Abstract
Nicandra physaloides, a common weed in South America, was found to be infected by an isolate of Tomato severe rugose virus (ToSRV), a bipartite begomovirus. The plants developed severe yellow rugose mosaic and were collected in São Paulo State, Brazil. This isolate of ToSRV was transmitted by Bemisia tabaci B biotype from infected plants of N. physaloides to healthy plants of N. physaloides and tomato in a glasshouse. This is the first report of natural infection of N. physaloides by ToSRV in Brazil.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Tomato severe rugose virus (ToSRV) is one of several species of begomovirus (family Geminiviridae) infecting solanaceous crops in Brazil during the last 15 years. ToSRV was first described infecting tomato crops (Solanum lycopersicum) in Uberlandia County, Minas Gerais State, Brazil, in 1999 causing yield losses up to 80% (L. R. Goulart, Uberlandia Federal University, personal communication). Later, it was identified from potato (Souza-Dias et al. 2008) and chili pepper (Bezerra-Agasie et al. 2006). Currently, ToSRV has become the predominant virus found in tomato crops in the country (Fernandes et al. 2008).
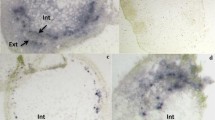
The virus has a genome composed of two circular ssDNA and is transmitted by the whitefly Bemisia tabaci (Gennadius) (Homoptera: Aleyrodidae) in a persistent and circulative manner. During 2007 and 2008, studies were carried out to evaluate the temporal and spatial progress of epidemics caused by ToSRV in tomato crops under field conditions at Sumaré County, State of São Paulo, Brazil, which will be published elsewhere. Nicandra physaloides (family Solanaceae) with severe yellow rugose mosaic (Fig. 1), characteristic of a virus infection, were frequently found around tomato fields. Identical symptomatic plants were also found scattered within maize and sorghum fields in areas previously planted with tomato in the same region. N. physaloides is a common weed (known as apple of Peru) in most vegetable-growing areas in South America, mainly in Brazil, where it is sometimes used as popular medicine (Agra et al. 1994).
Leaf samples obtained from five symptomatic plants randomly selected in the field were analyzed in the laboratory for viruses previously reported in tomato crops in Brazil (Kurozawa and Pavan 2005). We first used a plate trapped antigen–enzyme-linked immunosorbent assay (PTA-ELISA) to detect the presence of Cucumber mosaic virus (CMV), Potato leafroll virus (PLRV), Pepper yellow mosaic virus (PepYMV), Potato virus Y (PVY) and Tomato spotted wilt virus (TSWV) with polyclonal specific antisera against the coat protein of each virus. All five samples were negative for these antisera, while positive and negative controls confirmed the specificity of the tests (data not shown).
Symptomatic leaves, ground in 0.02 M potassium phosphate buffer (pH 7.0) were used to mechanically inoculate the following test plants in the glasshouse: Chenopodium amaranticolor, C. quinoa, Cucurbita pepo ‘Caserta’, Datura stramonium, Gomphrena globosa, Nicotiana clevelandii, N. benthamiana, N. glutinosa, N. physaloides, N. tabacum ‘Havana’, N. tabacum ‘TNN’ and S. lycopersicum. None of the plants developed typical symptoms of mechanically transmitted tomato-infecting viruses. Together, these results suggested that the plants could be infected by another virus.
The samples were further analyzed for the presence of begomoviruses. Initially, total DNA was separately extracted from each sample (Dellaporta et al. 1983) and analyzed with a PCR reaction using the universal primer pairs PAL1v1978/PAR1c496 and PBL1v2040/PCRc1 for begomovirus (Rojas et al. 1993). Two fragments of approximately 1,300 and 600 bp, respectively, were amplified from all five samples (Fig. 2), suggesting infection with a bipartite begomoviruses. These fragments were directly sequenced and compared with other begomovirus deposited in GenBank (http://www.ncbi.nlm.nih.gov/BLAST). The consensus sequence of 1,059 nucleotides (nt) of the begomovirus DNA-A component, comprising the 5′-region of the replication-associated protein (Rep) gene, the entire intergenic region (IR), and the 5′-region of the coat protein (CP) gene, shared 98% nucleotide identity with the corresponding sequence of the DNA-A of ToSRV pepper isolate [BR:PG1:Pep:03] (accession DQ207749). The 501 nt consensus sequence of the DNA-B component, comprising the 5′-region of the MP gene, shared 95% nucleotide identity with the DNA-B of the same isolate of ToSRV (accession EF534708).
