Abstract
Microplastics contamination is becoming a major concern worldwide. More than 1 million seabirds and 100,000 sea animals have died due to plastic contamination. In addition, plastic particles have been found in juvenile turtles. Statistical data on plastic pollution indicate that this is a serious issue. Due to their small size, microplastics have a large surface area and have more ability to absorb into biological cells. The hydrophobic surface of microplastics attracts co-contaminants such as heavy metals, pharmaceutical toxicants, flame retardants, and other plasticizers, which can then enter biological organisms. Microplastics are usually recalcitrant in the environment, causing microplastics to be transported along the food chain, with humans as the final consumer. Research has been conducted to evaluate the best way to treat and remediate microplastic pollution. Research on microplastic degradation is focused on biological and non-biological approaches. To date, microorganisms such as algae, fungi, and bacteria have attracted the attention of scientists as a tool for microplastic treatment. The degradation of microplastics is closely related to the enzymatic reactions produced by the microorganisms. Here we review microplastics degradation through enzymes from the microorganism’s perspective. We present the enzymes that have been isolated from microorganisms for specific microplastics; the mechanisms of microplastics degradation by various enzymes; and the types of microplastics for which degradation mechanisms remain unclear.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
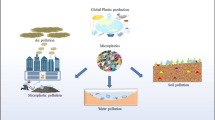
Human activities are major contributors towards global contamination (López-Pedrouso et al. 2020). Contamination covers various sectors, such as air, water, and soil. Approximately 2.01 billion metric tons of municipal waste are produced each year, with a projected increase to 3.40 billion metric tons by 2050 (Ellis 2018). As previously stated, humans can impact environmental quality in many ways, such as through deforestation, agriculture, mining, industrial activities, and urbanization (Haddaway et al. 2019; Campbell 2019; Balogh and Jámbor 2020; Arshad et al. 2020). These human activities lead to various types of contamination, which can be categorized into two major groups, namely organic pollutants and inorganic pollutants (Bharagava et al. 2020). Inorganic pollution by definition is contamination that arises from the inorganic by-products of inorganic matter due to radiant energy and noise, light, or heat (Borah et al. 2020). Inorganic pollutants primarily consist of metals/metalloids (arsenic, mercury, cadmium, and lead) and radioactive elements (Wen et al. 2021). While inorganic pollutants are primarily grouped under metal contaminants, organic pollutants consist of various types of contaminants, such as phenols, azo dyes, polyaromatic hydrocarbon pesticides, plastics, and plasticizers (Bharagava et al. 2018).
Plastics are organic polymers synthesized from non-renewable resources, including natural gas, coal, and crude oil. They are easy to mold, making them suitable for a variety of uses (Rios et al. 2010; Worm et al. 2017). In general, plastics are categorized into thermoplastics and thermosets (Ayodeji et al. 2020). Plastic consumption has been increasing yearly. Within 10 years, global plastic production increased from 254 million tons to 359 million tons between 2008 and 2018, with an expected threefold increase by 2050 (Chia et al. 2020). Plastic waste is known for its stability and recalcitrance in the environment. Due to this, plastic waste is commonly assumed to be non-degradable. Some state that plastic degradation varies in the environment, ranging between 10 and 1000 years depending on the environmental condition, the type of plastic monomer, and the treatment applied to the plastic waste. For example, treatment analysis using thermooxidative and photodegradation toward microplastics pollutants is claimed to degrade microplastics approximately within 50 years (Mohammed et al. 2019; Ward et al. 2019; Qi et al. 2020; Chamas et al. 2020). The degradation of plastic waste in the environment will lead to the formation of secondary microplastic pollution. The presence of microplastics in the environment has been shown to cause numerous hazardous effects on vast flora and fauna species. In addition, microplastics act as carriers or chelators to various types of co-contaminants, such as heavy metals, brominated flame retardants and other types of plasticizers, and pharmaceutical toxicants. These co-contaminants easily bind to the microplastic surface due to their hydrophobicity (Chatterjee and Sharma 2019).
Microplastics
Definition
The term microplastic itself was first mentioned by an African scientist in the 1990s in his article entitled “Plastic and other artifact on South African beaches: temporal trends in abundance and composition.” The term was then recognized worldwide and has been widely used to describe small plastic particles (Alimi et al. 2021). The characteristics of microplastics, also known as tiny plastic particles, are still under debate, but most researchers agree that plastic particles ranging between 100 and 5 mm in size are considered microplastics. Plastic particles that are > 25 mm in size are known as macroplastics, those that are 5–25 mm in size are classified as mesoplastics, and those that are < 100 nm in size are classified as nanoplastics (Löder et al. 2017; Budi Kurniawan et al. 2020; Jaafar et al. 2020; Khalid et al. 2021; Yang et al. 2021b). In the environment, microplastics can be categorized into two major groups: primary microplastics and secondary microplastics. These two types of microplastics are distinguished by the source point. Primary microplastics are derived from manufacturing activity. Millimeter plastic particles are synthesized and designed for commercial products, such as personal care products, e.g., toothpaste, facial cleanser, and shower gel. Primary microplastics can be generated from the air-blasting industry due to the abrasion of materials during the preproduction of resin pellets (Suardy et al. 2020; Nava and Leoni 2020; Khalid et al. 2021). On the other hand, secondary microplastics are derived from chemical (i.e., UV radiation, the freeze–thaw cycle), physical (abrasion, wave strike, water disturbance), and biological (degradation) activities involving fragmentation and degradation of large plastics into micro-sized particles (Khalid et al. 2021; Dong et al. 2021). Microplastics are built by the polymerization of plastic monomers. Table 1 shows the monomer structure of each major microplastic present in the environment (Table 1).
Microplastics formation route
Microplastic contamination is commonly caused by human anthropogenic activities. Since plastics are used daily by humans, this pollution comes from a wide range of sources, from industry sectors to domestic activities. Industries such as raw plastic manufacturing and textile and laundry services are some of the major industries that have been reported to contribute to the presence of microplastics in the environment (Lechner and Ramler 2015; De Falco et al. 2019; Henry et al. 2019; Cai et al. 2020b; Tang et al. 2020). Besides industry, wastewater treatment plants (WWTPs) have been mentioned in numerous articles as the main source of microplastic contamination in the environment (Mintenig et al. 2017; Wolff et al. 2019; Funck et al. 2020; Frehland et al. 2020; Sol et al. 2020). In addition, the agricultural sector has also been reported to significantly contribute to microplastic pollution (Mohajerani and Karabatak 2020; Zhou et al. 2020; van den Berg et al. 2020; Ding et al. 2020; Crossman et al. 2020; Zurier and Goddard 2020; Kumar et al. 2020). Domestic activities such as improper littering and leachate from runoff surface water are reported to contribute to microplastic contamination (Gandara e Silva et al. 2016; Green et al. 2018; He et al. 2019; Kalnasa et al. 2019; Esquinas et al. 2020; Li et al. 2020; Shi et al. 2020). The activities mentioned above are considered to be major anthropogenic activities that cause microplastic contamination, but many human activities also contribute to this contamination.
Microplastics toxicity
Microplastics circulate in soil and aquatic ecosystems and have been proven to impact flora and fauna. Numerous reports in ecotoxicology toward plants were mentioned in several plant species. In summary, microplastics can affect plants directly and indirectly. Microplastics have been shown to directly affect plants by blocking nutrient uptake and accumulating in roots, shoots, and leaves, while microplastics have been found to indirectly alter the properties of soil, such as the presence of soil-dwelling microorganisms and physicochemical properties (Khalid et al. 2020). A growth rate analysis in tomato plants using sludge containing microplastics indicated that the growth rate decreased significantly after exposure to microplastic sludge for 109 days. The study stated that the growth rate was significantly affected due to the alteration of the C:N ratio in the soil system, which affected the nutrient availability in the soil (Hernández-Arenas et al. 2021). An analysis using the aquatic plant Utricularia vulgaris showed that microplastics accumulate in several regions in plants. Microplastics were found in the root, leaves, and bladders of plants. Microplastics accumulations in plants are shown to cause oxidative damage through increment in plant antioxidative enzyme activity (Yu et al. 2020). A similar report mentioned the microplastic effect in the plant Vicia faba. Exposure to microplastics leads to various oxidative enzymatic responses such as catalase, superoxide dismutase, and peroxidase. These enzymes are known to be closely related to oxidative stress, which can be caused by the presence of microplastics and co-contaminants, such as heavy metals and plasticizers. Laser confocal scanning microscopy analysis showed that microplastics accumulate in the root parts of plants (Jiang et al. 2019; Abbasi et al. 2020). The ecotoxicity of microplastics has not only been observed in plants, but microplastics have also been reported to affect animals. Fish are among the common biological models used to investigate the toxicity of microplastics. Laboratory-based environments have indicated that microplastics affect nutrients by accumulating in the fish intestine. Microplastics were also found to induce inflammatory responses, reduce innate immunity, reduce reproduction rate, and promote organ failure in fish. However, data from laboratory-based environments do not provide an actual environment scenario in microplastic ecotoxicity. Reports have shown that microplastics in the environment also contain other co-contaminants, such as plasticizers and heavy metals. An analysis mimicking actual environmental conditions showed a greater effect of toxicity on fish with the toxicity effect increased with 30 times higher compared to the laboratory-based environment (Rainieri et al. 2018; Cheng et al. 2020; Wang et al. 2020b). Aquatic gastropods, bivalve, crustacean, amphipods, and insect larvae were among the biological models used for accessing the impact of microplastics on ecosystems. The same rate of toxicity was observed, with significant effects on the growth and reproductive systems of the animals tested. In addition, microplastics have been found to significantly affect regulatory enzymes, such as acetylcholinesterase, catalase, and glutathione-s-transferase (Jaikumar et al. 2019; Chagas et al. 2020; Trestrail et al. 2020). Humans are considered the top consumers in the food chain. Therefore, humans are prone to microplastic contamination. The source or point of contamination can be almost anything, such as the water source, food source, and even air. Ingestion and inhalation of microplastics from the environment promote a wide range of toxicities in humans. Microplastics have been shown to promote the inflammatory response in humans by activating the mitogen-activated protein kinase pathway. They have also been shown to have neurotoxic effects by inhibiting acetylcholinesterase activity, the inflammatory response, which can lead to the development of cancer development. The interaction between microplastics and humans has also been found to affect cell function at a molecular level (Hwang et al. 2019; Huang et al. 2020; Ju et al. 2020; Llorca et al. 2020; Wang et al. 2020a). Due to the long list of toxicities toward flora and fauna, microplastic treatment or removal from the environment is compulsory. Degradation can be separated into biological and non-biological approaches. In this review, we focus on biological degradation, especially the enzyme-based approach.
