incidence. Plants were evaluated using the 6-point scale of Islam et al. (2010) described later.
Screening in insect-proof net houses after inoculation
The disease response of the 10 most-resistant genotypes and five susceptible checks in the field screening were evaluated during January–June 2018. Thirty seedlings of each genotype were inoculated using viruliferous whiteflies or sap as described later.
Virus identification and confirmation
Although several viruses cause leaf curling and stunting in cucurbits (Mitra and Nariani 1965; Singh et al. 2001), yellow mosaic on young leaves is typical of ToLCNDV, the predominant species of begomovirus in southern India (Patil et al. 2017). To verify that young leaves with yellow mosaic were infected with ToLCNDV, inoculum was prepared from infected young leaves of a ridge gourd plants to inoculate healthy ridge gourd seedlings grown in pots. Viral DNA from infected tissue of the inoculated seedlings was then isolated to confirm the presence of ToLCNDV with polymerase chain reaction (PCR) (Swarnalatha et al. 2013) and sequencing of amplified products. The sequence was analysed and used in a BLAST search of the NCBI GenBank database. The sequence of the ridge gourd virus isolate shared 92–97% similarity with several ToLCNDV isolates such as ToLCNDV-ridge gourd isolate RG3. Thus, the sequences were submitted to GenBank using Bankit for verification and registration of sequences (GenBank accession MT981253). These infected ridge gourd plants were maintained in an insect-proof net house and used as an inoculum source.
Whitefly-mediated inoculation
Whiteflies (Bemesia tabaci Genn.; Hemiptera, Aleyrodidae) were reared on brinjal (Solanum melongena L.) variety Arka Anand. Fifteen genotypes of Luffa species were screened using a whitefly inoculation method to revalidate their resistance. Thirty seeds per genotype were directly sown in poly bags (10 × 8 cm) filled with farm-yard manure and soil mixture. Avirulent whiteflies were allowed to feed overnight on infected twigs to acquire ToLCNDV. Ten viruliferous whiteflies per seedling were then released on test seedlings for 24 h and covered in an insect-proof cage (Patil et al. 2017; Sohrab et al. 2013). Test seedlings were inoculated at the two-true-leaf (fully expanded) stage in an insect-proof net house. Plants were evaluated for up to 2 months using the 6-point scale of Islam et al. (2010) described later.
Mechanical inoculation
The primary inoculum was collected from the infected ridge gourd plants maintained in an insect-proof net house. Inoculation buffer was. One gram of young infected leaves were ground in inoculation buffer prepared as described by López et al. (2015). The extract was filtered through a non-absorbent cotton pad, and the resultant homogenate was used as inoculum. Cotyledons of test plants (7–10 days after germination) were dusted with carborundum and inoculated, then kept in an insect-proof net house and evaluated using the 6-point scale of Islam et al. (2010) described later.
Disease diagnosis
ToLCNDV transmission in test plants was analyzed using the PCR amplification profile and sequencing of the random samples. Apical leaves were collected from inoculated plants 30 days post-inoculation (dpi). Total genomic DNA was isolated using a cetyltrimethyl ammonium bromide (CTAB) method (Swarnalatha et al. 2013). DNA was quantified using a spectrophotometer and diluted with sterile deionised water to give final concentration of 100 ng/l. For ToLCNDV detection, 1.2 µl of total DNA was used as the template for PCR reactions in 25 µl reaction volume with 3 U of Taq DNA polymerase (Fermentas, Germany), 2 mM dNTP (Fermentas, Baden-Wurttemberg, Germany), 25 mM MgCl2 (Fermentas, Germany), and 100 pmol of virus-specific primer (Ashwathappa et al. 2020). The PCR thermocycling conditions were 94 °C for 3 min; 35 cycles of 94 °C for 45 s, 55 °C for 1 min, and 72 °C for 1 min; 20 min at 72 °C. PCR products were electrophoresed in 0.8% (w/v) agarose gel stained with ethidium bromide (10 mg/ml), viewed with a gel documentation system (Alpha Innotech, San Leandro, CA, USA), then purified and sequenced in both directions at the Medaxin DNA Sequencing facility, Bangalore, Karnataka, India.
