Abstract
Geminiviruses are single-stranded (ss) DNA viruses that not only cause devastating diseases of important food and fiber crops worldwide, but also are important models to study fundamental aspects of virus-induced gene silencing and RNA interference. As a counterdefense mechanism, viruses have evolved various antisilencing strategies that are being progressively unraveled. The geminiviruses, ssDNA molecules that replicate inside the nucleus and therefore have no dsRNA phase during replication, can both induce and become targets of gene silencing. Proteins AC2 (encoding the transcriptional activator protein) and AC4 of bipartite geminiviruses and protein C2, a positional homolog of AC2 of monopartite viruses, have been identified as suppressors of posttranscriptional gene silencing. The majorities of geminiviral suppressors characterized to date do not share any obvious structural or sequence similarity across families and groups except that they have been identified as pathogenicity determinants. This review mainly focuses on the geminivirus-encoded suppressors of RNA-silencing—the βC1 and V2 proteins—and their possible role in the interference of silencing at different steps in the pathways.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
RNA-silencing, including posttranscriptional gene silencing (PTGS in plants) and RNA interference in animals and gene quelling in fungi, represents a sequence-specific RNA degradation mechanism directed against invasive nucleic acid molecules (Napoli et al. 1990; Cogoni and Macino 1997; Fire et al. 1998). RNA-silencing, a robust host defense mechanism against plant viruses, is generally countered by virus-encoded silencing suppressors. PTGS, a sequence-specific defense mechanism that can target both cellular and viral mRNA for degradation, is widely used as a tool for inactivating gene expression. Three initially unrelated lines of research led to the recognition of RNA-silencing as an important means of defense against viruses. The first clue came from studies of transgene-induced RNA-silencing in which attempts to overexpress endogenous genes by introducing additional copies resulted instead in turning off the endogenous gene as well as the transgene (Napoli et al. 1990). The second line of research led to the discovery of pathogen-derived resistance in that RNA-silencing directed against a viral transgene provided resistance to any virus carrying the target sequence (Baulcombe 1996; Dougherty and Parks 1995). Thus viruses could be targets of RNA-silencing. The third clue came from studies of synergistic viral diseases (Kasschau and Carrington 1998; Fondong et al. 2000; Vanitharani et al. 2004).
A common feature of RNA-silencing involves structured or double-stranded (ds) RNA that is processed into small interfering (si) RNAs of 21–25 nucleotides by the enzyme Dicer, a member of the RNase III family of dsRNA-specific endonucleases. The siRNAs become incorporated into an RNA-induced silencing complex (RISC) via a Dicer-associated protein R2D2 that links the initiation and execution of RNA-silencing. There are at least three different pathways in the gene silencing mechanism: cytoplasmic siRNA silencing, silencing of endogenous mRNAs by microRNAs (miRNAs), and DNA methylation and suppression of transcription (Baulcombe 2004).
The recent development of molecular techniques has led to significant advances in our knowledge of geminiviruses, their genome and role in disease etiology. Geminiviridae family is divided into four genera: Mastrevirus, Curtovirus, Topocuvirus and Begomovirus, depending on the genomic, host and vector characteristics (Fauquet and Stanley 2005). Detailed genome organizations of the family Geminiviridae are shown in Fig. 1. They have geminate (twinned) particles approximately 18–20 nm in diameter and 30 nm long, consisting of two incomplete T = 1 icosahedra joined together in a structure with 22 pentameric capsomers and 110 identical protein subunits. There are now 200 officially recognized geminivirus species, of which 147 belong to the genus Begomovirus, and there are almost 592 complete nucleotide sequences deposited in databases (Fauquet et al. 2007), reflecting their economic importance and enormous diversity resulting from their widespread geographic distribution and host adaptation. Geminivirus DNA components vary in size between 2,500 and 3,100 nucleotides depending on the virus; each encodes two or more genes that are distributed between both the virion-sense and the complementary-sense DNA strands and are transcribed bidirectionally from an intergenic region that also contains the origin of replication (Hanley-Bowdoin et al. 1999). Although the majority of begomoviruses have bipartite genomes, an increasing number are being identified that have only a single DNA component equivalent to the DNA-A component of the bipartite viruses. Tomato yellow leaf curl virus (TYLCV) and Cotton leaf curl virus (CLCuV) are the most notable and economically most significant examples of a monopartite begomovirus (Navot et al. 1991; Sharma and Rishi 2007). Genome replication occurs in the nucleus (Fig. 2) by a rolling circle mechanism that employs circular, double-stranded (dsDNA) replicative-form (RF) intermediates. The dsDNA molecules, which serve as replication and transcription templates, are associated with histones and assembled into minichromosomes.
Genomic organization of Geminiviridae family. Open reading frames (ORFs) are denoted (black arrows) as either being encoded on the virion sense (V) or complementary sense (C) strands, preceded by component designated (A or B) in case of bipartite and DNAβ in case of monopartite begomoviruses. That part of the intergenic region (IR) whose sequence is identical in bipartite begomovirus components is called the common region (CR). The complementary strand origin of replication in Mastrevirus in the short intergenic region (SIR) and the position of the stem loop motif containing the conserved nanonucleotide sequence in the large intergenic region (LIR) are shown. Introns (open boxes) occur in ORF V1 and at the overlap between ORFs C1 and C2. In Curtovirus, the C2-encoded protein does not seem to have transcriptional activator protein (TrAP) activity. The position of the stem loop motif is shown at the top of each genomic component. Rep replication-associated protein; CP coat protein; Ren replication enhancer; MP movement protein; NSP nuclear shuttle protein; SCR satellite conserved region; A-rich adenine rich region
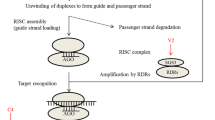
RNA-silencing model encompassing geminivirus suppressor features. Geminivirus replication cycle as shown in the nucleus, where dsDNA (RF) serves as a potential target of methyltransferses, which modify the DNA and histone proteins. Cytoplasmic RNA-silencing (PTGS) ultimately degrades the target mRNA, and siRNA-directed methylation leads to TGS. During trans-methylation AC2 and C2 (Curtovirus) proteins interfere with the methyl cycle by inhibiting ADK. Dicer cleaves dsRNA into siRNA, and RISC then distinguishes different strands of siRNA forms. The sense strand is degraded (not shown), while the anti-sense strand is used to target the genes for silencing. AC4 protein binds single-stranded siRNA forms. βC1 of TYLCCNV-[Y10] suppresses silencing by acting in the nucleus, while AC2 protein of MYVMV-[Vig] inactivates transcription of host genes (WEL 1), which suppress silencing by an unknown manner. In contrast, begomoviruses like βC1 protein of ToLCJAV DNAβ and V2 protein of TYLCV-[IL] suppress silencing and are localized in the cytoplasm. Still precise mechanisms of these suppressors are not known
Geminiviruses do not encode DNA or RNA polymerases and so depend on cellular replication and transcription machinery to express their genes and amplify their genomes (Hanley-Bowdoin et al. 1999). Consequently, they are good models for the study of host replication, transcription, and how these processes can be affected by epigenetic modification. The bipartite begomoviruses infect dicotyledonous plants, are whitefly (Bemisia tanaci Genn.) transmissible such as Bean golden mosaic virus (BGMV), Tomato golden mosaic virus (TGMV), African cassava mosaic virus (ACMV) and Mungbean yellow mosaic virus (MYMV), have genomes consisting of two components. These bipartite viruses are further divided into Old World (ACMV, MYMV) and New World (BGMV), having DNA-A and DNA-B genome components of similar size that differ in sequence except for an identical common region (CR) of ∼250 bp that differs in viruses such as BGMV (Morinaga et al. 1987) and MYMV (Morinaga et al. 1993). All geminiviruses, regardless of genomes, have a similar intergenic region (IR), which contains a stem loop structure, RNA II promoters and a nucleosome-free region in the minichromosomes (Pilartz and Jeske 2003). Monopartite begomoviruses, which are transmitted by whitefly are confined to the Old World, and some of them are associated with a satellite DNAβ required for the induction of disease symptoms (Briddon and Stanley 2006). Recently, begomoviruses and the curtoviruses were shown to encode proteins that are suppressors of RNA-silencing (Vanitharani et al. 2004; Trinks et al. 2005; Avi et al. 2007; Kon et al. 2007; Bisaro 2006). There has been no report of a silencing suppressor of mastreviruses. So far, 29 proteins that inhibit RNA-silencing and counter antiviral RNA-silencing have been identified in several plant and animal viruses (Kasschau and Carrington 1998; Li et al. 2002; Anandalakshmi et al. 1998). Three distinct phases have been identified in RNA-silencing: initiation, maintenance and systemic signaling or the effectors step (Llave et al. 2000). However, these suppressor proteins do not share homology at either the sequence or viral functional levels. These identified suppressor proteins might target similar or different steps of the RNA-silencing pathways.