Enzymes in microplastic degradation
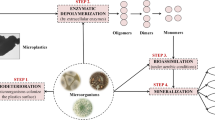
Microplastic pollution is currently becoming a worldwide concern. This is due to its toxicity, which contributes to numerous diseases, especially those in humans. Anthropogenic microplastic pollution is one of the environmental stressors that is causing ecotoxicology. The technologies and methodologies in removing microplastics in the environment were extensively reviewed by Padervand et al. (2020). Chemical, physical and biological approaches were critically discussed of their advantages and disadvantages toward microplastics removal. This article did mention the microplastics removal by microorganisms through their adaptation to the microplastics-existence ecosystem (Padervand et al. 2020). The existence of microplastics in the environment leads to the adaptation of microorganisms to survive stressful conditions caused by this pollutant (Oberbeckmann and Labrenz 2020; Yang et al. 2020a). Microorganisms respond to this stress in several aspects, such as growth rate, energy reproduction (metabolism rate), and the synthesis of new macromolecules for cellular protection purposes (NicAogáin and O’Byrne 2016; Guan et al. 2017; Guan and Liu 2020). These stress responses are closely related to enzyme activity since such enzymes play a major role in regulating cell functions (Cooper 2000; Cheng et al. 2011; Winkel 2017). Enzymes are not only involved in cell function and cell regulation, but they are also involved in the degradation of anthropogenic pollutants, including microplastics. For example, the degrading enzyme from microorganism can specifically target the polymer structure of microplastic and degrade it into its monomer, later will be used as a carbon source in the microorganism energy production cycle (Fig. 1) (Gong et al. 2018; Islam et al. 2019; Kawai et al. 2019; Ganesh Kumar et al. 2020). Each enzyme shows unique interaction mechanisms when degrading microplastics. In general, the mechanism is divided into two major mechanisms. Enzyme surface modification mechanisms by enzyme hydrolases (lipases, carboxylesterases, cutinases, and proteases) were said to be responsible for modifying microplastic polymer surfaces prone to the degradation process (Vertommen et al. 2005). This situation was intensively reviewed by Kawai et al. (2019) who claimed that certain microplastic hydrolases only react as a surface modifier to a microplastic. This type of enzyme is called a surface modifying enzyme, as suggested by the author. As such, this enzyme increases the hydrophilicity of the microplastic surface and does not degrade the building blocks of the microplastic (Kawai et al. 2019). The interaction mechanism between the enzyme and microplastic surface has been proven and explained by elemental spectroscopy chemical analysis (ESCA). Changes in C–O bonding were observed when comparing the control and the enzyme-treated microplastic. From this analysis, the author concluded that the bonding change was due to the enzyme surface interaction (Vertommen et al. 2005). As a consequence of the surface modification activity, only a few cutinase enzymes were reported to be able to degrade the inner block of the microplastic (Austin et al. 2018).
To find a novel enzyme that can degrade microplastics, a large amount of research has been conducted in the last decade. Several groups of enzymes are claimed to have the ability to degrade polymers into their monomer forms. Oxidases, amidases, laccases, hydrolases, and peroxidases are groups of enzymes that are involved in polymer degradation (Álvarez-Barragán et al. 2016; Ashter 2016; Gómez-Méndez et al. 2018). Due to the enzyme-specific characteristic towards its substrate, the next subchapter will discuss the identified enzyme that is responsible for degrading specific microplastic polymers based on the types of major microplastic presence in the environment.
Microplastic-degrading enzyme
Polyethylene-group-degrading enzyme
Polyethylene (PE) group microplastics can be categorized into the following two major groups based on their density: high-density polyethylene (HDPE) and low-density polyethylene (LDPE) (Restrepo-Flórez et al. 2014; Wu and Montalvo 2020; Patel et al. 2020). Chosen due to its excellent chemical and physical properties, PE is the largest plastic commodity presence in the various types of industries. Therefore, microplastic pollution caused by PE is common in terrestrial environments (Chen et al. 2015; Nuawi et al. 2016; Gao et al. 2021).
The PE microplastic group commonly contaminates the ecosystem in the form of HDPE and LDPE. It is commonly known that the PE microplastic is associated with numerous illnesses and significant toxicity effect on animals, plants, and humans (Kalčíková et al. 2017; Shaikh et al. 2018; Mateos-Cárdenas et al. 2019; Bellas and Gil 2020; Silva et al. 2021; Abbasi and Turner 2021). Therefore, treatment of PE contaminant in the environment is crucial. Biodegradation using the enzyme activity of microorganisms is a current research trend. LDPE is commonly found in plastic bags. Its low density property is mainly due to the small branching molecule in the polymer backbone (Kumar Sen and Raut 2015). The biodegradation of LDPE using microorganisms has been performed for more than 50 years. The degradation mechanism is related to the enzyme degradation mechanism. SDS-PAGE analysis of Staphylococcus epidermis supernatant exposed to LDPE for three months revealed that the degradation of LDPE is enzyme mediated (Chatterjee et al. 2010). Later, in 2015, a review on an LDPE-degrading enzyme explained the mechanism involved. This review suggests that enzyme-based degradation is divided into two stages. First, depolymerization of the polymer takes place extracellularly, where extracellular enzymes act as a key player in the process. In this stage, the LDPE polymer is broken down into shorter chains (oligomer, dimer, and monomer). The depolymerization stage mainly facilitates the absorption of LDPE into the cell through the permeable membrane lipid. The second stage of the process is called mineralization. In this stage, the shorter LDPE chain is mineralized to the end product, such as CO2, H2O, and CH4. These end products will be used as a carbon source for microorganism metabolism in general. In the same review article, two enzymes (laccase and alkane hydrolase) that showed significant reaction towards LDPE are mentioned by the author. Laccase and alkane hydrolase are from the AlkB family enzyme. Between these two enzymes, alkane hydrolase gets more attention in the discussion due to its novel activity in the degradation of PE (Kumar Sen and Raut 2015; Ghatge et al. 2020; Montazer et al. 2020).
HDPE is one of the microplastics that is commonly found in the environment. HDPE density is between 0.94 and 0.97 g cm3 which is lower than water density. Therefore, HDPE floats in the water environment and contributes to almost 46% of total microplastics contamination worldwide (Lee and Chae 2021). Being the most abundant microplastics in the environment, the degradation of HDPE has attracted the attention of scientists looking for potential natural degraders of this pollutant. Over the last few years, numerous microorganisms capable of degrading HDPE in the environment were isolated. Fungi and bacteria are the most common microorganisms reported to degrade this microplastic, with bacteria phyla divided into three major groups. Proteobacteria, Firmicutes, and Actinobacteria are the common phyla reported to be related to microplastics degradation (Sangeetha Devi et al. 2015, 2019; Ojha et al. 2016; Bonilla et al. 2020; Matjašič et al. 2021). Laccase is the most reported enzyme associated with HDPE degradation. Categorized under the oxidase group enzyme, laccase is found to depolymerize polymers through oxidative cleavage of the amorphous region of HDPE, providing an easily accessible carbonyl region within the polymer chain (Kang et al. 2019; Ghatge et al. 2020). Physical analysis using scanning electron microscopy (SEM) revealed that in the presence of the laccase enzyme, the HDPE surface developed pits and cracks after 90 days of incubation (Kang et al. 2019). Several other enzymes were reported to be involved in microplastic degradation through either direct or indirect degradation. Manganese peroxidase from the fungi Phanerochaete chrysosporium was reported to be able to reduce and decrease the tensile strength and total molecular weight of PE. In addition, the enzyme soybean peroxidase with the presence of hydrogen peroxide has been shown to reduce the hydrophobicity of the PE surface Although the ability of certain enzymes involved in microplastic degradation has been discussed, the actual mechanism is still unclear (Ghatge et al. 2020). The degradation scheme of LDPE and HDPE is simulated in Fig. 2. In summary, few enzymes were identified by researchers related to LDPE and HDPE degradation. Those enzymes were identified as laccase and alkane hydrolase. Manganese peroxidase and soybean peroxidase were claimed to reduce tensile strength and reduce hydrophobicity of PE surface, respectively. Although depolymerization and intake of the microplastics were discussed, the complete mineralization of microplastics monomer inside the microorganisms is still unclear and unexplored. In author opinion, LDPE and HDPE degradation will undergo almost similar mechanism due to the similar monomer structure of these two microplastics. Thus, this can provide a potential research gap in understanding the metabolism pathway involved in microplastics biodegradation.
Degradation scheme of low-density polyethylene (LDPE) and high-density polyethylene (HDPE) microplastics. An extracellular enzyme secreted by microorganisms will degrade microplastics polymer and produce monomer. Microplastic monomer uptake by the microorganism is facilitated by a permeable membrane bilayer. Microplastics mineralization inside the cell through unknown metabolic pathway produces CO2, H2O and CH4. An The scanning electron microscopy (SEM) image shows the formation of pits and cracks on the HDPE surface after exposure to the microorganism’s enzyme
Polyethylene terephthalate-degrading enzyme
Polyethylene terephthalate (PET) is categorized under thermoplastics. Linked by an ester bond, this plastic polymer is used in various industries, such as fiber, bottle, and film industries. The PET structure consists of an amorphous semi-crystalline structure. PET melts at high temperatures (260 °C). The PET half-life is approximately 700 years in the normal environment (Abdelaal et al. 2008; Zulkifley et al. 2014; Horvath et al. 2018). Similar to other types of microplastics, PET has been shown to have toxicity toward living cells. PET has been shown to significantly reduce the zooplankton population and egg production when exposed to PET from the environment (Heindler et al. 2017). A toxicity effect has been observed in higher organisms, such as benthic grazer organisms (Parolini et al. 2020). PET toxicity in humans has raised concerns on the effect of PET leachate from bottles on the human endocrine system. Research has shown that exposure to PET that has leached from containers affects the human endocrine system, such as human estrogenic regulation. Additionally, there has been a 78% increase in breast cancer development after exposure to PET (Sax 2010). Several steps have been taken to reduce the negative impact of PET on the environment, including reducing usage quantity, recycling, and degradation PET from the environment. Of these, PET degradation has received more attention from researchers since it is believed to solve PET contamination in the environment (Chowdhury et al. 2018; Sang et al. 2020).