Disease scoring
Plants were scored using the 6-point interaction phenotype scale of Islam et al. (2010) where 0 = no symptoms, 1 = mild mosaic on young leaves covering > 10% of leaf area; 2 = mosaic on young leaves covering > 25% of leaf area; 3 = mosaic on young leaves covering > 50% of leaf area, leaves blistered and puckered of leaves; 4 = mosaic on young leaves covering > 75% of leaf area, leaves distorted; and 5 = mosaic on young leaves covering > 75% area, leaves distorted, plants stunted. For a better comparison between different genotypes, the scores of individual plants thus recorded were used to calculate the vulnerability index (VI) value described by Silbernagel and Jafari (1974) and modified by Bos (1982):
where n0, n1, n2…n5 is the number of plants in score 0, 1, 2…5, nt is the total number of plants, and nc is the total number of categories. Symptoms on each plant were scored each week for 6 or 8 weeks post inoculation to determine VI.
On the basis of the mean VI, the genotypes were classified into five categories: immune, VI = 0; resistant, VI = 1–25; moderately resistant, VI = 26–50; moderately susceptible, VI = 51–75; susceptible, VI = 76–100 (Islam et al. 2011). The area under disease progress curve (AUDPC) was determined using the formula of Cambell and Madden (1990):
where yi is the percentage of diseased plants (VI, 1 to 6 weeks) on the ith observation, ti is the time (days) of observation expressed as dpi, and N is the total number of observations during the experiment.
Results
Field screening after natural infection
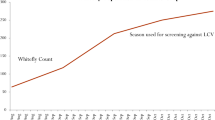
The reactions of the cultivated varieties and advanced breeding lines of Luffa in the field are summarized in Tables 1, 2. The susceptible genotype, Arka Prasan had yellow spots, yellow mosaic, leaf curling, vein thickening, darkening of leaf margins, puckering, and severe plant stunting (Fig. 1a). L. cylindrica L. genotype IIHR-137 was highly resistant to ToLCNDV; no symptoms developed through maturity regardless of the experimental techniques during both years (Tables 1, 2; Fig. 1b). IIHR-137 and IIHR-138 had a low frequency of whitefly visits, which can be attributed to their lack of preference for these genotypes. L. acutangula genotype IIHR-Sel-1 (Fig. 1c) was also fairly resistant in the field and thus classified as resistant (Table 2). Disease development was delayed and very slow in all these genotypes when compared with highly susceptible lines. Among the 52 genotypes, the asymptomatic genotypes IIHR-137 and IIHR-138 (VI 0) and the 16 genotypes with only mild symptoms were classified as resistant (VI 0–25) (Tables 1, 2). Fifteen genotypes were moderately resistant (VI 26–50), and 19 genotypes were moderately susceptible to susceptible (VI > 50) (Tables 1, 2). Disease progress was very high in the susceptible checks Arka Prasan, IIHRDM-16-129-3 and IIHRRG-117 and reached a maximum 5 weeks after inoculation. Disease progress was slow in resistant and moderately resistant lines. Genotype IIHR 138 had mild virus symptoms at the end of the experiment, whereas IIHR 137 was completely symptom free throughout the crop period (Fig. 2).
Among 52 tested genotypes, 15 genotypes, including some highly susceptible lines (as susceptible checks), were further screened using whiteflies or mechanical inoculation with viruliferous sap.
Insect-proof net house screening after inoculations
Whitefly-mediated inoculation
Virus transmission was more certain after whitefly-mediated inoculation. Mild yellow mosaic to severe curling symptoms started appearing 10 dpi. Among the 15 genotypes screened, all tested plants of L. cylindrica IIHR-137 were symptom-free throughout the growing period (VI 0.00). Symptoms from virus infection developed late, and progression was slow in resistant L. cylindrica line IIHR-138 (VI 3.75) and L. acutangula line IIHR-Sel-1 (VI 24.07) (Table 3). Five of the 15 lines were moderately resistant (VI 26–50), whereas seven lines were moderately susceptible to susceptible (VI ≥ 50) (Table 3).
Mechanical sap inoculation
ToLCNDV was transmitted through mechanical sap inoculation to test seedlings in seedling trays in the insect-proof net house. Yellow mosaic was observed on young leaves, and severe curling and stunting of plants started appearing on susceptible entries 30 dpi. Among 15 genotypes, L. cylindrica IIHR-137 was symptom-free (VI 0.00). Two other lines, IIHR-138 (VI 5.00) and IIHR-Sel-1 (VI 22.00), were resistant (Table 3). Disease progression in the resistant genotypes was delayed and very slow. Five genotypes were moderately resistant (VI 26–50); the other seven genotypes were moderately susceptible (VI ≥ 50) (Table 3).