A few reviews on RNA-silencing suppressors have been published on viral-encoded suppressors (Baulcombe 2004; Ding et al. 2004; Voinnet 2005; Vanitharani et al. 2005; Bisaro 2006). In this review, however, we discuss recent information regarding geminiviruses that encode silencing suppressors and how geminiviruses can encode RNA-silencing proteins and the mechanism of action of these proteins. We will highlight interesting features shared by a few of the better-studied suppressors with the intention of generating new ideas for future research.
Molecular basis of RNA-silencing machinery
Recently, tremendous progress has been made in understanding the various silencing pathways. At least three basic silencing pathways have been identified, all of which are potentially antiviral: (1) siRNA-mediated degradation of abundant or aberrant mRNAs, including viral mRNAs and RNA genomes (PTGS or RNAi); (2) miRNA-mediated silencing involved in translational inhibition or degradation of cellular mRNAs, and viral mRNAs and RNA genomes; (3) siRNA-directed de novo methylation of DNA and histone proteins (e.g. H3K9), leading to transcriptional gene silencing (TGS). The third pathway supports methylation of DNA virus genomes, inhibiting virus replication and/or transcription. That is, RNA-directed methylation is a novel form of defense against DNA viruses. At least one geminivirus-silencing suppressor protein has been hypothesized to counter this defense by inhibiting methylation reactions (Wang et al. 2003). RNA-silencing in multicellular plants and animals is mediated by 21–24 nt small RNAs (sRNAs) that guide sequence-specific gene regulation, chromatin modification, and defense against viruses. These sRNAs are broadly classified into miRNAs and siRNAs, which have similar chemical structures but differ in function and mode of biogenesis. Production of both types of sRNAs depends on the activity of Dicer proteins. Plants such as Arabidopsis have evolved a diversity of RNA-silencing pathways, sRNA classes and Dicer-like (DCL) genes that are unmatched in other eukaryotes (Meins et al. 2005; Vaucheret 2006).
Since the discovery of RNA-silencing in animal model systems (Fire et al. 1998; Li et al. 2002), work on dissecting the RNA-silencing machinery has been progressing rapidly. Though the RNA-silencing mechanism in plants is the major focus of research, knowledge about the RNA-silencing machinery in plants also has built on information gathered from several animal model systems. Parts of the conserved RNA-silencing machinery have been studied in many organisms ranging from plants to insects to mammals and back to protozoans. A comprehensive model of RNA-silencing that encompasses many features of geminiviruses is illustrated in Fig. 2. The key action of RNA-silencing involves sequence-specific cytoplasmic degradation of RNA molecules. It can be induced in a variety of ways. For instance, plant viral RNAs can be targeted after the transgenic expression of over-abundant or dsRNA. The key intermediary element in the RNA-silencing pathway is dsRNA, which is recognized and cleaved by the dsRNA-specific nuclease Dicer to yield 21–24 nucleotides (nt) (Hamilton et al. 2002). These siRNAs subsequently serve as guides for cleavage of homologous RNA molecules, mediated by RISC. Endogenous gene silencing can occur at the transcriptional and the posttranscriptional levels. In PTGS, mRNA is degraded or repressed translationally; in TGS, DNA and/or histones are modified, leading to heterochromatization and transcriptional repression. PTGS and TGS are often correlated with the appearance of siRNAs 21–24 nt long, derived from silenced sequences. The miRNA and trans-acting siRNA (ta-siRNA) pathways, which play a crucial role in developmental gene regulation in plants (Bartel 2004; Meins et al. 2005), are PTGS-related processes, in which the respective small RNAs, miRNAs and ta-siRNAs are derived from separate genetic loci and act in trans to silence their target genes. One of the siRNA strands is channeled to the RISC or an RNA-induced initiation of TGS (RITS) complex to guide these effectors to their respective targets. Both RISC and RITS appear to contain a distinct Argonaute (AGO) protein as an active component (Zilberman et al. 2003; Baumberger and Baulcombe 2005). In plants, nematodes and fungi, an RNA-dependent RNA polymerase (RdRP) plays an important role in RNA-silencing, most likely by converting single-stranded transcripts into dsRNA (Meins et al. 2005). Indeed, virus-derived siRNAs have been detected in plants infected with various RNA viruses (Szittya et al. 2002; Xie et al. 2004; Molnar et al. 2005) and DNA geminiviruses (Chellappan et al. 2004; Kon et al. 2007). Furthermore, both RNA and DNA viruses encode distinct suppressors of RNA-silencing that target different components of this system (Voinnet 2005). In particular, tombusvirus p19 protein selectively sequesters 21-nt siRNA duplexes (Lakatos et al. 2004). Geminivirus suppressor protein AC4 appears to selectively bind single-stranded sRNAs (Fig. 2) including miRNAs (Chellappan et al. 2005). The latter observation is consistent with the hypothesis that not only the siRNA but also the miRNA pathway might restrict virus replication, as demonstrated for a mammalian retrovirus (Lecellier et al. 2005). In line with this idea, most viral silencing suppressors, when overexpressed in transgenic plants, interfere with production and/or action of miRNAs, thus leading to various abnormalities of plant development, often resembling viral symptoms (Voinnet 2005).
Very little is known about the biogenesis and possible modification of virus-derived siRNAs. On the basis of biochemical studies of RNAi in animal systems (Elbashir et al. 2001) and wheat germ extracts (Tang et al. 2003), viral siRNAs are assumed to be duplexes with 2 nt in 3′-overhangs produced from longer perfect dsRNA by Dicer activity. However, predominantly the positive strand of RNA virus-derived siRNAs accumulates, suggesting that at least some sRNAs are produced as miRNA-like duplexes from secondary structure elements of the single-stranded viral genomes (i.e., imperfect dsRNA), rather than from the replicative intermediates (i.e., perfect dsRNA) (Szittya et al. 2002; Xie et al. 2004). Genetic evidence suggests that the four Arabidopsis DCL genes have diversified (Xie et al. 2004) partially redundant functions (Gasciolli et al. 2005). DCL1 is involved in the production of primarily 21- and 22-nt miRNAs from hairpin-like precursor transcripts (Kurihara and Watanabe 2004). DCL3 produces larger 24–26 nt repeat-associated siRNAs (ra-siRNAs), involved in TGS of the respective repetitive DNA loci, presumably from dsRNA precursors generated in a Pol IV- and RDR2-dependent pathway (Xie et al. 2004; Gasciolli et al. 2005; Herr et al. 2005). The miRNA negatively regulate their target mRNAs, either by inhibiting translation or by degradation. In plants, miRNAs are usually perfectly complementary to their target mRNAs and direct RISC cleavage in essentially the same manner as siRNAs (Llave et al. 2002). DCL4 produces 21 nt ta-siRNAs from perfect dsRNA substrates generated by RDR6 on the miRNA-cleaved transcripts of ta-siRNA genes (Gasciolli et al. 2005; Allen et al. 2005; Xie et al. 2005). The function of DCL2 is still unclear, but it seems to be a redundant DCL in the production of endogenous sRNAs. It is still unclear which DCLs are involved in producing virus-derived siRNAs. Neither the DCL3 null mutation dcl3-1 nor the DCL1 weak mutation dcl1-7 compromised accumulation of RNA virus-derived siRNAs (Xie et al. 2004). Although Turnip crinkle virus (TCV) siRNA production was compromised in the Arabidopsis DCL2 mutant dcl2-1 early in infection, at late stages TCV siRNAs did accumulate to wild-type levels. Moreover, two other RNA viruses Cucumber mosaic virus (CMV) and Turnip mosaic virus (TuMV) produced wild-type levels of siRNAs in dcl2-1 plants at both early and late stages of infection (Xie et al. 2004).