The degradation of PET can be classified into two groups: abiotic degradation and biotic degradation. Abiotic degradation, such as hydrolysis, thermal degradation, and chemical degradation, is commonly mentioned in articles (Arhant et al. 2019; Wu et al. 2019; Das and Tiwari 2019). Biotic degradation, or biodegradation, attracts researcher’s attention due to its complete mineralization, especially in microorganism degradation. A long list of bacteria and fungi are associated with PET degradation (Herrero Acero et al. 2011; Ribitsch et al. 2011; Kawai et al. 2014; Yoshida et al. 2016; Sangale et al. 2019; Danso et al. 2019; Bollinger et al. 2020; da Costa et al. 2020; Denaro et al. 2020). Out of the microorganisms associated with PET degradation, Ideonella sakaiensis 201-F6 bacterium has been shown to successfully express the enzyme related to PET degradation. Isolated in 2016 by Yoshida et al., this bacterium strain led to further research in understanding the mechanism of the reaction (Yoshida et al. 2016). The PET-degrading enzyme, known as PETase, became the center of attention related to PET degradation. The early mechanism explained the involvement of two major enzymes. First, PETase converts PET into mono(2-hydroxyethyl) terephthalic acid (MHET), with trace amounts of terephthalic acid (TPA) and bis(2-hydroxyethyl)-TPA (BHET) as secondary products. Second, the involvement of a secondary enzyme, known as MHETase, converts MHET to terephthalic acid and ethylene glycol (EG). Ethylene glycol, on the other hand, can be used as a precursor in the tricarboxylic acid (TCA) cycle substrate depending on the metabolic route. Ethylene glycol can be converted either to acetate via acetyl CoA or converted to isocitrate. The terephthalic acid molecule will undergo a series of reactions, including protocatechuate (PCA) synthesis from terephthalic acid. Protocatechuate then undergoes a typical metabolic pathway for toxic and recalcitrant aromatic molecules, known as the β-ketoadipate pathway (Fig. 3) (Yoshida et al. 2016; Chen et al. 2018; Salvador et al. 2019). Further analysis focused on the PETase structure and mechanism was done in 2018. Austin et al. 2018 through X-ray crystallography analysis, showed that PETase structure is expressed by the common α/β hydrolases, which consists of six α-helicase and eight β-sheets. A comparison between the cutinase enzyme structure (previously mentioned as the PET-degrading enzyme) indicated that PETase and cutinase are distinctively different, with PETase having a polarized surface compared to cutinase and isoelectric point (pI) values of 9.6 and 6.3, respectively. The interaction between the PETase enzyme and its substrate was determined in this analysis. PETase was found to interact with PET through an induced fit mechanism. Due to this, the author suggested that the PETase mechanism could be wider, with other types of polyaromatic microplastics is possible for PETase substrate. This was later proven by the analysis that PETase can bind and degrade polyethylene-2,5-furandicarboxylate (PEF) (Austin et al. 2018). Furthermore, the PETase structure was determined using higher X-ray crystallography resolution (2.02 Å). The PETase active site showed high flexibility due to the presence of a disulfide bond (Fecker et al. 2018). The understanding of PETase structure and mechanism leads to an improvement of PETase performance. Several analyses including mutation and overexpression were conducted to improve PETase performance. Microalgae are one of the microorganisms chosen for overexpression of PETase since microalgae are considered the best model organism due to several advantages, such as being safe and eco-friendly toward the environment, easy to cultivate and capture CO2 from the environment through their photosynthetic ability. The transformation of PETase in microalgae has been successfully expressed and proved to be able to degrade PET through overexpression of PETase enzyme and its activity (Moog et al. 2019; Kim et al. 2020b). The PETase mutation was evaluated and compared with the wild-type performance. The mutant PETase was found to increase the activity rate up to 2.5-fold (Ma et al. 2018). PET degradation is regulated by two major enzymes. MHETase acts as a secondary enzyme that uptakes MHET and converts it to terephthalic acid and ethylene glycol. MHETase structure reminisces of feruloyl esterases. Similar to PETase, MHETase consists of an α/β hydrolase domain in addition to a lid domain structure, differentiating between MHETase and PETase. The lid domain is also crucial in MHET hydrolysis. MHETase has been shown to share the same mechanism (induced fit) with PETase toward its substrate. A comprehensive analysis of the MHETase structure through structure modification is needed to enhance its performance (Palm et al. 2019; Knott et al. 2020). PET mainly originated from food container was found to be completely mineralized by the microorganisms. PETase and MHETase were two enzymes involved in degrading PET polymer to its monomer. Complete mineralization of PET involving two major pathways (TCA cycle and β-ketoadipate pathway) depends on the metabolic substrates formed from the degradation. Though PET-degrading enzymes (PETase and MHETase) have been extensively explored, few other enzymes have been reported to contribute to PET degradation, such as lipase and esterase, which are still unexplored. The potential showed by other enzymes can be a new baseline for PET degrading enzyme analysis in the future.
Enzymatic degradation mechanism toward the polyethylene terephthalate monomer. Mineralization of polyethylene terephthalate (PET) produces acetate and isocitrate, which are used in the tricarboxylic acid (TCA) cycle. TPA: terephthalic acid, BHET: bis(2-hydroxyethyl)-terephthalic acid, MHET: mono(2-hydroxyethyl) terephthalic acid
Polystyrene-degrading enzyme
Polystyrene (PS) was first synthesized by BASF in the 1930s. Constructed from an aromatic styrene monomer, a liquid hydrocarbon, which originated from a petroleum base, polystyrene is considered an aromatic polymer. Having unique characteristics, such as hard, rigid, and solid at room temperature, transparently makes this plastic a major plastic used in food and packaging industries (Koerner et al. 2006; Dağ et al. 2019). Polystyrene presence in the environment or water body leads to numerous toxicities. Polystyrene in a form of microparticle or nanoparticle is proved to be toxic towards animals, humans and plants. Animals, especially aquatic animals are one of the common species studied for polystyrene toxicity. Polystyrene is shown to significantly affect fish reproduction systems over time (Wang et al. 2019; Zhu et al. 2020; Qiang and Cheng 2021). The absorption of polystyrene is also reported to affect the molecular level. Absorption of polystyrene in the cell promotes DNA damage in erythrocytes and brain tissue (Zhang et al. 2011; Farrelly and Shaw 2017; Sökmen et al. 2020; Guimarães et al. 2021). Although polystyrene is considered less toxic or not harmful towards humans, excess exposure to certain particle sizes may lead to certain immune responses in human cells. Depending on the site of exposure, polystyrene can enter human systems through air, food and skin contact. Polystyrene is found to accumulate in human alveoli tissue through inhalation, penetration through skin and food consumption result in polystyrene accumulation in cells and bloodstream. Analysis in the human bloodstream showed that excess of polystyrene in the red blood cells (RBCs) can promote hemolysis. Polystyrene also is proved to promote the expression of local proinflammatory cytokine (Interleukin-6) which indicates local inflammation in human cells (Farrelly and Shaw 2017; Kik et al. 2020; Hwang et al. 2020). Different from animals and humans, absorption of polystyrene in plants is assessed through the root part. Root in plants is a major nutrient uptake point. Numerous analysis reports, plants that were exposed to polystyrene in the environment showed deficiency in their biomass. This situation is due to the accumulation of polystyrene in plant root tissue. The accumulation promotes blockage in the nutrient transportation in the plants. In addition to that, the accumulation of polystyrene in plant tissue leads to inhibition of seed germination, gene expression and induce cytogenotoxicity (Jiang et al. 2019; Maity and Pramanick 2020; Taylor et al. 2020; Gao et al. 2020). Polystyrene in the environment is proved not only to affect higher organisms, a group of protist phyla or known as algae are reported to be affected by the presence of polystyrene. Several analyses in different types of microalgae showed that interaction between polystyrene and microalgae reduced microalgae growth rate. Polystyrene is found to agglomerate at the microalgae cell wall in general. This situation is reported to take place when exposed to small size (micro) polystyrene (Sjollema et al. 2016; Nolte et al. 2017; Libralato et al. 2017; Reynolds et al. 2021).
Adaptation towards microplastic environments promotes the biodegradation process in general. Degradation study by living organisms covers a broad range of organisms’ types and species. Numerous studies report on the association of decomposer animals towards polystyrene degradation. Removal of microplastics by organisms such as zooplankton was associated with their uptake and ingestion factors. A high concentration of zooplankton in the environment able to remove polystyrene (Padervand et al. 2020) Recently, the utilization of snail and larvae is used to determine the biodegradation rate of polystyrene in the environment. Achatina fulica land snail is reported to reduce approximately 30.7% of ingested polystyrene in 4 weeks (Song et al. 2020). Additional to this, various types of mealworm larvae are reported to be able to degrade polystyrene from the environment. Each of these larvae showed different degradation rates with Zophobas atratus larva claimed to be the super worm by degrading polystyrene in less than 1 month. Even though these studies report in different organisms related to polystyrene degradation, one similar fact that these reports are that polystyrene degradation takes place in the gut of these organisms and this condition is regulated by the presence of intestine microbiota (bacteria and fungi) in the gut (Yang et al. 2020b, 2021a; Billen et al. 2020; Cucini et al. 2020; Peng et al. 2020). High throughput next gene sequencing analysis of gut microbiota in Tenebrio molitor and Alphitobius diaperinus larva showed that several bacteria and fungi strain identified able to use diverse types of plastics as a sole carbon source. Those strains belong to Klebsiella, Pseudomonas, Serratia and Trichoderma (Urbanek et al. 2020; Cucini et al. 2020). Although polystyrene degradation by microorganisms is abundantly mentioned (O’Leary et al. 2002; Oikawa et al. 2003; Hwang et al. 2008; Mor and Sivan 2008; Atiq et al. 2010; Atiq 2011; Bhardwaj et al. 2013), data on the related enzyme involved in the degradation mechanism are still scarce. In 1997, the first polystyrene degrading enzyme was reported. Hydroquinone peroxidase from Azotobacter beijerinckii HM121 is claimed to degrade polystyrene. Dichloromethane is used to convert water-insoluble polystyrene into a small water-soluble molecule. Water-soluble polystyrene is later degraded by the hydroquinone peroxidase (Nakamiya et al. 1997). More than two decades later, a study report on polystyrene degradation involving sets of enzymes reaction. Mineralization of polystyrene is reported to be used as a substrate in TCA cycle (Fig. 4). The mechanism is initiated by degrading the polystyrene backbone by hydrolytic activity into styrene monomer. Styrene monomer is oxidized to styrene oxide in the presence of styrene monooxygenase. Next, styrene oxide undergoes isomerization into 3-phenyl acetaldehyde by styrene oxide isomerase. 3-phenyl acetaldehyde was then converted to 4-phenylacetic acid by phenylacetaldehyde dehydrogenase. Lastly, 4-phenylacetic acid is converted to 5-phenylacetyl coenzyme A in the presence of phenylacetyl coenzyme A ligase enzyme. 5-phenylacetyl coenzyme A undergoes β-oxidation to yield acetyl-CoA which is fed to TCA cycle (Ho et al. 2018; Danso et al. 2019). To date, another enzyme-mediated mechanism is reported performed by bacteria Pseudomonas sp. DSM 50,071 isolated from Zophobas atratus guts. Serine hydrolase (SH) is claimed to show a significant effect in polystyrene degradation. Analysis using serine hydrolase inhibitor (SH inhibitor) indicates that at low SH inhibitor concentration (10 µM), polystyrene degradation is dropped from 2.6% (control) to 1.3% after 15 days of incubation. Incubation at high SH inhibitor concentration (50 µM) showed complete inhibition SH activity with no polystyrene degradation observed (Kim et al. 2020a). In summary, polystyrene degradation is proved by enzymatic degradation. The polystyrene monomer mineralization will produce acetyl-coenzyme A (CoA) through β-oxidation. Acetyl-CoA produced will be used as a feeder for TCA cycle in microorganism metabolism. Another enzyme known as serine hydrolase is proven to be involved in polystyrene degradation. Though enzyme mechanisms aforementioned can be a baseline for researchers to understand polystyrene degradation, further extend analysis is needed especially for serine hydrolase to support and provide mass knowledge related to the enzyme mechanism in polystyrene degradation.