However, all the inoculated plants tested by PCR were positive for amplification of ~ 1.1-kb DNA bands (Fig. 3), and all non-inoculated control plants were negative. Hence, the symptom-free line IIHR-137 was also classified as resistant instead of immune.
Polymerase chain reaction detection of ToLCNDV in inoculated genotypes classified as resistant. M, Lambda DNA/EcoRI + HindIII Marker; C, Control (uninoculated, healthy plant). Lane 1, highly susceptible check Arka Prasan showing high viral accumulation; lanes 2–4, virus is present in asymptomatic plant of IIHR-Sel-1, IIHR-137 and IIHR-138, respectively
Correlation studies between different methods of screening
The variables used to assess disease, VI and AUDPC, for the natural infection and the inoculation screenings were positively correlated, and correlation coefficient values for VI and AUDPC after natural infection, mechanical inoculation, and whitefly-mediated inoculation were 0.968, 0.0.814, and 0.943 (P < 0.001), respectively (Table 4). Results after natural infection in the field were significantly correlation with those after the two inoculation methods with correlation coefficient value of 0.629 and 0.708, respectively. Results after mechanical sap inoculation and whitefly-mediated inoculation in the insect-proof net house were also directly correlated with a correlation coefficient of 0.762 (P < 0.001) (Table 4).
Discussion
The use of ToLCNDV-resistant varieties is key to an integrated disease management programme to reduce crop losses from ToLCNDV. To date, no commercial varieties of any vegetable crop have resistance to ToLCNDV; thus, thorough screening of numerous germplasm sources is needed to identify resistant sources. Here, we screened Luffa germplasm in the field after natural infection and in insect-proof net houses after inoculation (mechanical and whitefly-mediated inoculation) as in previous screening tests for resistance (Islam et al. 2011; López et al. 2015; Sáez et al. 2016; Sohrab et al. 2003, 2014). Mechanical inoculation with viruliferous sap is a simple method to screen large numbers of germplasm stock. Symptoms were observed in all the inoculated plants of Luffa germplasm 15–20 dpi, similar to previous studies on different cucurbit crops (López et al. 2015; Sáez et al. 2016; Sohrab et al. 2014). We observed that successful mechanical sap inoculation required precise climatic conditions, as shown for sponge gourd (Sohrab et al. 2014). Whitefly-mediated inoculation is quick and the most effective method for virus transmission (Pico et al. 1998), and test seedlings developed severe symptoms and had yellow mosaic by 10 dpi as reported for sponge gourd (Islam et al. 2010) and Cucurbita accessions (Sáez et al. 2016). The correlation between the vulnerability index for the natural infection and the inoculation methods (whitefly and mechanical sap) showed significant positive correlation, complimenting each other. Hence, the use of a controlled screening system allows initiating the disease screening trials of ToLCNDV at any time in the season, regardless of the weather. Mechanical sap inoculation is a more recently reported method (López et al. 2015) for screening, and we confirm its correlation with other standard methods. Screening results from the two inoculation methods were positively correlated. Also, the positive correlation between the disease variables indicates the significance and authenticity of the mechanical inoculation in the study. Thus, the mechanical sap inoculation screening should be effective for screening numerous accessions of large ridge gourd germplasm within a short time to identify sources of resistance.
Some of the genotypes screened responded differently after the different inoculation methods. After field infection, all Arka Prasan plants had severe symptoms such as yellow mosaic, curling, and stunting, while plants of IIHRRV-2-5-10, IIHRRV-5-2-4, IIHRRV-8-4-6, IIHRRV-9-1-1, IIHRRV-Sel-3, IIHRRV-9-3-3, and IIHRRV-9-5-5 had mild symptoms, indicating that they had escaped infection in these conditions. Disease intensity also varied significant among resistant and susceptible genotypes (Fig. 2). Hence, integrated management strategies for controlling the disease should be specifically applied during the period identified in the investigation as shown by the rate of disease progress. IIHR-137, IIHR-138, and IIHR-Sel-1 were resistant after field infection; and none of the plants of IIHR-137 had symptoms and a few plants of IIHR-138 and IIHR-Sel-1 had mild symptoms. These three genotypes were also resistant after the two inoculation methods. However, the virus was detected by PCR in the asymptomatic plants of the resistant genotypes; hence, these genotypes are considered resistant to ToLCNDV. Other work to identify ToLCNDV-resistant sources for different vegetable crops has been started. For example, Prasanna et al. (2015) identified a set of tomato ToLCNDV-resistant lines originally derived from wild species Solanum habrochaites, S. chilense, S. peruvianum, and S. pimpinellifolium. Tomato cultivars were also screened for ToLCNDV resistance in Bangladesh (Maruthi et al. 2005). Islam et al. (2011) identified two sponge gourd lines, DSG-6 and DSG-7 (VI 3.3 and 6.0, respectively), with resistance to ToLCNDV after inoculation with a purified virion in an insect-proof net house. López et al. (2015) reported that ToLCNDV can be readily mechanically transmitted via sap to the two most important Cucumis crops, C. melo, and C. sativus; tolerance to ToLCNDV after mechanical sap transmission was also identified for C. melo—within Cucumis melo subsp. agrestis var. momordica and in wild agrestis accessions.