In addition to DCL1, two more Arabidopsis genes have been implicated in the biogenesis of miRNAs such as HEN1 and HYL1 (Vazquez et al. 2004; Xie et al. 2004). The processing of the primary miRNA (pri-miRNA) into the miRNA duplex most probably occurs in the nucleus, but it is also guided by DCL1. The HYL1 (Han et al. 2004) product has a dsRNA-binding motif and can physically interact with the DCL1 protein; other members of HYL1 family dsRNA-binding protein have also been proposed to interact with distinct DCLs (Hiraguri et al. 2005). Interestingly, HEN1 is involved not only in miRNA biogenesis, but also in transgene silencing and natural virus resistance as shown by a CMV-based sensitivity assay (Boutet et al. 2003). HEN1 encodes a methyl transferase that methylates the last nucleotide of miRNAs at the 2′-O- or 3′-O position (Yu et al. 2005), with the 2′-OH claimed to be the major target of the modification (Ebhardt et al. 2005).
Compared to plants, processing of miRNA precursors in animals is different. The pri-miRNAs, synthesized by the RNA polymerase II, are first processed by a nucleus-specific enzyme, Drosha, initially discovered in Drosophila (Filippov et al. 2000), into precursor miRNAs (pre-miRNAs) (Lee et al. 2003). These pre-miRNAs, imperfect hairpins of approximately 70 nt, are then exported to the cytoplasm and processed into miRNAs by cytoplasmic Dicer.
Recent evidence suggests that all endogenous sRNAs in Arabidopsis are methylated by HEN1, which protects them from a 3′-end uridylation activity (Li et al. 2005). So far, HEN1 has not been found to methylate virus-derived sRNAs, albeit the bulk signal of CMV-derived siRNAs in Nicotiana benthamiana was shown to be resistant to β-elimination (Ebhardt et al. 2005), suggesting a 3′-terminal nucleotide modification.
Another fascinating feature of RNA-silencing is its movement from cell to cell and systemically throughout the plant (Hamilton et al. 2002; Bernstein et al. 2001). The patterns of systemic silencing suggest that the signal moves from cell to cell and through the phloem, resembling viral movement through the plant (Mlotshwa et al. 2002), and the fact that many viral silencing suppressors are typically required for long-distance spread in the infected plant (Voinnet et al. 2000; Li and Ding 2001; Kasschau and Carrington 2001; Ding et al. 2004) suggests that the signal is a crucial component of the antiviral defense system. Perhaps more importantly, viral suppressors of silencing also provide unique tools to understand the mechanism of RNA-silencing. Much of what is currently known about the RNA-silencing pathway comes from elegant in vitro and genetic studies in organisms other than plants (Tijsterman et al. 2002). In fact, traditional genetic approaches have led to the identification of a number of cellular genes required for RNA-silencing (Dalmay et al. 2000; Mourrain et al. 2000). Interestingly, all these genes are required for sense but not for amplicon transgene-induced silencing (Boutet et al. 2003). The plant viral suppressors, many of which appear to work downstream of dsRNA, provide a novel means of entry into parts of the silencing pathway that are not easily accessible by genetic means. The currently known suppressors appear to work a number of points in the pathway where silencing can be controlled.
Functions of RNA-silencing suppressor proteins
Mainly four types of viral suppressors of RNA-silencing have been distinguished using different assays (Table 1). The assay involves transgenic N. benthamiana plants carrying a highly expressed GFP transgene (Vanitharani et al. 2004; Gopal et al. 2007; Avi et al. 2007; Kon et al. 2007), systemic RNA-silencing of the GFP transgene is induced to completion by agroinfilteration with 35S GFP before the plants are infected with viruses carrying a suppressor or by using a Potato virus X (PVX)-based assay (Brigneti et al. 1998). Geminiviruses are single-stranded DNA (ssDNA)-containing plant viruses that replicate in the nuclei of host cells by a rolling circle mechanism that involves dsDNA intermediates that associate with cellular histone proteins (Hanley-Bowdoin et al. 1999). In a comprehensive model of RNA-silencing, geminiviral-encoded suppressors are envisioned to act at different steps in the silencing pathways (Fig. 2). Like most viruses, geminiviruses are initiators and targets of RNA-silencing and encode proteins that suppress this adaptive host defense. In plants, PTGS acts as a natural antiviral defense system and plays a role in genome maintenance and development. During the past decade, there has been considerable evidence of PTGS suppression by viruses, which is often required to establish infection in plants. In particular, geminiviruses, which have no double-stranded RNA phase in their replication cycle, can induce and suppress the PTGS and become targets of PTGS. Hence, geminiviruses are of interest considering that these viruses replicate in the nucleus and their genomes consist of DNA and do not encounter a dsRNA phase in its replication cycle.
How do geminiviruses trigger PTGS in plants? Replicative forms serve as the template for both replication and transcription. The transcription is bidirectional with two major polycistronic transcripts in opposite orientations occurring from the CR that contains the bidirectional promoter sequences. The virion sense AV2-AV1 (CP) transcript and the complementary sense AC1–AC3 transcript overlap by 4 bp at their 3′ ends as demonstrated by Chellappan et al. (2004). It was therefore suggested that the overlapping transcripts in opposite polarity at the 3′ end might generate dsRNA due to complementary base pairing, which could induce PTGS (Voinnet 2001). Hence, geminivirus-derived dsRNA intermediates never occur during replication. It has, however, been reported that geminiviral mRNAs in the plant are targeted by RNA-silencing in a plant RdRP(RDR6, previously named SGS2/SDE1) dependent manner (Muangsan et al. 2004). None of the majority of plant viral suppressors characterized to date share structural or sequence homology across viral families and groups. The only feature shared by many suppressors is that they are often identified as pathogenicity determinants. Suppressor activity has been identified in structural as well as nonstructural proteins, replication enhancers, transcriptional activators and movement proteins. Therefore, researchers are faced with a plethora of potential mechanisms to unravel.
Bipartite begomoviruses possess an additional monodirectional promoter for the leftward gene AC2 (Shivaprasad et al. 2005) that codes for a transactivator protein that activates viral and host transcription and suppresses PTGS (Trinks et al. 2005). Geminiviruses do not obligatorily produce long dsRNA during their life cycle, and their processed leftward and rightward transcripts overlap only in a short region (Shivaprasad et al. 2005). However, aberrant RNA transcription on a circular viral DNA could potentially lead to production of longer antisense transcripts that might trigger RNA-silencing, and such aberrant transcripts derived from the “nontranscribed” promoter region of the Mungbean yellow mosaic virus-[Vig] (MYMV-[Vig] DNA-A were detected by Shivaprasad et al. (2005). Recently, Rashid et al. (2006) detected begomoviral sRNAs (21, 22 and 24 nt) of both polarities, representing both coding and intergenic regions. These viral sRNAs, similar to siRNAs derived from a dsRNA transgene, and endogenous ta-siRNAs and miRNAs were phosphorylated at the 5′ end and modified at the 3′-terminal nucleotide. Genetic evidence indicated that DCL3, DCL2, at least one additional DCL activity and HEN1 are involved in the biogenesis of begomoviral siRNAs. Genetic analysis suggests that both TGS- and PTGS-related silencing pathways are involved in plant geminivirus interactions.