Polypropylene degrading enzyme
Polypropylene (PP) can be derived from primary and secondary microplastic reactions. Primary polypropylene microplastic is commonly found in cosmetics and personal care products (Uheida et al. 2020). Polypropylene is considered as low-density plastic with an average density is 0.94 g/cm3. Building blocks for polypropylene are a straight chain of hydrocarbon structure with only carbon atoms in its main ring structure. Due to this hydrocarbon arrangement, polypropylene has a hydrophobic surface. There are three stereoisomers for polypropylene (isotactic, syndiotactic, and atactic) with isotactic polypropylene is the most abundant plastic used mainly in food and medical industries. Having hydrophobic property and rough surface, making it one of the resilient and recalcitrant in the environment (Khoironi et al. 2020). The distribution of polypropylene is reported to be ubiquitous and scattered around the world from east to west regions. A marine water sampling was conducted by two different groups, sampling site in Zhubi Reef from South China Sea and Chesapeake Bay in USA. Act as the main water basin in both regions, numbers of samples are taken from these areas and analyzed for microplastic content. In both regions, it is reported that polypropylene is the most abundant microplastic found in these regions (Huang et al. 2019; Bikker et al. 2020). Similar to other types of microplastic, cells exposed to polypropylene will show toxicity. Analysis using PBMCs, RAW 264.7, and HMC-1 human-derived cells showed that exposure to polypropylene stimulated immune response and enhanced hypersensitivity via increment in cytokines and histamine level in respected cells (Hwang et al. 2019).
The reports on microbial degradation associated with polypropylene are extensively discussed by Ru et al. (2020). In this report, the author mentioned the degradation of polypropylene polymer is not only targeting the backbone of the polypropylene but also targeting the plasticizers that exist on the surface of the polypropylene (Ru et al. 2020). Two species of bacteria are associated with polypropylene degradation. Rhodococcus sp. strain 36, Bacillus sp. strain 27 and Bacillus gottheilii are the bacteria reported to have the ability in degrading polypropylene from the environment (Auta et al. 2017, 2018). Although the research did mention bacteria species able to degrade polypropylene, no enzyme was identified with respect to the degradation mechanism (Chandra et al. 2020; Ganesh Kumar et al. 2020). Even though there is no clear explanation related to the enzyme and its mechanism in polypropylene degradation, the polypropylene degradation facilitated by the enzyme is proved in 2019, but the actual enzyme and its characteristic are never mentioned (Pires et al. 2019). Compared to other microplastics, information on polypropylene degradation and removal is still lacking. Through various sources of literature search, only three bacteria species were said to be able to degrade polypropylene from the environment. In the authors' opinion, the lack of information in microorganisms degrading polypropylene is due to the resilient characteristic showed by the polypropylene. The resilient characteristic might cause difficulty in the degradation process, with that, only certain microorganisms can show the ability in degrading polypropylene. Due to this, the research area for responsible enzymes from microorganisms that can degrade polypropylene from the environment is something that can be looked at in-depth in the future.
Polyvinyl chloride degrading enzyme
Polyvinyl-based microplastic consists of vinyl backbone polymer. The repetition of vinyl (ethenyls) monomers in the formation of polymer chains consist of variation in its branch that represents the uniqueness of polyvinyl-integrated β-diketonebased polymer. The incorporation of chlorine in its branch is known as polyvinyl chloride (PVC), as one of the most plastic produced in the industry. Changing in branch molecules will give different properties of the polyvinyl polymer, e.g. incorporation of acetate will give plastic material named polyvinyl acetate/polyvinyl alcohol (PVA). When a butyral molecule is added to the branch, it is known as polyvinyl butyral (PVB) (Akovali 2012). PVC toxicity is well reported in numerous sources including scientific articles and mainstream articles. PVC is known to cause angiosarcoma in the human liver, other than that PVC also proved in targeting lung, brain and lymphohematopoietic function (Wagoner 1983). As years passed and awareness related to PVC toxicity increased, PVA was introduced to the industry as an alternative plastic replacing PVC or as an additive to PVC, mainly to increase the hydrophilicity of PVC (Cai et al. 2020a). PVA is said to be less toxic compared to PVC. PVA toxicity is very low even if orally administered, PVA also showed poorly absorbed by the gastrointestinal tract (DeMerlis and Schoneker 2003).
Oxidases are a group of enzymes involved in catalyzing the oxidation of C–N and C–O bonds in the presence of the oxygen molecule. Three major principal substrate classes for oxidase enzymes are amino acids, amines, and alcohols. The end product for amino acids and amines is imine, while alcohol end product will be ketones and aldehydes (Turner 2012). One of the oxidase enzymes reported in PVA degradation is PVA oxidase which present in Gram-negative bacteria, Pseudomonas sp. including Pseudomonas sp. O-3, P. vesicularis PD and Pseudomonas sp. VM15C (Wilkes and Aristilde 2017). PVA oxidase activity is mentioned to correlate with PVA dehydrogenase enzyme. PVA oxidase oxidizes the PVA through its serine hydrolase active site. The mechanism is initiated by the product from PVA dehydrogenase reaction introducing β-diketone group into the PVA polymer molecule. Later, PVA oxidase hydrolyzes the integrated β-diketone group within the PVA molecule to produce PVA monomer (Shimao et al. 2000).
However, data on PVC degradation by enzymes are still scarce. A review article released in 2020 by Ru et al. and his colleagues concluded that no scientific report mentioned the ability of biological components in degrading PVC (Ru et al. 2020). However, when searching through wider sources of publications, we found that (to the very best that we can do) few articles report the ability of fungi in degrading PVC (Ali et al. 2014a,b; Sumathi et al. 2016). These articles were later reviewed in 2019 (Glaser 2019). Even though these reports claimed to succeed in isolating fungi that may be beneficial in degrading PVC, detail mechanisms, especially enzyme/s involved in the reaction and fate of the PVC monomer were never mentioned in detail. In the same review article, it is mentioned that the recalcitrant feature showed by PVC can be the major factor for PVC to be resistant to biodegradation process (Glaser 2019). Thus, this leaves a research gap that is worth paying attention to the scientist. Research specifically on the microorganism degrading PVC is a potential area in the future and can be beneficial in microplastics treatment.
Conclusion
As reviewed above, microplastics are categorized based on their monomer types. The enzymes responsible for their degradation from microorganisms were mentioned and discussed for their mechanism in this review based on previously published articles. Several conclusions can be extracted from this review and mentioned below.
-
(a)
PE can be grouped into two major groups: LDPE and HDPE. With the most widespread pollution in the environment, PE has attracted the interest of researchers trying to understand degradation mechanisms through enzymes. Laccase, alkane hydrolase, manganese peroxidase, and soybean peroxidase were the enzymes reported to be responsible for PE degradation in the environment.
-
(b)
The presence of PET in the environment is mostly from the leachate of plastic bottles. Two types of enzymes were closely related to the degradation of PET. PETase and MHETase work continuously after one another. PETase breaks the PET polymer into MHET. MHETase uptakes MHET and converts it into TPA and ethylene glycol. Nonetheless, the mechanism of PETase and MHETase is well understood. The existence of other enzymes that can degrade PET, such as lipase and esterase, remains unclear.
-
(c)
Polystyrene is mainly used in food packaging industries. The degradation of polystyrene has been widely explored. Numerous organisms have been reported to degrade this pollutant. A series of enzymes (styrene monooxygenase, styrene oxide isomerase, phenylacetaldehyde dehydrogenase, and phenylacetyl coenzyme A ligase) were reported to be associated with polystyrene degradation, with acetyl-CoA as a final monomer that is utilized in the TCA cycle. Another enzyme that can degrade polystyrene is serine hydrolase, with little knowledge known about the mechanism.
-
(d)
PP is a low-density plastic that is commonly found in cosmetics and personal care products. Numerous microorganisms are claimed to be able to degrade PP. With numerous reports related to the microorganism’s degradation, there are no data on the enzyme-based mechanism.
-
(e)
PVC is categorized under the thermoplastic group. PVC degradation data is considered to be the scarcest among those mentioned above, with fungi as a sole microorganism reported to be able in degrading PVC. In addition, the enzyme that is responsible for PVC degradation has not been mentioned (until this article was written).
Although some microplastics (PE, PET and PS) degradation by enzyme was fully understood, the information on other microplastics (PP and PVC) degradation through enzyme reaction is still unexplored. Therefore, further effort to understand the enzyme-based degradation of these microplastics is needed, such as those listed below:
-
(a)
The purification and identification of enzymes responsible for PP and PVC degradation.
-
(b)
Mechanism elucidation of each enzyme that has been reported to degrade microplastics.
-
(c)
Enhancing the performance of the identified enzymes through various advanced engineering approaches.
Based on the important points aforementioned in this review, understanding the enzymatic reaction plays great importance in microplastics degradation. Enzyme identification and elucidating the mechanism are challenging, current advancement in protein analysis through proteomic approach can be the potential technique in solving this obstacle. If we manage to address these issues, we can provide a novel finding in this research area and a sustainable solution for the current pollution issue.