PCR amplified fragments of DNA-A (~1300 bp, A) and DNA-B (~600 bp, B) of begomoviruses from total DNA extracted from five (lanes 1–5) field-collected, symptomatic Nicandra physaloides plants; −: healthy N. physaloides; +: tomato control infected with Tomato severe rugose virus; M: 1-kb ladder (Invitrogen, Carlsbad, CA, USA)
To confirm these results, we cloned and completely sequenced the DNA-A component. Viral DNA from three samples was amplified by a rolling circle procedure using the bacteriophage ϕ29 DNA polymerase (TempliPhi; GE Healthcare, Piscataway, NJ, USA). The amplified DNA was separately digested at a single site with restriction endonucleases BamHI, SacI, or EcoRI to generate 2.6-kb DNA linear fragments (Inoue-Nagata et al. 2004). These linearized DNAs were cloned into a pBluescript SK(+) vector (Stratagene, La Jolla, CA, USA) and introduced into Escherichia coli DH5α according to standard procedures (Sambrook et al. 1989). Seven clones representing randomly selected three DNA samples were completely sequenced. Sequences were assembled by the Staden Package program (Staden 1996), and analysis and comparisons were performed using the programs ORF finder (http://www.ncbi.nlm.nih.gov/gorf/gorf) and BLASTn (Altschul et al. 1997).
All seven clones were 2,590 nt long and shared over 99% nucleotide identity. Therefore, clone JR01 was selected for further analysis and herein called nicandra virus. The DNA-A sequence of nicandra virus (accession EU086591) shared 98% nucleotide identity with the DNA-A of ToSRV pepper isolate-[BR:PG1:Pep:03] (accession DQ207749), using DNAMAN (Lynnon BioSoft, Quebec, Canada), confirming its identification, hence designated as ToSRV-[BR:Sum1:Nic:07]. Five ORFs were identified: AV1, coding for the coat protein (CP); AC1, coding for the replication-associated protein (Rep); AC2, coding for the trans-activating protein (TrAP); and AC3, coding for the replication enhancer protein (REn). This genome organization is typical of bipartite New World begomoviruses (Stanley et al. 2005). An approximately 300-nt intergenic region (IR) containing the origin (ori) of replication was also identified. The deduced amino acid sequences of this virus shared identities of 99% for AV1, 99% for AC1, 98% for AC2, 98% for AC3, and 98% for AC4 with ToSRV-[Br:PG1:Pep:03].
A phylogenetic tree of DNA-A sequences (Fig. 3), constructed using the program MEGA 4.0 (Tamura et al. 2007) after multiple alignments obtained with the program Clustal_W (Thompson et al. 1994), confirmed the close evolutionary relationship of the nicandra virus with pepper ToSRV-[BR:PG1:Pep:03] (accession no. DQ207749) and tomato ToSRV-[BR:Ube1:00] (accession AY029750) isolates and with Tomato rugose mosaic virus (ToRMV-[BR:Ube1:96], accession AF291705).
Phylogenetic tree of DNA-A of nicandra virus with other begomoviruses reported in Brazil: Bean golden mosaic virus (BGMV, NC_004042); Blainvillea yellow spot virus (BlYSV-[BR:Coi25:07], EU710756); Sida common mosaic virus (SiCmMV-[BR:Coi4:07], EU710751); Sida micrantha mosaic virus (SimMV-[BR:A2B2], AJ557451); Sida mottle virus (SiMoV-rho[BR:Vic1:99], AY090555); Sida yellow leaf curl virus (SiYLCV-[BR:Coi3:07], EU710750); Sida yellow mosaic virus (SiYMV-[BR:Vic2:99], AY090558); Tomato chlorotic mottle virus (ToCMV, NC_003664); Tomato common mosaic virus (ToCmMV-[BR:Coi22:07], EU710754); Tomato golden mosaic virus (TGMV-[BR:Com:84], K02029); Tomato leaf distortion virus (ToLDV- [BR:Pda4:05], EU710749); Tomato mild mosaic virus (ToMIMV-[BR:Pda58:05], EU710752); Tomato rugose mosaic virus (ToRMV-[BR:Ube1:96], AF291705); Tomato severe rugose virus (ToSRV-[BR:Ube1:00], AY029750 and ToSRV-[BR:PG1:Pep:03], DQ207749); Tomato yellow spot virus (ToYSV-[BR:Bic2:99], DQ336350); Tomato yellow vein streak virus (ToYVSV-[Ba-3], EF417915). Tomato yellow leaf curl virus (ToYLCV, NC_004005), a monopartite begomovirus, was used as the outgroup. Phylogenetic tree generated in MEGA 4.0 using a neighbor-joining method with 1000 bootstrap replications and default parameters. The numbers at the nodes indicate the percentage bootstrap values. The scale indicates the number of substitutions per site. The nomenclature for the viruses follows the guidelines proposed by Castillo-Urquiza et al. (2008) and Fauquet et al. (2008)
Virus-free adults of B. tabaci biotype B were caged in a 50 ml polypropylene tube containing ToSRV infected leaves of N. physaloides for an acquisition access period of 48 h. Viruliferous whiteflies were then transferred to healthy tomato and N. physaloides test-plants for a transmission access period of 5 days. Approximately 25 whiteflies were confined on each pot containing two plants of each species, with three replicates. Twenty days after inoculation, all test plants of both species developed symptoms of virus infection (data not shown). The virus was further transmitted by whiteflies from infected tomato and N. physaloides test plants to healthy seedlings of both species (data not shown). The infection was evaluated by PCR with primers PAL1v1978/PAR1c496 (Rojas et al. 1993) and subsequent direct sequencing the 5′-region of the CP gene confirmed the identity of the transmitted viruses as ToSRV isolates.