Suppressors encoded by monopartite and bipartite begomoviruses and curtoviruses
Circular ssDNA begomoviruses are further divided on the basis of genome organization: bipartite begomoviruses, monopartite begomoviruses, monopartite begomoviruses with associated DNAβ satellites. AL2/L2 nomenclature is being used in some cases such as TGMV and Beet curly top curtovirus (BCTV), but following International Committee on Taxonomy of Viruses (ICTV) guidelines, we have denoted AL2/L2 as AC2/C2 followed by the virus names. We have listed the different suppressor categories of geminiviruses as mentioned in Table 1 and discuss them next.
Bipartite begomoviruses
Several begomoviruses encode suppressors of RNA-silencing. Mainly two proteins, AC2 and AC4, were shown to have suppressor activity that indicates PTGS (Gopal et al. 2007; Kon et al. 2007; Trinks et al. 2005; Vanitharani et al. 2004; van Wezel et al. 2003). The AC2, AL2 protein or TrAP (transcriptional activator protein), which suppresses RNA-silencing by controlling the expression of host genes coding for positive or negative effectors of RNA-silencing (Trinks et al. 2005; Vanitharani et al. 2005). ORF AC4 lies entirely within the Rep (AC1)-coding region of bipartite begomoviruses and is one of the least conserved among all geminiviruses, making its study difficult. The suppressor activity of these proteins differs significantly between species.
The RNA-silencing suppressor proteins AC2 and AC4, encoded by bipartite begomoviruses, were recently reviewed by Bisaro (2006), thus we have limited our discussion to information on recent studies on a silencing suppressors. AC2 suppressor of TGMV interacts with and inactivates adenosine kinase (ADK), an important cellular enzyme required for adenosine salvage and methyl cycle maintenance (Fig. 2), suggesting that ADK activity is required to suppress silencing (Wang et al. 2005). The AC2 resembles a typical transcription factor in several respects: it has a nuclear localization signal (NLS), a zinc finger-like domain composed of cysteine and histidine residues, and an acidic activation domain (Dong et al. 2003; Shivaprasad et al. 2005). However, dsDNA-binding activity is weak and sequence nonspecific, and AC2 is probably targeted to responsive promoters by its interactions with cellular proteins. The identities of these proteins and of those in contact with the activation domain are not yet known. Recently, Yang et al. (2007) demonstrated that TGMV AC2 self-interaction correlates with nuclear localization and efficient activation of transcription, whereas AC2 (TGMV) and C2 (BCTV) monomers can suppress local silencing by interacting with ADK in the cytoplasm. Several groups have independently demonstrated the potent silencing suppressor activity of AC2 and AC4 from a number of different geminiviruses using agroinfiltration assays (Voinnet et al. 1999; Vanitharani et al. 2004; Trinks et al. 2005). Chellappan et al. (2004) demonstrated that in case of African cassava mosaic virus-[CM] (ACMV-[CM])-infected plants; the presence of virus specific siRNA promotes the degradation of the corresponding miRNA in a sequence-specific manner, which in turn affects viral replication and transcription. As a result, virus titer and symptom development was greatly reduced in new leaves, indicative that the viral suppressor is important in determining recovery phenotypes (Szittya et al. 2002). In the case of ACMV-[KE], the AC2 is a mild suppressor of PTGS (Voinnet et al. 1999).
Transgenic expression of AC4 showed that it is responsible for symptom determination in several bipartite geminiviruses (Latham et al. 1997; Piroux et al. 2007), while disruption of the C4 ORF of monopartite begomoviruses results in attenuated symptoms and low infectivity, suggesting that it is involved in either symptom development, virus movement or both (Jupin et al. 1994). Recently, different research groups have shown that Sri Lankan cassava mosaic virus (SLCMV), East African cassava mosaic virus (EACMV), Indian cassava mosaic virus (ICMV), ACMV-[CM] suppresses PTGS (Vanitharani et al. 2004; Fondong et al. 2007). In contrast to the behaviour of the BCTV C4 protein, TGMV AC4 protein does not contribute to the disease phenotype (Pooma and Petty 1996), suggesting redundancy as a consequence of the second genomic component encoding factors responsible for intra- and intercellular virus movement. However, this is not always the case; the AC4 protein of the bipartite begomoviruses such as ACMV and EACMV can induce developmental abnormalities when expressed as transgenes, a phenomenon attributed to their ability to bind to miRNA and siRNA to suppress PTGS (Vanitharani et al. 2004; Chellappan et al. 2005).
Plant protein myristoylation is only now gaining great interest and is involved in disease resistance (de Vries et al. 2006), salt tolerance (Ishitani et al. 2000) and growth regulation (Raices et al. 2001). Fondong and associates (2007) thus examined the role of conserved, amino terminal consensus myristoylation and palmitoylation sites for the EACMV-[CM] AC4 protein is required for membrane binding and as a pathogenicity determinant. In their experimental system using confocal imaging analysis, they showed that AC4 protein of EACMCV binds preferentially to the plasma membrane as well to cytoplasmic membranes. Furthermore, replacement of gly-2 and cys-3 (sites of posttranslational attachment of myristic and palmatic acids, respectively) with alanine inhibited AC4 membrane binding and pathogenesis. This report was probably the first on a membrane protein involved in pathogenesis and the suppression of RNA-silencing. On the other hand, Vanitharani et al. (2004) reported that EACMV-[CM] AC4 does not suppress PTGS, perhaps because AC4 is efficient only at suppressing the systemic phase of RNA-silencing; indeed, AC4 does not block the production of siRNA but does interfere with its spread as demonstrated. Piroux et al. (2007) identified additional amino acids within a central domain that contribute to the pathogenicity and interaction with A. thaliana shaggy related protein kinase (AtSKα) indicates that BCTV C4 protein interacts with the brassinosteroid-signaling pathway.
Monopartite begomoviruses
A few truly monopartite begomoviruses with genomes that consist of only the homolog of the DNA A components of bipartite viruses, have been identified, and they occur almost exclusively in tomato in the Old World. The report that the V2 protein is not directly involved in movement or replication but is essential for Tomato yellow leaf curl virus-[Sardinia] (TYLCV-[Sar]) infection (Wartig et al. 1997) is rare. Although V2 has no homologs among proteins with known biological functions, V2 protein of Tomato yellow leaf curl virus-[Israel] (TYLCV-[IL]) recently has been identified as an RNA-silencing suppressor (Avi et al. 2007), which is unrelated to presently known viral suppressors. Only the V2 protein of TYLCV-[IL], inhibited RNA-silencing of a reporter transgene, GFP while Tomato yellow leaf curl China virus (TYLCCNV) C2 tested here, did not suppress RNA-silencing. This inhibition elevated the cellular levels of the GFP transcript and the GFP protein, but it had no apparent effect on the accumulation of GFP-specific siRNAs, suggesting that TYLCV-[IL] V2 protein targets a step in the RNA-silencing pathway that is subsequent to the Dicer-mediated cleavage of dsRNA.
Subcellular localization of TYLCV-[IL] V2 protein in plant protoplasts and tissues showed that this protein is associated with cytoplasmic strands and inclusion bodies in the cortical regions of the cell. The TYLCV-[IL] V2 cytoplasmic distribution is similar to that of TYLCV-[DO] V2 (Rojas et al. 2001) and of the p21 of Beet yellows virus (BYV) (Reed et al. 2003). In this regard, V2 is similar to other viral suppressors like ToLCJAV βC1, HC-Pro of Cowpea aphid borne mosaic virus (CABMV), P19 of Tomato bushy stunt virus (TBSV) which lies within the cytoplasm (Kon et al. 2007; Uhrig et al. 2004; Mlotshwa et al. 2002), but it is unlike AC2 of MYMV and βC1 of TYLCCNV-[Y10], which are localized inside nucleus (Cui et al. 2005; Trinks et al. 2005). However, functional subcellular distribution of different viral silencing suppressors remains to be elucidated. TYLCV-[IL] may exert the V2 suppressor effect by targeting a step in the RNA-silencing pathway that occurs after siRNA production (Fig. 2). Therefore, the experimental system in this case detected early events required for RNA-silencing, whereas the assay to study the TYLCCNV C2 protein (Dong et al. 2003) was better suited for detecting later silencing events, such as chromatin remodeling (Bisaro 2006). In TYLCCNV, C2 (a positional homolog of AC2), the zinc finger, and the ability to bind DNA were essential for mediating the PTGS suppressor (van Wezel et al. 2003). How a weak suppressor is initially recognized remains to be elucidated.