References
Abbasi S, Moore F, Keshavarzi B et al (2020) PET-microplastics as a vector for heavy metals in a simulated plant rhizosphere zone. Sci Total Environ 744:140984. https://doi.org/10.1016/j.scitotenv.2020.140984
Abbasi S, Turner A (2021) Human exposure to microplastics: a study in Iran. J Hazard Mater 403:123799. https://doi.org/10.1016/j.jhazmat.2020.123799
Abdelaal MY, Sobahi TR, Makki MSI (2008) Chemical degradation of poly(ethylene terephthalate). Int J Polym Mater Polym Biomater 57:73–80. https://doi.org/10.1080/00914030701329080
Akovali G (2012) Plastic materials: polyvinyl chloride (PVC). In: Pacheco-Torgal F, Jalali S, Fucic A (eds) Toxicity of building materials. Elsevier, Amsterdam, pp 23–53
Ali MI, Ahmed S, Javed I et al (2014a) Biodegradation of starch blended polyvinyl chloride films by isolated Phanerochaete chrysosporium PV1. Int J Environ Sci Technol 11:339–348. https://doi.org/10.1007/s13762-013-0220-5
Ali MI, Ahmed S, Robson G et al (2014b) Isolation and molecular characterization of polyvinyl chloride (PVC) plastic degrading fungal isolates. J Basic Microbiol 54:18–27. https://doi.org/10.1002/jobm.201200496
Alimi OS, Fadare OO, Okoffo ED (2021) Microplastics in African ecosystems: current knowledge, abundance, associated contaminants, techniques, and research needs. Sci Total Environ 755:142422
Álvarez-Barragán J, Domínguez-Malfavón L, Vargas-Suárez M et al (2016) Biodegradative activities of selected environmental fungi on a polyester polyurethane varnish and polyether polyurethane foams. Appl Environ Microbiol 82:5225–5235. https://doi.org/10.1128/AEM.01344-16
Arhant M, Le Gall M, Le Gac PY, Davies P (2019) Impact of hydrolytic degradation on mechanical properties of PET - Towards an understanding of microplastics formation. Polym Degrad Stab 161:175–182. https://doi.org/10.1016/j.polymdegradstab.2019.01.021
Arshad Z, Robaina M, Shahbaz M, Veloso AB (2020) The effects of deforestation and urbanization on sustainable growth in Asian countries. Environ Sci Pollut Res 27:10065–10086. https://doi.org/10.1007/s11356-019-07507-7
Ashter SA (2016) Introduction to bioplastics engineering. Elsevier, Amsterdam
Atiq N (2011) Biodegradability of synthetic plastics polystyrene and styrofoam by fungal isolates. Quaid-i-Azam University, Islamabad
Atiq N, Ahmed S, Ali MI et al (2010) Isolation and identification of polystyrene biodegrading bacteria from soil. African J Microbiol Res 4:1537–1541
Austin HP, Allen MD, Donohoe BS et al (2018) Characterization and engineering of a plastic-degrading aromatic polyesterase. Proc Natl Acad Sci U S A 115:E4350–E4357. https://doi.org/10.1073/pnas.1718804115
Auta HS, Emenike CU, Fauziah SH (2017) Screening of Bacillus strains isolated from mangrove ecosystems in Peninsular Malaysia for microplastic degradation. Environ Pollut 231:1552–1559. https://doi.org/10.1016/j.envpol.2017.09.043
Auta HS, Emenike CU, Jayanthi B, Fauziah SH (2018) Growth kinetics and biodeterioration of polypropylene microplastics by Bacillus sp. and Rhodococcus sp. isolated from mangrove sediment. Mar Pollut Bull 127:15–21. https://doi.org/10.1016/j.marpolbul.2017.11.036
Ayodeji A, Prashant B, Suren S, Santhosh P (2020) Plastic biodegradation: frontline microbes and their enzymes. Sci Total Environ. https://doi.org/10.1016/j.scitotenv.2020.143536
Balogh JM, Jámbor A (2020) The environmental impacts of agricultural trade: a systematic literature review. Sustain 12:1–16. https://doi.org/10.3390/su12031152
Bellas J, Gil I (2020) Polyethylene microplastics increase the toxicity of chlorpyrifos to the marine copepod Acartia tonsa. Environ Pollut 260:114059. https://doi.org/10.1016/j.envpol.2020.114059
Bharagava RN, Purchase D, Saxena G, Mulla SI (2018) Applications of metagenomics in microbial bioremediation of pollutants: from genomics to environmental cleanup. In: Das S, Dash HR (eds) Microbial Diversity in the genomic era. Elsevier, Amsterdam, pp 459–477
Bharagava RN, Saxena G, Mulla SI (2020) Introduction to industrial wastes containing organic and inorganic pollutants and bioremediation approaches for environmental management. In: Saxena G, Kishor R, Bharagava RN (eds) Bioremediation of industrial waste for environmental safety. Springer, Singapore, pp 1–18
Bhardwaj H, Gupta R, Tiwari A (2013) Communities of microbial enzymes associated with biodegradation of plastics. J Polym Environ 21:575–579
Bikker J, Lawson J, Wilson S, Rochman CM (2020) Microplastics and other anthropogenic particles in the surface waters of the Chesapeake Bay. Mar Pollut Bull 156:111257. https://doi.org/10.1016/j.marpolbul.2020.111257
Billen P, Khalifa L, Van Gerven F et al (2020) Technological application potential of polyethylene and polystyrene biodegradation by macro-organisms such as mealworms and wax moth larvae. Sci Total Environ 735:139521. https://doi.org/10.1016/j.scitotenv.2020.139521
Bollinger A, Thies S, Knieps-Grünhagen E et al (2020) A novel polyester hydrolase from the marine bacterium pseudomonas aestusnigri: structural and functional insights. Front Microbiol 11:1–16. https://doi.org/10.3389/fmicb.2020.00114
Bonilla J, Paiano RB, Lourenço RV et al (2020) Biodegradability in aquatic system of thin materials based on chitosan, PBAT and HDPE polymers: respirometric and physical-chemical analysis. Int J Biol Macromol 164:1399–1412. https://doi.org/10.1016/j.ijbiomac.2020.07.309
Borah P, Kumar M, Devi P (2020) Types of inorganic pollutants: metals/metalloids, acids, and organic forms. In: Inorganic pollutants in water. Elsevier, pp 17–31
Budi Kurniawan S, Rozaimah Sheikh Abdullah S, Fauzul Imron M, Ismail I (2020) Current state of marine plastic pollution and its technology for more eminent evidence: a review. J Clean Prod. https://doi.org/10.1016/j.jclepro.2020.123537
Cai H, Wang Y, Wu K, Guo W (2020a) Enhanced hydrophilic and electrophilic properties of polyvinyl chloride (PVC) biofilm carrier. Polymers (Basel). https://doi.org/10.3390/POLYM12061240
Cai Y, Mitrano DM, Heuberger M et al (2020b) The origin of microplastic fiber in polyester textiles: the textile production process matters. J Clean Prod 267:121970. https://doi.org/10.1016/j.jclepro.2020.121970
Campbell JF (2019) Human factors: the impact on industry and the environment. In: Natural resources management and biological sciences [Working Title]. IntechOpen
Chagas TQ, da Araújo APC, Malafaia G (2020) Biomicroplastics versus conventional microplastics: an insight on the toxicity of these polymers in dragonfly larvae. Sci Total Environ. https://doi.org/10.1016/j.scitotenv.2020.143231
Chamas A, Moon H, Zheng J et al (2020) Degradation rates of plastics in the environment. ACS Sustain Chem Eng 8:3494–3511. https://doi.org/10.1021/acssuschemeng.9b06635
Chandra P, Enespa, Singh DP (2020) Microplastic degradation by bacteria in aquatic ecosystem. In: Microorganisms for sustainable environment and health. Elsevier, pp 431–467
Chatterjee S, Roy B, Roy D, Banerjee R (2010) Enzyme-mediated biodegradation of heat treated commercial polyethylene by Staphylococcal species. Polym Degrad Stab 95:195–200. https://doi.org/10.1016/j.polymdegradstab.2009.11.025
Chatterjee S, Sharma S (2019) Microplastics in our oceans and marine health. F Actions Sci Reports 54–61
Chen C, Han X, Ko T et al (2018) Structural studies reveal the molecular mechanism of PETase. FEBS J 285:3717–3723. https://doi.org/10.1111/febs.14612
Chen Y, Wang L, Yu H et al (2015) Synthesis and application of polyethylene-based functionalized hyperbranched polymers. Prog Polym Sci 45:23–43
Cheng H, Feng Y, Duan Z et al (2020) Toxicities of microplastic fibers and granules on the development of zebrafish embryos and their combined effects with cadmium. Chemosphere. https://doi.org/10.1016/j.chemosphere.2020.128677
Cheng HC, Qi RZ, Paudel H, Zhu HJ (2011) Regulation and function of protein kinases and phosphatases. Enzyme Res. 2011
Chia WY, Ying Tang DY, Khoo KS et al (2020) Nature’s fight against plastic pollution: algae for plastic biodegradation and bioplastics production. Environ Sci Ecotechnol. https://doi.org/10.1016/j.ese.2020.100065
Chowdhury TU, Mahi MA, Haque KA, Mostafizur Rahman M (2018) A review on the use of polyethylene terephthalate (PET) as aggregates in concrete. Malaysian J Sci 37:118–136
Cooper GM (2000) The cell: a molecular approach, 2nd edn. Sinauer Associates Inc, Sunderland
Crossman J, Hurley RR, Futter M, Nizzetto L (2020) Transfer and transport of microplastics from biosolids to agricultural soils and the wider environment. Sci Total Environ 724:138334. https://doi.org/10.1016/j.scitotenv.2020.138334
Cucini C, Leo C, Vitale M et al (2020) Bacterial and fungal diversity in the gut of polystyrene-fed Alphitobius diaperinus (Insecta: Coleoptera). Anim Gene. https://doi.org/10.1016/j.angen.2020.200109
da Costa AM, de Oliveira Lopes VR, Vidal L et al (2020) Poly(ethylene terephthalate) (PET) degradation by Yarrowia lipolytica: Investigations on cell growth, enzyme production and monomers consumption. Process Biochem 95:81–90. https://doi.org/10.1016/j.procbio.2020.04.001
Dağ SE, Bozkurt PA, Eroğlu F, Çelik M (2019) Preparation, characterization, and properties of polystyrene/Na-montmorillonite composites. J Thermoplast Compos Mater 32:1078–1091. https://doi.org/10.1177/0892705718785691