The present study also confirmed ToLCNDV resistance in L. cylindrica IIHR-137 and IIHR-138 collected from ICAR-IARI in Bangalore, which is a hotspot for ToLCNDV screening of cucurbits. The L. acutangula advanced breeding line IIHR-Sel-1 was also found to have promising resistance to ToLCNDV and may be useful in breeding programmes for ToLCNDV resistance. In addition, resistance in the identified lines should be part of integrated disease management approaches such as vector management and cropping practices to prolong the durability of the available resistance.
Because the primary objective of the present study was to identify sources of resistance against ToLCNDV in available germplasm of genus Luffa, we did not measure tolerance to the disease, plant development or fruit yield. However, in general, most of the genotypes were susceptible to ToLCNDV, which caused flower abortion and thus resulted in lack of fruit set. Any fruit that did form were brittle and misshaped. The identified resistant genotypes will be evaluated for horticultural traits in future studies on incorporating resistance into the ridge gourd background. These numerous germplasm sources also need to be screened using a large population size to ensure that susceptible plants do not escape disease. Seed transmission of ToLCNDV in Luffa genus also needs to be studied in case quarantine measures are needed to prevent potential distribution of the virus into new geographical areas.
References
Ashwathappa KV, Venkataravanappa V, Reddy CNL, Krishna Reddy M (2020) Association of Tomato leaf curl New Delhi virus with mosaic and leaf curl disease of chrysanthemum and its whitefly cryptic species. Indian Phytopathol 73:533–542
Bos L (1982) Crop losses caused by viruses. Crop Prot 1:263–282
Cambell CL, Madden LV (1990) Introduction to plant epidemiology. Wiley, New York
Dhillon NPS, Sanguansil S, Singh SP, Masud MAT, Kumar P, Bharathi LK, Yetisir H, Huang R, Canh DX, McCreigh JD (2016) Gourds: bitter, bottle, wax, snake, sponge and ridge. In: Grumet R, Katzir N, Garcia-Mas J (eds) Genetics and genomics of Cucurbitaceae Plant genetics and genomics: crops and models, vol 20. Springer, Cham, pp 155–172
Islam S, Munshi AD, Mandal B, Kumar R, Behera TK (2010) Genetics of resistance in Luffa cylindrica Roem. against Tomato leaf curl New Delhi virus. Euphytica 174:83–89
Islam S, Munshi AD, Verma M, Arya L, Mandal B, Behera TK, Kumar R, Lal SK (2011) Screening of Luffa cylindrica Roem for resistance against Tomato leaf curl New Delhi virus, inheritance of resistance, and identification of SRAP markers linked to the single dominant resistance gene. J Horticult Sci Biotechnol 86:661–667
Ito T, Sharma P, Kittipakorn K, Ikegami M (2008) Complete nucleotide sequence of a new isolate of tomato leaf curl New Delhi virus infecting cucumber, bottle gourd and muskmelon in Thailand. Arch Virol 153:611–613
Karthik T, Ganesan P (2015) Characterization and analysis of ridge gourd (Luffa acutangula) fibres and its potential application in sound insulation. J Text Inst 107:1412–1425
Legg JP, Shirima R, Tajebe LS, Guastella D, Boniface S, Jeremiah S, Nsami E, Chikoti P, Rapisarda C (2014) Biology and management of Bemisia whitefly vectors of cassava virus pandemics in Africa. Pest Manag Sci 70:1446–1453
López C, Ferriol M, Picó MB (2015) Mechanical transmission of Tomato leaf curl New Delhi virus to cucurbit germplasm: selection of tolerance sources in Cucumis melo. Euphytica 204:679–691
Maruthi MN, Alam SN, Kader KA, Rekha A, Cork A, Colvin J (2005) Nucleotide sequencing, whitefly transmission, and screening tomato for resistance against two newly described begomoviruses in Bangladesh. Phytopathology 95:1472–1481