Thus, TYLCV-[IL] can be concluded to encode two types of RNA-silencing suppressors: the V2 protein for earlier silencing events and the C2 protein for later silencing events. This type of information may soon help us develop new strategies such as attenuating infection by TYLCV-[IL], a destructive pathogen worldwide, by interfering with viral suppression by the host. Preliminary evidence on V2 of ToLCJAV when expressed in a PVX-based vector showed severe downward leaf curling followed by necrosis 2 weeks after inoculation. After 4 weeks, the plant died (P. Sharma and M. Ikegami, unpublished data). Any suppressor activity or HR defense response encoded by ToLCJAV V2 needs to be further confirmed.
Monopartite begomoviruses with DNAβ satellite
Since the first report of DNAβ satellite associated with Ageratum yellow vein virus (AYVV) (Saunders et al. 2000), several DNAβs have been cloned and their sequences deposited in the GenBank database. Possibly, begomoviruses earlier assumed to be monopartite are actually satellite-requiring. The major suppressor involved in the PTGS phenomenon of the begomoviruses satellite complex is DNAβ-encoded βC1. Besides, weak suppressor activity was demonstrated for the genomic component of C2 and C4 proteins of monopartite Bhendi yellow vein mosaic virus (BYVMV) and ToLCJAV (Gopal et al. 2007; Kon et al. 2007).
Recently, DNA satellites associated with begomoviruses have come to our attention, they are widespread throughout the world and are associated with many diseases, causing huge losses to economically important crops particularly in developing countries. Some of the monopartite viruses with genomic DNA-A such as TYLCV-[IL] and Tomato leaf curl Philippines virus (ToLCPV) do produce symptomatic infections in their hosts. However, the genomes of others are not. In these cases, studies have recently demonstrated the existence of disease complexes consisting of the geminivirus and a satellite DNA known as DNAβ (Kon et al. 2006; Briddon et al. 2001; Mansoor et al. 2003; Saunders et al. 2000). DNAβ is about half the size (∼1.4 kb) of the helper virus (Fig. 1) on which it depends for replication, encapsidation, and systemic spread. It has been shown by mutational analysis that its single open reading frame encodes the pathogenicity determinant βC1, and transgenic expression of the 14 kDa βC1 protein or expression from a PVX vector results in severe developmental abnormalities (Kon et al. 2007; Cui et al. 2004; Saunders et al. 2004; Zhou et al. 2003). The molecular basis of βC1 pathogenicity can be explained by its suppression of silencing. Recently, BYMV C2 protein has been shown to act as suppressor (Gopal et al. 2007). The N-terminal of ToLCJAV C2 protein contains a stretch of arginine rich (RRRR) residues and the nuclear localization signal (NLS), which appears to be bipartite (Kon et al. 2007), whereas TGMV AC2 protein is located in both the nucleus and the cytoplasm (Wang et al. 2003). However, taken together, the observation that C2 of ToLCJAV, which has an NLS domain, the zinc finger domain may imply transcription-dependent activation mechanisms of silencing suppression, as has been demonstrated in other geminiviruses (van Wezel et al. 2002; Trinks et al. 2005). Identifying host proteins (like WEL1 protein) that interact with a viral suppressor of RNA-silencing is a very promising approach that is being used to take advantage of viral suppressors to elucidate the silencing pathway. Whether and how these inactivated endogenous proteins are involved in RNA-silencing is not known. AC2 of MYMV-[Vig] protein appears to target both cytoplasmic RNA-silencing (PTGS) and mRNA directed DNA methylation. Further, it would be interesting to see if any other begomoviruses can activate the WEL1 protein in their hosts.
In our initial work demonstrating the silencing suppressor activity of DNAβ, we did an experiment that hinted at a silencing suppression role for the βC1 protein of ToLCJAV. Infection of plants silenced for GFP expression showed that ToLCJAV plus DNAβ, but not ToLCJAV alone, could prevent silencing in newly emerging leaves of infected plants. A βC1 with gene frame shift mutant of ToLCJAV DNAβ02 failed to induce symptoms when co-inoculated with ToLCJAV and consequently did not play a role in silencing suppression (Kon et al. 2007). Clearly, this data suggest that the βC1 protein appears to be similar to many silencing suppressor proteins like HC-Pro, CMV 2b and TBSV P19, which function as pathogenicity determinants in RNA viruses (Anandalakshmi et al. 1998; Kasschau and Carrington 1998; Roth et al. 2004). Further, inoculation with TYLCCNV-[Y10] alone failed to reverse the established GFP silencing. In contrast, infections in the bipartite ACMV lead to suppression of RNA-silencing in fully expanded and newly developed leaves 3 days after inoculation (Voinnet et al. 1999). This difference suggests that the suppressor encoded by TYLCCNV-[Y10] might be weak. Consequently, the suppressor genes could not overcome host RNA-silencing, and TYLCCNV-[Y10] failed to induce symptoms, while TYLCCNV induced leaf curling (Dong et al. 2003). In the study by van Wezel et al. (2003), the C4 gene of TYLCCNV, was found to counter the Rep induction of the hypersensitive response in N. benthamiana. Therefore, these two isolates from China would be expected to have different activities in terms of inducing symptoms and to act at different steps in the RNA-silencing pathways. Expression of βC1 TYLCCNV-[Y10] protein also interfered with local silencing in transient Agrobacterium-based assays, while the recombinant protein binds ssDNA and dsDNA in vitro in a sequence-nonspecific fashion, and the βC1 fusion proteins are primarily localized in the nucleus in insect and plant cells. The putative NLS is required to suppress silencing (Cui et al. 2005). In contrast, the ToLCJAV βC1 protein does not encode any putative NLS and is localized in cytoplasmic stands. Although reminiscent of the C2 protein with respect to size, DNA-binding properties, and nuclear localization, the βC1 protein lacks a zinc finger and shares little or no homology with the begomovirus genomic protein. In addition, TGMV AC2 and BCTV C2 proteins do not generate developmental defects when expressed in transgenic plants (Chellappan et al. 2005; Sunter et al. 2001). Thus, the developmental defects observed with βC1 expression suggest that it targets a different step in the silencing process (Fig. 2) and most likely one that overlaps the miRNA pathway. However, there is not enough information at present to separate the activities of the AC4 and βC1 proteins. Again, because related monopartite begomoviruses, including TYLCV-[IL] and TYLCCNV (Dong et al. 2003), can cause disease on their own and encode functional silencing suppressors, it is logical to assume that a requirement of βC1 protein for pathogenicity reflects the attenuated function of any other suppressors associated with DNAβ in viruses. The precise mechanism of action of the βC1 protein is presently not known. The transgenic expression of βC1 protein elicited virus-like symptoms in the absence of viral infection suggesting that these proteins may also play role in developmental regulation by interfering with miRNA pathways (Kon et al. 2007). Thus, it may be possible that βC1 protein may affect the activity of the Dicer-like proteins in plants that function in silencing suppression and could either downregulate transcription of a host protein that acts in the PTGS of a pathway in the cytoplasm or could activate transcription of a host PTGS inhibitor. It is worth noting that the ToLCJAV C4 protein does not code for suppressor activity. But C4 of ToLCJAV might have lost its silencing activity during an evolutionary step (Kon et al. 2007). In the case of monopartite begomoviruses associated with DNAβ satellites, genomic DNA C2 and C4 have mild suppressive activity, while βC1 encoded by the DNAβ molecule has strong suppression activity (Gopal et al. 2007). A possible reason for different suppressive activities of monopartite begomoviruses associated with satellites molecules could be that the βC1 suppressor stops the silencing signal of other suppressors encoded by its helper virus.