Danso D, Chow J, Streita WR (2019) Plastics: environmental and biotechnological perspectives on microbial degradation. Appl Environ Microbiol 85
Das P, Tiwari P (2019) Thermal degradation study of waste polyethylene terephthalate (PET) under inert and oxidative environments. Thermochim Acta 679:178340. https://doi.org/10.1016/j.tca.2019.178340
De Falco F, Di Pace E, Cocca M, Avella M (2019) The contribution of washing processes of synthetic clothes to microplastic pollution. Sci Rep 9:1–11. https://doi.org/10.1038/s41598-019-43023-x
DeMerlis CC, Schoneker DR (2003) Review of the oral toxicity of polyvinyl alcohol (PVA). Food Chem Toxicol 41:319–326
Denaro R, Aulenta F, Crisafi F et al (2020) Marine hydrocarbon-degrading bacteria breakdown poly(ethylene terephthalate) (PET). Sci Total Environ. https://doi.org/10.1016/j.scitotenv.2020.141608
Ding L, Zhang S, Wang X et al (2020) The occurrence and distribution characteristics of microplastics in the agricultural soils of Shaanxi Province, in north-western China. Sci Total Environ 720:137525. https://doi.org/10.1016/j.scitotenv.2020.137525
Dong H, Chen Y, Wang J et al (2021) Interactions of microplastics and antibiotic resistance genes and their effects on the aquaculture environments. J Hazard Mater 403:123961
Ellis C (2018) World Bank: Global waste generation could increase 70% by 2050 | Waste Dive. In: https://www.wastedive.com/. https://www.wastedive.com/news/world-bank-global-waste-generation-2050/533031/. Accessed 19 Oct 2020
Esquinas GGMS, Mantala AP, Atilano MG et al (2020) Physical characterization of litter and microplastic along the urban coast of Cagayan de Oro in Macajalar Bay. Philippines Mar Pollut Bull 154:111083. https://doi.org/10.1016/j.marpolbul.2020.111083
Farrelly TA, Shaw IC (2017) Polystyrene as hazardous household waste. In: Household hazardous waste management. InTech
Fecker T, Galaz-Davison P, Engelberger F et al (2018) Active site flexibility as a hallmark for efficient PET degradation by I. sakaiensis PETase. Biophys J 114:1302–1312. https://doi.org/10.1016/j.bpj.2018.02.005
Frehland S, Kaegi R, Hufenus R, Mitrano DM (2020) Long-term assessment of nanoplastic particle and microplastic fiber flux through a pilot wastewater treatment plant using metal-doped plastics. Water Res 182:115860. https://doi.org/10.1016/j.watres.2020.115860
Funck M, Yildirim A, Nickel C et al (2020) Identification of microplastics in wastewater after cascade filtration using Pyrolysis-GC–MS. MethodsX 7:100778. https://doi.org/10.1016/j.mex.2019.100778
Gandara e Silva PP, Nobre CR, Resaffe P et al (2016) Leachate from microplastics impairs larval development in brown mussels. Water Res 106:364–370. https://doi.org/10.1016/j.watres.2016.10.016
Ganesh Kumar A, Anjana K, Hinduja M et al (2020) Review on plastic wastes in marine environment: biodegradation and biotechnological solutions. Mar Pollut Bull 150:110733
Gao M, Liu Y, Dong Y, Song Z (2021) Effect of polyethylene particles on dibutyl phthalate toxicity in lettuce (Lactuca sativa L.). J Hazard Mater 401:123422. https://doi.org/10.1016/j.jhazmat.2020.123422
Gao M, Xu Y, Liu Y et al (2020) Effect of polystyrene on di-butyl phthalate (DBP) bioavailability and DBP-induced phytotoxicity in lettuce. Environ Pollut 268:115870. https://doi.org/10.1016/j.envpol.2020.115870
Ghatge S, Yang Y, Ahn J-H, Hur H-G (2020) Biodegradation of polyethylene: a brief review. Appl Biol Chem 63:27. https://doi.org/10.1186/s13765-020-00511-3
Glaser JA (2019) Biological degradation of polymers in the environment. In: Gomiero A (ed) Plastics in the environment. IntechOpen, London
Gómez-Méndez LD, Moreno-Bayona DA, Poutou-Piñales RA et al (2018) Biodeterioration of plasma pretreated LDPE sheets by Pleurotus ostreatus. PLoS ONE 13:e0203786. https://doi.org/10.1371/journal.pone.0203786
Gong J, Kong T, Li Y et al (2018) Biodegradation of microplastic derived from poly(ethylene terephthalate) with bacterial whole-cell biocatalysts. Polymers (Basel). https://doi.org/10.3390/polym10121326
Green DS, Kregting L, Boots B et al (2018) A comparison of sampling methods for seawater microplastics and a first report of the microplastic litter in coastal waters of Ascension and Falkland Islands. Mar Pollut Bull 137:695–701. https://doi.org/10.1016/j.marpolbul.2018.11.004
Guan N, Li J, Shin H, et al (2017) Microbial response to environmental stresses: from fundamental mechanisms to practical applications. Appl Microbiol Biotechnol 101:3991–4008
Guan N, Liu L (2020) Microbial response to acid stress: mechanisms and applications. Appl Microbiol Biotechnol 104:51–65
Guimarães ATB, Estrela FN, Pereira PS et al (2021) Toxicity of polystyrene nanoplastics in Ctenopharyngodon idella juveniles: a genotoxic, mutagenic and cytotoxic perspective. Sci Total Environ 752:141937. https://doi.org/10.1016/j.scitotenv.2020.141937
Haddaway NR, Cooke SJ, Lesser P et al (2019) Evidence of the impacts of metal mining and the effectiveness of mining mitigation measures on social-ecological systems in Arctic and boreal regions: a systematic map protocol. Environ Evid 8:9. https://doi.org/10.1186/s13750-019-0152-8
He P, Chen L, Shao L et al (2019) Municipal solid waste (MSW)landfill: a source of microplastics? -Evidence of microplastics in landfill leachate. Water Res 159:38–45. https://doi.org/10.1016/j.watres.2019.04.060
Heindler FM, Alajmi F, Huerlimann R et al (2017) Toxic effects of polyethylene terephthalate microparticles and Di(2-ethylhexyl)phthalate on the calanoid copepod, Parvocalanus crassirostris. Ecotoxicol Environ Saf 141:298–305. https://doi.org/10.1016/j.ecoenv.2017.03.029
Henry B, Laitala K, Klepp IG (2019) Microfibres from apparel and home textiles: prospects for including microplastics in environmental sustainability assessment. Sci Total Environ 652:483–494. https://doi.org/10.1016/j.scitotenv.2018.10.166
Hernández-Arenas R, Beltrán-Sanahuja A, Navarro-Quirant P, Sanz-Lazaro C (2021) The effect of sewage sludge containing microplastics on growth and fruit development of tomato plants. Environ Pollut 268:115779. https://doi.org/10.1016/j.envpol.2020.115779
Herrero Acero E, Ribitsch D, Steinkellner G et al (2011) Enzymatic surface hydrolysis of PET: effect of structural diversity on kinetic properties of cutinases from Thermobifida. Macromolecules 44:4632–4640. https://doi.org/10.1021/ma200949p
Ho BT, Roberts TK, Lucas S (2018) An overview on biodegradation of polystyrene and modified polystyrene: the microbial approach. Crit Rev Biotechnol 38:308–320
Horvath T, Kalman M, Szabo T, et al (2018) The mechanical properties of polyethylene-terephthalate (PET) and polylactic-acid (PDLLA and PLLA), the influence of material structure on forming. In: IOP conference series: materials science and engineering. Institute of Physics Publishing, p 012018
Huang W, Song B, Liang J et al (2020) Microplastics and associated contaminants in the aquatic environment: a review on their ecotoxicological effects, trophic transfer, and potential impacts to human health. Elsevier, Amsterdam
Huang Y, Yan M, Xu K et al (2019) Distribution characteristics of microplastics in Zhubi Reef from South China Sea. Environ Pollut 255:113133. https://doi.org/10.1016/j.envpol.2019.113133
Hwang J, Choi D, Han S et al (2019) An assessment of the toxicity of polypropylene microplastics in human derived cells. Sci Total Environ 684:657–669. https://doi.org/10.1016/j.scitotenv.2019.05.071
Hwang J, Choi D, Han S et al (2020) Potential toxicity of polystyrene microplastic particles. Sci Rep 10:1–12. https://doi.org/10.1038/s41598-020-64464-9
Hwang JW, Choi CY, Park S, Lee EY (2008) Biodegradation of gaseous styrene by Brevibacillus sp. using a novel agitating biotrickling filter. Biotechnol Lett 30:1207–1212. https://doi.org/10.1007/s10529-008-9670-0
Islam S, Apitius L, Jakob F, Schwaneberg U (2019) Targeting microplastic particles in the void of diluted suspensions. Environ Int 123:428–435. https://doi.org/10.1016/j.envint.2018.12.029
Jaafar N, Musa SM, Azfaralariff A et al (2020) Improving the efficiency of post-digestion method in extracting microplastics from gastrointestinal tract and gills of fish. Chemosphere. https://doi.org/10.1016/j.chemosphere.2020.127649
Jaikumar G, Brun NR, Vijver MG, Bosker T (2019) Reproductive toxicity of primary and secondary microplastics to three cladocerans during chronic exposure. Environ Pollut 249:638–646. https://doi.org/10.1016/j.envpol.2019.03.085
Jiang X, Chen H, Liao Y et al (2019) Ecotoxicity and genotoxicity of polystyrene microplastics on higher plant Vicia faba. Environ Pollut 250:831–838. https://doi.org/10.1016/j.envpol.2019.04.055
Ju P, Zhang Y, Zheng Y et al (2020) Probing the toxic interactions between polyvinyl chloride microplastics and Human Serum Albumin by multispectroscopic techniques. Sci Total Environ 734:139219. https://doi.org/10.1016/j.scitotenv.2020.139219
Kalčíková G, Žgajnar Gotvajn A, Kladnik A, Jemec A (2017) Impact of polyethylene microbeads on the floating freshwater plant duckweed Lemna minor. Environ Pollut 230:1108–1115. https://doi.org/10.1016/j.envpol.2017.07.050
Kalnasa ML, Lantaca SMO, Boter LC et al (2019) Occurrence of surface sand microplastic and litter in Macajalar Bay. Philippines Mar Pollut Bull 149:110521. https://doi.org/10.1016/j.marpolbul.2019.110521