This is the first record of the occurrence of ToSRV infected N. physaloides under field conditions, although natural infection of this species with other begomoviruses have been reported earlier in Brazil (Fernandes et al. 2006; Inoue-Nagata et al. 2003). Together, these data suggest that N. physaloides can serve as natural reservoirs of ToSRV for primary infection of tomato crops. Therefore, management strategies for begomoviruses in tomato crops should seriously consider the elimination of N. physaloides before transplanting tomato seedlings.
References
Agra MF, Rocha EA, Formiga SC, Locatelli EM (1994) Plantas medicinais dos Cariris Velhos, Paraíba. Parte I: subclasse Asterideae (in Portuguese). Rev Bras Farm 75:61–64
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search program. Nucleic Acids Res 25:3389–3402
Bezerra-Agasie IC, Ferreira GB, Ávila AC, Inoue-Nagata AK (2006) First report of Tomato severe rugose virus in chilli pepper in Brazil. Plant Dis 90:114
Castillo-Urquiza GP, Beserra JEA Jr, Bruckner FP, Lima ATM, Varsani A, Alfenas-Zerbini P, Zerbini FM (2008) Six novel begomoviruses infecting tomato and associated weeds in southeastern Brazil. Arch Virol 153:1985–1989
Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA minipreparation: Version II. Plant Mol Biol Rep 1:19–21
Fauquet CM, Briddon RW, Brown JK, Moriones E, Stanley J, Zerbini M, Zhou X (2008) Geminivirus strain demarcation and nomenclature. Arch Virol 153:783–821
Fernandes JJ, Carvalho MG, Andrade EC, Brommonschenkel SH, Fontes EPB, Zerbini FM (2006) Biological and molecular properties of Tomato rugose mosaic virus (ToRMV), a new tomato-infecting begomovirus from Brazil. Plant Pathol 55:513–522
Fernandes FR, Albuquerque LC, Giordano LB, Boiteux LS, Ávila AC, Inoue-Nagata AK (2008) Diversity and prevalence of Brazilian bipartite begomovirus species associated to tomatoes. Virus Genes 36:251–258
Inoue-Nagata AK, Ribeiro SG, Rocha WB, Albuquerque LC, Ávila AC, Giordano LB (2003) A new begomovirus isolated from Nicandra physaloides. Virus Rev Res 116:186
Inoue-Nagata AK, Albuquerque LC, Rocha WB, Nagata T (2004) A simple method for cloning the complete begomovirus genome using the bacteriophage phi 29 DNA polymerase. J Virol Methods 116:209–211
Kurozawa C, Pavan MA (2005) Doenças do Tomateiro. In: Kimati H, Amorin L, Rezende JAM, Bergamin Filhjo A, Camargo LEA (eds) Manual de Fitopatologia - Doenças da Plantas Cultivadas (in Portuguese). Editora Agronômica Ceres Ltda, São Paulo, pp 607–626
Rojas MR, Gilbertson RL, Russell DR, Maxwell DP (1993) Use of degenerate primers in the polymerase chain reaction to detect whitefly-transmitted geminiviruses. Plant Dis 77:340–347
Sambrook J, Fritsch EF, Maniatis T (eds) (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Press, New York
Souza-Dias JAC, Sawazaki HE, Pernambuco-Fo PCA, Elias LM, Maluf H (2008) Tomato severe rugose virus: another begomovirus causing leaf deformation and mosaic symptoms on potato in Brazil. Plant Dis 92:487
Staden R (1996) Indexing and using sequence databases. Methods Enzymol 266:105–114
Stanley J, Bisaro DM, Briddon RW, Brown JK, Fauquet CM, Harrison BD, Rybicki EP, Stenger DC (2005) Family Geminiviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA (eds) Virus taxonomy. Eighth Report of the International Committee on Taxonomy of Viruses. Elsevier-Academic Press, San Diego, pp 301–326
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL_W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Barbosa, J.C., Barreto, S.S., Inoue-Nagata, A.K. et al. Natural infection of Nicandra physaloides by Tomato severe rugose virus in Brazil. J Gen Plant Pathol 75, 440–443 (2009). https://doi.org/10.1007/s10327-009-0198-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10327-009-0198-5