Curtoviruses
The C2 protein of BCTV does not code for the expression of late viral genes (Sunter et al. 1994), unlike AC2 of bipartite TGMV, and is required for the expression of late viral gene (Sunter and Bisaro 2003). However, the AC2 protein of begomoviruses has a functional activation domain, which is lacking in curtoviruses. Wang et al. (2005) also showed that C2 of BCTV suppresses silencing in a TGS manner, and ADK is needed for silencing (reviewed by Bisaro 2006).
Suppression of RNA-silencing and synergism
In many cases, mixed infection results in an increase in the virus titer and produces symptoms that are more severe than those caused by infection by a single virus. For example, a mixed infection with PVX and potyviruses led to the identification of the HC-Pro protein is both a synergy determinant and strong PTGS suppressor (Kasschau and Carrington 1998). Most often, synergism occurs between two genera or two families, implying a very different nature and origin of involvement of viral proteins, but recently synergism has been found within the same genus, i.e., in begomoviruses (Fondong et al. 2000; Pita et al. 2001). Likewise, the capacity of proteins AC2 of ACMV-[KE] and AC4 of East African cassava mosaic Cameroon virus (EACMCV) to suppress PTGS indicates that each virus is different and thus explains why the observed synergism is rare even though infections with multiple geminiviruses are frequent (Vanitharani et al. 2004). The SLCMV AC4 protein and ICMV AC2 protein have been identified as PTGS suppressors, which strengthens the argument that AC2 and AC4 proteins have different roles (Vanitharani et al. 2004) and target different steps in the silencing pathway. In GFP-silenced plants, the bipartite geminivirus ACMV was shown to efficiently suppress RNA-silencing, and the AC2 protein was identified as its suppressor of RNA-silencing (Vanitharani et al. 2004; Voinnet et al. 1999). However, for EACMCV, the unrelated AC4 protein encodes a suppressor of RNA-silencing. Similar to the synergism observed for PVX and Potato virus Y (PVY), mixed infections of ACMV and EACMCV revealed enhanced virulence. AC2 and AC4 proteins were shown to be involved in this synergism (Pita et al. 2001). AC2 protein of ACMV could enhance EACMCV DNA accumulation, and reciprocally, the AC4 protein increased the accumulation of ACMV DNA. Although RNA-silencing was originally regarded as entirely cytoplasmic, there is evidence that elements of the mechanism also have effects in the nucleus. The fact that AC2 protein requires both a DNA-binding domain and an NLS for its activity as a suppressor of RNA-silencing might fit this notion (Dong et al. 2003). Considering their range of activities and lack of sequence homology, RNA-silencing suppressors of the geminiviruses appear to have evolved independently even within the genus. It remains to be discovered, whether this is a mere reflection of the renowned plasticity of geminivirus genomes or an indication of a powerful selection pressure (even on DNA viruses) to be able to counteract RNA-silencing. Previously, we had isolated ToLCJAV and AYVV and their associated DNAβ satellites (Kon et al. 2006) from the same infected tomato plants in Indonesia.
Concluding remarks
This homology-dependent silencing has established a novel paradigm with far-reaching consequences in the field of transcription regulation. The regulatory mechanism offers cellular protection against parasitic nucleic acid sequences, carries out epigenetic as well as genetic alterations on the one hand, and governs organism’s architecture and development on the other. The stepwise detailed mechanism of RNA-silencing and miRNA related processes are waiting to be explored as an antiviral counterdefense. Geminiviruses being ssDNA with no dsRNA phase in their replication cycle have been shown to be involved in the gene silencing. They encode or can be associated with as many as four distinct silencing suppressors (AC2/C2, βC1, AC4/C4 and V2 proteins), emphasizing the importance of silencing as a cellular host defense. The presence of geminiviral PTGS suppressor proteins such as AC2/C2 and AC4/C4 implies that these proteins play different roles in the interaction with the host, and as a consequence might target different steps in the silencing pathway or might interact with different host proteins. AC2/C2 protein appears to target both cytoplasmic RNA-silencing and siRNA-directed DNA methylation. The AC4/C4 and possibly the V2 and βC1 suppressor proteins appear to interfere with a step common to both the cytoplasmic and miRNA pathways. Still many questions remain unanswered. How do the different suppressor proteins regulate PTGS? What kinds of proteins interact with viral suppressors and interfere with the miRNA pathways? Whether the inhibition of miRNA function by RNA-silencing suppressors, which leads to enhanced virulence, is a genuine role of these suppressor proteins in virus infection or a mere side effect of their inhibition of siRNA-mediated RNA-silencing remains to be established. In future, the miRNA and siRNA pathways need to receive serious consideration as antiviral defense mechanisms against emerging geminivirus disease complexes. Identification of host proteins that interact with a viral suppressor of RNA-silencing is proving to be a very useful approach to take advantage of viral suppressors to elucidate the silencing pathway.
References
Allen E, Xie Z, Gustafson AM, Carrington JC (2005) MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121:207–221
Anandalakshmi R, Pruss GJ, Ge X, Marathe R, Smith TH, Vance VB (1998) A viral suppressor of gene silencing in plants. Proc Natl Acad Sci USA 95:13079–13084
Avi Z, Efrat G, Levy Y, Arazi T, Citovsky V, Gafni Y (2007) Suppressor of RNA silencing encoded by Tomato yellow leaf curl virus-Israel. Virology 358: 159–165
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297
Baulcombe DC (1996) RNA as a target and an initiator of post transcriptional gene silencing in transgenic plants. Plant Mol Biol 32:79–88
Baulcombe DC (2004) RNA silencing in plants. Nature 431:356–363
Baumberger N, Baulcombe DC (2005) Arabidopsis ARGONAUTE1 is an RNA slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA 102:11928–11933
Bernstein E, Caudy AA, Hammond SM, Hannon GJ (2001) Role for a ribonuclease in the initiation step of RNA interference. Nature 409:363–366
Bisaro DM (2006) Silencing suppression by geminivirus proteins. Virology 344:158–168
Boutet S, Vazquez F, Liu J, Beclin C, Fagard M, Gratias A, Morel JB, Crete P, Chen X, Vaucheret H (2003) Arabidopsis HEN1: a genetic link between endogenous miRNA controlling development and siRNA controlling transgene silencing and virus resistance. Curr Biol 13:843–848
Briddon RW, Stanley J (2006) Subviral agents associated with plant single stranded DNA viruses. Virology 344:198–210
Briddon RW, Mansoor S, Bedford ID, Pinner MS, Saunders K, Stanley J, Zafar Y, Malik KA, Markham PG (2001) Identification of DNA components required for induction of cotton leaf curl disease. Virology 285:234–243
Brigneti G, Voinnet O, Li WX, Ji LH, Ding SW, Baulcombe DC (1998) Viral pathogenicity deteriminants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J 17:6739–6746
Chellappan P, Vanitharani R, Fauquet CM (2004) Short interfering RNA accumulation correlates with host recovery in DNA virus-infected hosts, and gene silencing targets specific viral sequences. J Virol 78:7465–7477
Chellappan P, Vanitharani R, Fauquet CM (2005) MicroRNA-binding viral protein interferes with Arabidopsis development. Proc Natl Acad Sci USA 102:10381–10386