Kang BR, Bin KS, Song HA, Lee TK (2019) Accelerating the biodegradation of high-density polyethylene (Hdpe) using bjerkandera adusta tbb-03 and lignocellulose substrates. Microorganisms. https://doi.org/10.3390/microorganisms7090304
Kawai F, Kawabata T, Oda M (2019) Current knowledge on enzymatic PET degradation and its possible application to waste stream management and other fields. Appl Microbiol Biotechnol 103:4253–4268
Kawai F, Oda M, Tamashiro T et al (2014) A novel Ca2+-activated, thermostabilized polyesterase capable of hydrolyzing polyethylene terephthalate from Saccharomonospora viridis AHK190. Appl Microbiol Biotechnol 98:10053–10064. https://doi.org/10.1007/s00253-014-5860-y
Khalid N, Aqeel M, Noman A et al (2021) Linking effects of microplastics to ecological impacts in marine environments. Chemosphere 264:128541. https://doi.org/10.1016/j.chemosphere.2020.128541
Khalid N, Aqeel M, Noman A (2020) Microplastics could be a threat to plants in terrestrial systems directly or indirectly. Environ Pollut 267:115653. https://doi.org/10.1016/j.envpol.2020.115653
Khoironi A, Hadiyanto H, Anggoro S, Sudarno S (2020) Evaluation of polypropylene plastic degradation and microplastic identification in sediments at Tambak Lorok coastal area, Semarang. Indonesia Mar Pollut Bull 151:110868. https://doi.org/10.1016/j.marpolbul.2019.110868
Kik K, Bukowska B, Sicińska P (2020) Polystyrene nanoparticles: Sources, occurrence in the environment, distribution in tissues, accumulation and toxicity to various organisms. Environ Pollut 262:114297
Kim HR, Lee HM, Yu HC et al (2020a) Biodegradation of Polystyrene by Pseudomonas sp. Isolated from the Gut of Superworms (Larvae of Zophobas atratus). Environ Sci Technol 54:6987–6996. https://doi.org/10.1021/acs.est.0c01495
Kim JW, Bin PS, Tran QG et al (2020b) Functional expression of polyethylene terephthalate-degrading enzyme (PETase) in green microalgae. Microb Cell Fact 19:97. https://doi.org/10.1186/s12934-020-01355-8
Knott BC, Erickson E, Allen MD et al (2020) Characterization and engineering of a two-enzyme system for plastics depolymerization. Proc Natl Acad Sci 117:25476–25485. https://doi.org/10.1073/pnas.2006753117
Koerner GR, Hsuan YG, Koerner RM (2006) The durability of geosynthetics. In: Geosynthetics in civil engineering. Elsevier Ltd., pp 36–65
Kumar M, Xiong X, He M et al (2020) Microplastics as pollutants in agricultural soils. Environ Pollut 265:114980
Kumar Sen S, Raut S (2015) Microbial degradation of low density polyethylene (LDPE): A review. J Environ Chem Eng 3:462–473
Lechner A, Ramler D (2015) The discharge of certain amounts of industrial microplastic from a production plant into the River Danube is permitted by the Austrian legislation. Environ Pollut 200:159–160. https://doi.org/10.1016/j.envpol.2015.02.019
Lee J, Chae KJ (2021) A systematic protocol of microplastics analysis from their identification to quantification in water environment: a comprehensive review. J Hazard Mater 403:124049
Li L, Li Z, Liu D, Song K (2020) Evaluation of partial nitrification efficiency as a response to cadmium concentration and microplastic polyvinylchloride abundance during landfill leachate treatment. Chemosphere 247:125903. https://doi.org/10.1016/j.chemosphere.2020.125903
Libralato G, Galdiero E, Falanga A, et al (2017) Toxicity effects of functionalized quantum dots, gold and polystyrene nanoparticles on target aquatic biological models: a review. Molecules 22
Llorca M, Álvarez-Muñoz D, Ábalos M et al (2020) Microplastics in Mediterranean coastal area: toxicity and impact for the environment and human health. Trends Environ Anal Chem 27:e00090. https://doi.org/10.1016/j.teac.2020.e00090
Löder MGJ, Imhof HK, Ladehoff M et al (2017) Enzymatic purification of microplastics in environmental samples. Environ Sci Technol 51:14283–14292. https://doi.org/10.1021/acs.est.7b03055
López-Pedrouso M, Varela Z, Franco D et al (2020) Can proteomics contribute to biomonitoring of aquatic pollution? A critical review. Environ Pollut. https://doi.org/10.1016/j.envpol.2020.115473
Ma Y, Yao M, Li B et al (2018) Enhanced Poly(ethylene terephthalate) hydrolase activity by protein engineering. Engineering 4:888–893. https://doi.org/10.1016/j.eng.2018.09.007
Maity S, Pramanick K (2020) Perspectives and challenges of micro/nanoplastics-induced toxicity with special reference to phytotoxicity. Glob Chang Biol 26:3241–3250. https://doi.org/10.1111/gcb.15074
Mateos-Cárdenas A, Scott DT, Seitmaganbetova G et al (2019) Polyethylene microplastics adhere to Lemna minor (L.), yet have no effects on plant growth or feeding by Gammarus duebeni (Lillj.). Sci Total Environ 689:413–421. https://doi.org/10.1016/j.scitotenv.2019.06.359
Matjašič T, Simčič T, Medvešček N et al (2021) Critical evaluation of biodegradation studies on synthetic plastics through a systematic literature review. Sci Total Environ 752:141959
Mintenig SM, Int-Veen I, Löder MGJ et al (2017) Identification of microplastic in effluents of waste water treatment plants using focal plane array-based micro-Fourier-transform infrared imaging. Water Res 108:365–372. https://doi.org/10.1016/j.watres.2016.11.015
Mohajerani A, Karabatak B (2020) Microplastics and pollutants in biosolids have contaminated agricultural soils: an analytical study and a proposal to cease the use of biosolids in farmlands and utilise them in sustainable bricks. Waste Manag 107:252–265
Mohammed SA, Yusop RM, Mohammed MA et al (2019) Additives aid switch to protect the photodegradation of plastics in outdoor construction. Al-Nahrain J Eng Sci 22:277–282. https://doi.org/10.29194/njes.22040277
Montazer Z, Habibi Najafi MB, Levin DB (2020) Challenges with verifying microbial degradation of polyethylene. Polymers (Basel) 12:123. https://doi.org/10.3390/polym12010123
Moog D, Schmitt J, Senger J et al (2019) Using a marine microalga as a chassis for polyethylene terephthalate (PET) degradation. Microb Cell Fact 18:1–15. https://doi.org/10.1186/s12934-019-1220-z
Mor R, Sivan A (2008) Biofilm formation and partial biodegradation of polystyrene by the actinomycete Rhodococcus ruber: biodegradation of polystyrene. Biodegradation 19:851–858. https://doi.org/10.1007/s10532-008-9188-0
Nakamiya K, Sakasita G, Ooi T, Kinoshita S (1997) Enzymatic degradation of polystyrene by hydroquinone peroxidase of Azotobacter beijerinckii HM121. J Ferment Bioeng 84:480–482. https://doi.org/10.1016/S0922-338X(97)82013-2
Nava V, Leoni B (2020) A critical review of interactions between microplastics, microalgae and aquatic ecosystem function. Water Res 188:116476. https://doi.org/10.1016/j.watres.2020.116476
NicAogáin K, O’Byrne CP (2016) The role of stress and stress adaptations in determining the fate of the bacterial pathogen listeria monocytogenes in the food chain. Front Microbiol 7:1865. https://doi.org/10.3389/fmicb.2016.01865
Nolte TM, Hartmann NB, Kleijn JM et al (2017) The toxicity of plastic nanoparticles to green algae as influenced by surface modification, medium hardness and cellular adsorption. Aquat Toxicol 183:11–20. https://doi.org/10.1016/j.aquatox.2016.12.005
Nuawi MZ, Fadli Ahmad MA, Mohamed NF et al (2016) The study of polymer material characterisation using M-Z-N statistical analysis method. J Kejuruter 28:9–18. https://doi.org/10.17576/jkukm-2016-28-02
O’Leary ND, O’Connor KE, Dobson ADW (2002) Biochemistry, genetics and physiology of microbial styrene degradation. FEMS Microbiol Rev 26:403–417. https://doi.org/10.1111/j.1574-6976.2002.tb00622.x
Oberbeckmann S, Labrenz M (2020) Marine microbial assemblages on microplastics: diversity, adaptation, and role in degradation. Ann Rev Mar Sci 12:209–232. https://doi.org/10.1146/annurev-marine-010419-010633
Oikawa E, Linn KT, Endo T et al (2003) Isolation and Characterization of polystyrene degrading microorganisms for zero emission treatment of expanded polystyrene. Environ Eng Res 40:373–379. https://doi.org/10.11532/PROES1992.40.373
Ojha N, Pradhan N, Singh S et al (2016) Evaluation of HDPE and LDPE degradation by fungus, implemented by statistical optimization. Sci Rep. https://doi.org/10.1038/srep39515
Padervand M, Lichtfouse E, Robert D, Wang C (2020) Removal of microplastics from the environment: a review. Environ Chem Lett 18:807–828
Palm GJ, Reisky L, Böttcher D et al (2019) Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate. Nat Commun 10:1–10. https://doi.org/10.1038/s41467-019-09326-3
Parolini M, Ferrario C, De Felice B et al (2020) Interactive effects between sinking polyethylene terephthalate (PET) microplastics deriving from water bottles and a benthic grazer. J Hazard Mater 398:122848. https://doi.org/10.1016/j.jhazmat.2020.122848
Patel K, Chikkali SH, Sivaram S (2020) Ultrahigh molecular weight polyethylene: catalysis, structure, properties, processing and applications. Prog Polym Sci 109:101290
Peng BY, Li Y, Fan R et al (2020) Biodegradation of low-density polyethylene and polystyrene in superworms, larvae of Zophobas atratus (Coleoptera: Tenebrionidae): broad and limited extent depolymerization. Environ Pollut 266:115206. https://doi.org/10.1016/j.envpol.2020.115206
Pires JP, Miranda GM, de Souza GL et al (2019) Investigation of degradation of polypropylene in soil using an enzymatic additive. Iran Polym J 28:1045–1055. https://doi.org/10.1007/s13726-019-00766-8