Mitra DK, Nariani TK (1965) A mosaic disease of tori (Luffa acutangula). Indian Phytopathol 18:233–236
Moriones E, Praveen S, Chakraborty S (2017) Tomato leaf curl New Delhi virus: an emerging virus complex threatening vegetable and fiber crops. Viruses 9:264
Padidam M, Beachy RN, Fauquet CM (1995) Tomato leaf curl geminivirus from India has a bipartite genome and coat protein is not essential for infectivity. J Gen Virol 76:25–35
Patil CV, Ramdas SV, Premchand U, Shankarappa KS (2017) Survey, symptomatology, transmission, host range and characterization of begomovirus associated with yellow mosaic disease of ridge gourd in southern India. Virusdisease 28:146–155
Pico B, Diez MJ, Nuez F (1998) Evaluation of whitefly-mediated inoculation techniques to screen Lycopersicon esculentum and wild relatives for resistance to Tomato yellow leaf curl virus. Euphytica 101:259–271
Prasanna HC, Sinha DP, Rai GK, Krishna R, Kashyap SP, Singh NK, Singh M, Malathi VG (2015) Pyramiding Ty-2 and Ty-3 genes for resistance to monopartite and bipartite tomato leaf curl viruses of India. Plant Pathol 64:256–264
Sáez C, Martínez C, Ferriol M, Manzano S, Velasco L, Jamilena M, López C, Picó B (2016) Resistance to Tomato leaf curl New Delhi virus in Cucurbita spp. Ann Appl Biol 169:91–105
Sangeetha B, Malathi VG, Alice D, Suganthy M, Renukadevi P (2018) A distinct seed-transmissible strain of Tomato leaf curl New Delhi virus infecting chayote in India. Virus Res 258:81–91
Silbernagel MJ, Jafari AM (1974) Temperature effects on curly top resistance in Phaseolus vulgaris. Phytopathology 64:825–827
Singh RP, Mohan J, Singh DP (2001) Symptomatology and distribution of ridge gourd mosaic virus. Agric Sci Digest 21:149–152
Sohrab SS, Mandal B, Pant RP, Varma A (2003) First reports of association of Tomato leaf curl New Delhi virus with yellow mosaic disease of Luffa cylindrica in India. Plant Dis 87:1148
Sohrab SS, Karim S, Varma A, Abuzenadah AM, Chaudhary AG, Damanhouri GA, Mandal B (2013) Characterization of Tomato leaf curl New Delhi virus infecting cucurbits: Evidence for sap transmission in a host specific manner. Afr J Biotechnol 12:5000–5009
Sohrab SS, Karim S, Varma A, Azhar EI, Abuzenadah AM, Mandal B (2014) Sap transmission of Tomato leaf curl New Delhi virus infecting sponge gourd in northern India. J Plant Interact 9:241–248
Swarnalatha P, Mamatha M, Manasa M, Singh RP, Krishnaeddy M (2013) Molecular identification of Ageratum enation virus (AEV) associated with leaf curl disease of tomato (Solanum lycopersicum) in India. Australas Plant Dis Notes 8:67–71
Swetha MP, Muthukumar SP (2016) Characterization of nutrients, amino acids, polyphenols and antioxidant activity of ridge gourd (Luffa acutangula) peel. J Food Sci Technol 53:3122–3128
Zaidi SS, Martin DP, Amin I, Farooq M, Mansoor S (2017) Tomato leaf curl New Delhi virus: a widespread bipartite begomovirus in the territory of monopartite begomoviruses. Mol Plant Pathol 18:901–911
Acknowledgements
The authors thank Dr A. D. Munshi for providing seeds of sponge gourd lines DSG-6 and DSG-7. The first author gratefully acknowledges Dr M. K. Reddy for his valuable guidance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare they have no conflict of interest. The manuscript was prepared in compliance with ethical standards.
Animal studies and human participants
This article does not contain any studies with human participants or animal.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kaur, M., Varalakshmi, B., Pitchaimuthu, M. et al. Screening Luffa germplasm and advanced breeding lines for resistance to Tomato leaf curl New Delhi virus. J Gen Plant Pathol 87, 287–294 (2021). https://doi.org/10.1007/s10327-021-01010-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10327-021-01010-z