Cogoni C, Macino G (1997) Isolation of quelling-defective (qde) mutants impaired in post transcriptional transgene induced gene silencing in Neurospora crassa. Proc Natl Acad Sci USA 94:10233–10238
Cui X, Tao X, Xie Y, Fauquet CM, Zhou X (2004) A DNAβ associated with Tomato yellow leaf curl China virus is required for symptom induction. J Virol 78:13966–13974
Cui X, Li G, Wang D, Hu D, Zhou X (2005) A begomovirus DNAβ encoded protein binds DNA, functions as a suppressor of RNA silencing, and targets the cell nucleus. J Virol 79:10764–10775
Dalmay T, Hamilton AJ, Rudd S, Angell S, Baulcombe DC (2000) An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101:543–553
de Vries JS, Andriotis VM, Wu AJ, Rathjen JP (2006) Tomato Pto encodes a functional N-myristoylation motif that is required for signal transduction in Nicotiana benthamiana. Plant J 45:31–45
Ding SW, Li H, Lu R, Li F, Li WX (2004) RNA silencing: a conserved antiviral immunity of plants and animals. Virus Res 102:109–115
Dong X, van Wezel R, Stanley J, Hong Y (2003) Functional characterization of the nuclear localization signal for a suppressor of posttranscriptional gene silencing. J Virol 77:7026–7033
Dougherty WG, Parks TD (1995) Transgenes and gene suppression: telling us something new? Curr Opin Cell Biol 7:399–405
Ebhardt HA, Thi EP, Wang MB, Unrau PJ (2005) Extensive 3′ modification of plant small RNAs is modulated by helper component-proteinase expression. Proc Natl Acad Sci USA 102:13398–13403
Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K (2001) Duplexes of 21- nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494–498
Fauquet CM, Stanley J (2005) Revising the way we conceive and name viruses below the species level: a review of geminivirus taxonomy calls for new standardized isolate descriptors. Arch Virol 150:2151–2179
Fauquet CM, Briddon RW, Brown JK, Moriones E, Stanley J, Zerbini M, Zhou X (2008) Geminivirus strain demarcation and nomenclature. Arch Virol 153:783–821
Filippov V, Solovyev V, Filippova M, Gill SS (2000) A novel type of RNase III family proteins in eukaryotes. Gene 245:213–221
Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806–811
Fondong VN, Pita JS, Rey MEC, de Kochko A, Beachy RN, Fauquet CM (2000) Evidence of synergism between African cassava mosaic virus and a new double-recombinant geminivirus infecting cassava in Cameroon. J Gen Virol 81:287–297
Fondong VN, Chowda Reddy RV, Lu C, Hankoua B, Felton C (2007) The consensus N-myristoylation motif of a geminivirus AC4 protein is required for membrane binding and pathogenicity. Mol Plant Microbe Interact 20:380–391
Gasciolli V, Mallory AC, Bartel DP, Vaucheret H (2005) Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr Biol 15:1494–1500
Gopal P, Kumar PP, Sinilal B, Jose J, Yadunandam A, Usha R (2007) Differential roles of C4 and βC1 in mediating suppression of post-transcriptional gene silencing: evidence for transactivation by the C2 of Bhendi yellow vein mosaic virus, a monopartite begomovirus. Virus Res 123:9–18
Hamilton A, Voinnet O, Chappell L, Baulcombe D (2002) Two classes of short interfering RNA in RNA silencing. EMBO J 21:4671–4679
Han MH, Goud S, Song L, Fedoroff N (2004) The Arabidopsis double stranded RNA binding protein HYL1 plays a role in microRNA mediated gene regulation. Proc Natl Acad Sci USA 101:1093–1098
Hanley-Bowdoin L, Settlage SB, Orozco BM, Nagar S, Robertson D (1999) Geminiviruses: models for plant DNA replication, transcription, and cell cycle regulation. Crit Rev Plant Sci 18:71–106
Herr AJ, Jensen MB, Dalmay T, Baulcombe DC (2005) RNA polymerase IV directs silencing of endogenous DNA. Science 308:118–120
Hiraguri A, ltoh R, Kondo N, Nomura Y, Aizawa D, Murai Y, Koiwa H, Seki M, Shinozaki K, Fukuhara T (2005) Specific interactions between Dicer like proteins and HYL1/DRB family dsRNA binding proteins in Arabidopsis thaliana. Plant Mol Biol 57:173–188
Ishitani M, Liu J, Halfter U, Kim CS, Shi W, Zhu JK (2000) SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. Plant Cell 12:1667–1678
Jupin I, de Kouchkovsky F, Jouanneau F, Gronenborn B (1994) Movement of tomato yellow leaf curl geminivirus (TYLCV): involvement of the protein encoded by ORF C4. Virology 204:82–90
Kasschau KD, Carrington JC (1998) A counterdefensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell 95:461–470
Kasschau KD, Carrington JC (2001) Long-distance movement and replication maintenance functions correlate with silencing suppression activity of potyviral HC-Pro. Virology 285:71–81
Kon T, Hidayat SH, Hase S, Takahashi H, Ikegami M (2006) The natural occurrence of two distinct begomoviruses associated with DNAβ and a recombinant DNA in a tomato plant from Indonesia. Phytopathology 96:517–525
Kon T, Sharma P, Ikegami M (2007) Suppressor of RNA silencing encoded by the monopartite tomato leaf curl Java begomovirus. Arch Virol 152:1273–1282
Kurihara Y, Watanabe Y (2004) Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc Natl Acad Sci USA 101:12753–12758
Lakatos L, Szittya G, Silhavy D, Burgyan J (2004) Molecular mechanism of RNA silencing suppression mediated by the p19 protein of tombusviruses. EMBO J 23:876–884
Latham JR, Saunders K, Pinner MS, Stanley J (1997) Induction of plant cell division by beet curly top virus gene C4. Plant J 11:1273–1283
Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, Saib A, Voinnet O (2005) A cellular microRNA mediates antiviral defense in human cells. Science 308:557–560
Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN (2003) The nuclear RNase III Drosha initiates microRNA processing. Nature 425:415–419
Li WX, Ding SW (2001) Viral suppressors of RNA silencing. Curr Opin Biotechnol 12:150–154
Li H, Li WX, Ding SW (2002) Induction and suppression of RNA silencing by an animal virus. Science 296:1319–1321
Li J, Yang Z, Yu B, Liu J, Chen X (2005) Methylation protects miRNAs and siRNAs from a 3′ end uridylation activity in Arabidopsis. Curr Biol 15:1501–1507
Llave C, Kasschau KD, Carrington JC (2000) Virus-encoded suppressor of posttranscriptional gene silencing targets a maintenance step in the silencing pathway. Proc Natl Acad Sci USA 97:13401–13406
Llave C, Xie Z, Kasschau KD, Carrington JC (2002) Cleavage of scarecrew-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297:2053–2056
Mansoor S, Briddon RW, Zafar Y, Stanley J (2003) Geminivirus disease complexes: an emerging threat. Trends Plant Sci 8:128–134
Meins FJ, Si-Ammour A, Blevins T (2005) RNA silencing systems and their relevance to plant development. Annu Rev Cell Dev Biol 21:297–318
Mlotshwa S, Voinnet O, Mette MF, Matzke M, Vaucheret H, Ding SW, Pruss G, Vance VB (2002) RNA silencing and the mobile silencing signal. Plant Cell 14:2289–2301
Molnar A, Csorba T, Lakatos L, Varallyay E, Lacomme C, Burgyan J (2005) Plant virus-derived small interfering RNAs originate predominantly from highly structured single-stranded viral RNAs. J Virol 79:7812–7818
Morinaga T, Ikegami M, Shimotohno K, Miura K (1987) Total nucleotide sequences of the infectious cloned DNAs of bean golden mosaic virus. Microbiol Immunol 31:147–154
Morinaga T, Ikegami M, Miura K (1993) The nucleotide sequence and genome structure of mung bean yellow mosaic geminivirus. Microbiol Immunol 37:471–476
Mourrain P, Beclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Jouette D, Lacombe AM, Nikic S, Picault N, Remoue K, Sanial M, Vo TA, Vaucheret H (2000) Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101:533–542
Muangsan N, Beclin C, Vaucheret H, Robertson D (2004) Geminivirus VIGS of endogenous genes requires SGS2/SDE1 and SGS3 and defines a new branch in the genetic pathway for silencing in plants. Plant J 38:1004–1014
Napoli C, Lemieux C, Jorgensen R (1990) Introduction of a chimeric chalcone synthase gene into petunia results in reversible cosuppression of homologous genes in transgenic plant. Plant Cell 2:279–289
Navot N, Pichersky E, Zeidan M, Zamir D, Czosnek H (1991) Tomato yellow leaf curl virus: a whitefly-transmitted geminivirus with a single genomic component. Virology 185:151–161
Pilartz M, Jeske H (2003) Mapping of Abutilon mosaic geminivirus minicromosomes. J Virol 77:10808–10818
Piroux N, Keith S, Page A, Stanley J (2007) Geminivirus pathogenicity protein C4 interacts with Arabidopsis thaliana shaggy related protein kinase AtSKn, a component of the brassinosteroid singalling pathway. Virology 362:428–440
Pita JS, Fondong VN, Sangare A, Otim-Nape GW, Ogwal S, Fauquet CM (2001) Recombination, pseudorecombination and synergism of geminiviruses are determinant keys to the epidemic of severe cassava mosaic disease in Uganda. J Gen Virol 82:655–665
Pooma W, Petty ITD (1996) Tomato golden mosaic virus open reading frame AL4 is genetically distinct from its C4 analogue in monopartite geminiviruses. J Gen Virol 77:1947–1951
Raices M, Chico JM, Tellez-Inon MT, Ulloa RM (2001) Molecular characterization of StCDPK1, a calcium-dependent protein kinase from Solanum tuberosum that is induced at the onset of tuber development. Plant Mol Biol 46:591–601
Rashid A, Si-Ammour Z, Blevins T, Amin I, Kutter C, Vanderschuren H, Zhang P, Gruissem W, Meins JF, Hon T, Poggin MM (2006) Molecular characterization of geminiviruses derived small RNAs in different plant species. Nucleic Acids Res 34:462–471
Reed JC, Kasschau KD, Prokhnervsky AI, Gopinath K, Pogue GP, Carrington JC, Dolja VV (2003) Suppressor of RNA silencing encoded by Beet yellows virus. Virology 306:203–209
Rojas MR, Jiang H, Salati R, Xoconostle-Cázares B, Sudarshana MR, Lucas WJ, Gilbertson RL (2001) Functional analysis of proteins involved in movement of the monopartite begomovirus, Tomato yellow leaf curl virus. Virology 291:110–125
Roth BM, Pruss GJ, Vance VB (2004) Plant viral suppressors of RNA silencing. Virus Res 102:97–108
Saunders K, Bedford ID, Briddon RW, Markham PG, Wong SM, Stanley J (2000) A unique virus complex causes Ageratum yellow vein disease. Proc Natl Acad Sci USA 97:6890–6895
Saunders K, Norman A, Gucciardo S, Stanley J (2004) The DNAß satellite component associated with Ageratum yellow vein disease encodes an essential pathogenicity determinant (βC1). Virology 324:37–47
Sharma P, Rishi N (2007) Cotton leaf curl disease, an emerging whitefly transmissible begomovirus complex. Plant Viruses 1:127–134
Shivaprasad PV, Akbergenov R, Trinks D, Rajeswaran R, Veluthambi K, Hohn T, Pooggin MM (2005) Promoters, transcripts, and regulatory proteins of Mungbean yellow mosaic geminivirus. J Virol 79:8149–8163
Sunter G, Bisaro DM (2003) Identification of a minimal sequence required for activation of the tomato golden mosaic virus coat protein promoter in protoplast. Virology 305:452–462
Sunter G, Stenger DC, Bisaro DM (1994) Heterologous complementation by geminivirus AL2 and AL3 genes. Virology 203:203–210
Sunter G, Sunter J, Bisaro DM (2001) Plants expressing Tomato golden mosaic virus AL2 or beet curly top virus L2 transgenes show enhanced susceptibility to infection by DNA and RNA viruses. Virology 285:59–70
Szittya G, Molnar A, Silhavy D, Hornyik C, Burgyan J (2002) Short defective interfering RNAs of tombusviruses are not targeted but trigger post-transcriptional gene silencing against their helper virus. Plant Cell 14:359–372
Tang G, Reinhart BJ, Bartel DP, Zamore PD (2003) A biochemical framework for RNA silencing in plants. Genes Dev 17:49–63
Tijsterman M, Ketting RF, Plasterk RH (2002) The genetics of RNA silencing. Annu Rev Genet 36:489–519
Trinks D, Rajeswaran R, Shivaprasad PV, Akbergenov R, Oakeley EJ, Veluthambi K, Hohn T, Pooggin M (2005) Suppression of RNA silencing by a geminivirus nuclear protein, AC2, correlates with transactivation of host genes. J Virol 79:2517–2527
Uhrig JF, Canto T, Marshall D, MacFarlane SA (2004) Relocalization of nuclear ALY protein to the cytoplasm by the tomato bushy stunt virus P19 pathogenicity protein. Plant Physiol 135:2411–2423
van Wezel R, Liu H, Tien P, Stanley J, Hong Y (2002) Mutation of three cysteine residues in tomato yellow leaf curl virus-China C2 protein causes dysfunction in pathogenesis and posttranscriptional gene silencing suppression. Mol Plant Microbe Interact 15:203–208
van Wezel R, Liu H, Wu Z, Stanley J, Hong Y (2003) Contribution of the zinc finger to zinc and DNA binding by a suppressor of posttranscriptional gene silencing. J Virol 77:696–700
Vanitharani R, Chellappan P, Pita JS, Fauquet CM (2004) Differential roles of AC2 and AC4 of cassava geminiviruses in mediating synergism and suppression of posttranscriptional gene silencing. J Virol 78:9487–9498
Vanitharani R, Chellappan P, Fauquet CM (2005) Geminiviruses and RNA silencing. Trends Plant Sci 10:144–151
Vaucheret H (2006) Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev 20:759–771
Vazquez F, Gasciolli V, Crete P, Vaucheret H (2004) The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr Biol 14:346–351
Voinnet O (2001) RNA silencing as a plant immune system against viruses. Trends Genet 17:449–459
Voinnet O (2005) Induction and suppression of RNA silencing: insights from viral infections. Nat Rev Genet 6:206–221
Voinnet O, Pinto YM, Baulcombe DC (1999) Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc Natl Acad Sci USA 96:14147–14152
Voinnet O, Lederer C, Baulcombe DC (2000) A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103:157–167
Wang H, Hao L, Shung CY, Sunter G, Bisaro DM (2003) Adenosine kinase is inactivated by genimivirus AL2 and L2 proteins. Plant Cell 15:3020–3032
Wang H, Buckley KJ, Yang X, Buchmann RC, Bisaro DM (2005) Adenosine kinase inhibition and suppression of RNA silencing by geminivirus AL2 and L2 proteins. J Virol 79:7410–7418
Wartig L, Kheyr-Pour A, Noris E, Dekonchkovsky F, Jouanneau F, Gronrnborn B, Jupin I (1997) Genetic analysis of the monopartite tomato yellow leaf curl geminivirus: Roles of V1, V2 and C2 ORFs in viral pathogenesis. Virology 228:132–140
Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC (2004) Genetic and functional diversification of small RNA pathways in plants. PLOS Biol 2:E104
Xie Z, Allen E, Wilken A, Carrington JC (2005) DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc Natl Acad Sci USA 102:12984–12989
Yang X, Baliji S, Buchmann RC, Wang H, Lindbo JA, Sunter G, Bisaro DM (2007) Functional modulation of the geminivirus AL2 transcription factor and silencing suppression by self interaction. J Virol 81:11972–11981
Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X (2005) Methylation as a crucial step in plant microRNA biogenesis. Science 307:932–935
Zhou XP, Xie Y, Tao XR, Zhang ZK, Li ZH, Fauquet CM (2003) Characterization of DNAβ associated with begomovirus in China and evidence for co-evolution with their cognate DNA-A. J Gen Virol 84:237–247
Zilberman D, Cao X, Jacobsen SE (2003) ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299:716–719
Acknowledgments
Our research cited in this review was funded by Grant-in-Aid supports from Academic Frontier Promotion program and Scientific Research of the Ministry of Education, Culture, Sports, Science and Technology of Japan. P.S. was supported by the Japan Society for the Promotion of Science Grant in Aid from JSPS Post Doc fellowship for foreign researchers program.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sharma, P., Ikegami, M. RNA-silencing suppressors of geminiviruses. J Gen Plant Pathol 74, 189–202 (2008). https://doi.org/10.1007/s10327-008-0085-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10327-008-0085-5