Qi R, Jones DL, Li Z et al (2020) Behavior of microplastics and plastic film residues in the soil environment: a critical review. Sci Total Environ 703:134722
Qiang L, Cheng J (2021) Exposure to polystyrene microplastics impairs gonads of zebrafish (Danio rerio). Chemosphere 263:128161. https://doi.org/10.1016/j.chemosphere.2020.128161
Rainieri S, Conlledo N, Larsen BK et al (2018) Combined effects of microplastics and chemical contaminants on the organ toxicity of zebrafish (Danio rerio). Environ Res 162:135–143. https://doi.org/10.1016/j.envres.2017.12.019
Restrepo-Flórez JM, Bassi A, Thompson MR (2014) Microbial degradation and deterioration of polyethylene: a review. Int Biodeterior Biodegrad 88:83–90
Reynolds A, Giltrap DM, Chambers PG (2021) Acute growth inhibition & toxicity analysis of nano-polystyrene spheres on Raphidocelis subcapitata. Ecotoxicol Environ Saf 207:111153. https://doi.org/10.1016/j.ecoenv.2020.111153
Ribitsch D, Heumann S, Trotscha E et al (2011) Hydrolysis of polyethyleneterephthalate by p-nitrobenzylesterase from Bacillus subtilis. Biotechnol Prog 27:951–960. https://doi.org/10.1002/btpr.610
Rios LM, Jones PR, Moore C, Narayan UV (2010) Quantitation of persistent organic pollutants adsorbed on plastic debris from the Northern Pacific Gyre’s “eastern garbage patch.” J Environ Monit 12:2226–2236. https://doi.org/10.1039/c0em00239a
Ru J, Huo Y, Yang Y (2020) Microbial degradation and valorization of plastic wastes. Front Microbiol 11:442
Salvador M, Abdulmutalib U, Gonzalez J et al (2019) Microbial genes for a circular and sustainable bio-PET economy. Genes (Basel) 10:1–15. https://doi.org/10.3390/genes10050373
Sang T, Wallis CJ, Hill G, Britovsek GJP (2020) Polyethylene terephthalate degradation under natural and accelerated weathering conditions. Eur Polym J 136:109873
Sangale MK, Shahnawaz M, Ade AB (2019) Potential of fungi isolated from the dumping sites mangrove rhizosphere soil to degrade polythene. Sci Rep 9:1–11. https://doi.org/10.1038/s41598-019-41448-y
Sangeetha Devi R, Rajesh Kannan V, Nivas D et al (2015) Biodegradation of HDPE by Aspergillus spp. from marine ecosystem of Gulf of Mannar. India Mar Pollut Bull 96:32–40. https://doi.org/10.1016/j.marpolbul.2015.05.050
Sangeetha Devi R, Ramya R, Kannan K et al (2019) Investigation of biodegradation potentials of high density polyethylene degrading marine bacteria isolated from the coastal regions of Tamil Nadu, India. Mar Pollut Bull 138:549–560. https://doi.org/10.1016/j.marpolbul.2018.12.001
Sax L (2010) Polyethylene terephthalate May yield endocrine disruptors. Environ Health Perspect 118:445–448. https://doi.org/10.1289/ehp.0901253
Shaikh MM, Al Suhaimi AO, Hanafiah MM et al (2018) Study on migration of phenolic and volatile organic compounds from plastic pipes used in plumbing home networks into tap water. Desalin Water Treat 112:344–350. https://doi.org/10.5004/dwt.2018.22341
Shi J, Wu D, Su Y, Xie B (2020) Selective enrichment of antibiotic resistance genes and pathogens on polystyrene microplastics in landfill leachate. Sci Total Environ. https://doi.org/10.1016/j.scitotenv.2020.142775
Shimao M, Tamogami T, Kishida S, Harayama S (2000) The gene pvaB encodes oxidized polyvinyl alcohol hydrolase of Pseudomonas sp. strain VM15C and forms an operon with the polyvinyl alcohol dehydrogenase gene pvaA. Microbiology 146:649–657. https://doi.org/10.1099/00221287-146-3-649
Silva CJM, Patrício Silva AL, Campos D et al (2021) Oxidative damage and decreased aerobic energy production due to ingestion of polyethylene microplastics by Chironomus riparius (Diptera) larvae. J Hazard Mater 402:123775. https://doi.org/10.1016/j.jhazmat.2020.123775
Sjollema SB, Redondo-Hasselerharm P, Leslie HA et al (2016) Do plastic particles affect microalgal photosynthesis and growth? Aquat Toxicol 170:259–261. https://doi.org/10.1016/j.aquatox.2015.12.002
Sökmen TÖ, Sulukan E, Türkoğlu M et al (2020) Polystyrene nanoplastics (20 nm) are able to bioaccumulate and cause oxidative DNA damages in the brain tissue of zebrafish embryo (Danio rerio). Neurotoxicology 77:51–59. https://doi.org/10.1016/j.neuro.2019.12.010
Sol D, Laca A, Laca A, Díaz M (2020) Approaching the environmental problem of microplastics: importance of WWTP treatments. Sci Total Environ 740:140016
Song Y, Qiu R, Hu J et al (2020) Biodegradation and disintegration of expanded polystyrene by land snails Achatina fulica. Sci Total Environ 746:141289. https://doi.org/10.1016/j.scitotenv.2020.141289
Suardy NH, Tahrim NA, Ramli S (2020) Analysis and characterization of microplastic from personal care products and surface water in Bangi, Selangor. Sains Malaysiana 49:2237–2249. https://doi.org/10.17576/jsm-2020-4909-21
Sumathi T, Viswanath B, Sri Lakshmi A, Saigopal DVR (2016) Production of laccase by cochliobolus sp. isolated from plastic dumped soils and their ability to degrade low molecular weight PVC. Biochem Res Int. https://doi.org/10.1155/2016/9519527
Tang Y, Liu Y, Chen Y et al (2020) A review: research progress on microplastic pollutants in aquatic environments. Sci Total Environ. https://doi.org/10.1016/j.scitotenv.2020.142572
Taylor SE, Pearce CI, Sanguinet KA et al (2020) Polystyrene nano-A nd microplastic accumulation at Arabidopsis and wheat root cap cells, but no evidence for uptake into roots. Environ Sci Nano 7:1942–1953. https://doi.org/10.1039/d0en00309c
Trestrail C, Walpitagama M, Hedges C et al (2020) Foaming at the mouth: ingestion of floral foam microplastics by aquatic animals. Sci Total Environ 705:135826. https://doi.org/10.1016/j.scitotenv.2019.135826
Turner NJ (2012) 7.12 Oxidation: oxidases. In: Comprehensive chirality. Elsevier Ltd, pp 256–274
Uheida A, Mejía HG, Abdel-Rehim M et al (2020) Visible light photocatalytic degradation of polypropylene microplastics in a continuous water flow system. J Hazard Mater. https://doi.org/10.1016/j.jhazmat.2020.124299
Urbanek AK, Rybak J, Wróbel M et al (2020) A comprehensive assessment of microbiome diversity in Tenebrio molitor fed with polystyrene waste. Environ Pollut 262:114281. https://doi.org/10.1016/j.envpol.2020.114281
van den Berg P, Huerta-Lwanga E, Corradini F, Geissen V (2020) Sewage sludge application as a vehicle for microplastics in eastern Spanish agricultural soils. Environ Pollut 261:114198. https://doi.org/10.1016/j.envpol.2020.114198
Vertommen M, Nierstrasz V, Van Der VM, Warmoeskerken MMCG (2005) Enzymatic surface modification of poly(ethylene terephthalate). J Biotechnol 120:376–386. https://doi.org/10.1016/j.jbiotec.2005.06.015
Wagoner JK (1983) Toxicity of vinyl chloride and poly(vinyl chloride): a critical review. Environ Health Perspect 52:61–66. https://doi.org/10.1289/ehp.835261
Wang C, Zhao J, Xing B (2020a) Environmental source, fate, and toxicity of microplastics. J Hazard Mater. https://doi.org/10.1016/j.jhazmat.2020.124357
Wang J, Li Y, Lu L et al (2019) Polystyrene microplastics cause tissue damages, sex-specific reproductive disruption and transgenerational effects in marine medaka (Oryzias melastigma). Environ Pollut 254:113024. https://doi.org/10.1016/j.envpol.2019.113024
Wang W, Ge J, Yu X (2020b) Bioavailability and toxicity of microplastics to fish species: a review. Ecotoxicol Environ Saf 189:109913. https://doi.org/10.1016/j.ecoenv.2019.109913
Ward CP, Armstrong CJ, Walsh AN et al (2019) Sunlight converts polystyrene to carbon dioxide and dissolved organic carbon. Environ Sci Technol Lett. https://doi.org/10.1021/acs.estlett.9b00532
Wen D, Fu R, Li Q (2021) Removal of inorganic contaminants in soil by electrokinetic remediation technologies: a review. J Hazard Mater 401:123345
Wilkes RA, Aristilde L (2017) Degradation and metabolism of synthetic plastics and associated products by Pseudomonas sp.: capabilities and challenges. J Appl Microbiol 123:582–593. https://doi.org/10.1111/jam.13472
Winkel BSJ (2017) When an enzyme isn’t just an enzyme anymore. J Exp Bot 68:1387–1389
Wolff S, Kerpen J, Prediger J et al (2019) Determination of the microplastics emission in the effluent of a municipal waste water treatment plant using Raman microspectroscopy. Water Res X 2:100014. https://doi.org/10.1016/j.wroa.2018.100014
Worm B, Lotze HK, Jubinville I et al (2017) Plastic as a persistent marine pollutant. Annu Rev Environ Resour 42:1–26
Wu H, Lv S, He Y, Qu JP (2019) The study of the thermomechanical degradation and mechanical properties of PET recycled by industrial-scale elongational processing. Polym Test 77:105882. https://doi.org/10.1016/j.polymertesting.2019.04.029
Wu S, Montalvo L (2020) Repurposing waste plastics into cleaner asphalt pavement materials: a critical literature review. J Clean Prod 280:124355. https://doi.org/10.1016/j.jclepro.2020.124355
Yang L, Gao J, Liu Y et al (2021a) Biodegradation of expanded polystyrene and low-density polyethylene foams in larvae of Tenebrio molitor Linnaeus (Coleoptera: Tenebrionidae): broad versus limited extent depolymerization and microbe-dependence versus independence. Chemosphere 262:127818. https://doi.org/10.1016/j.chemosphere.2020.127818
Yang L, Zhang Y, Kang S et al (2021b) Microplastics in freshwater sediment: a review on methods, occurrence, and sources. Sci Total Environ 754:141948
Yang Y, Liu W, Zhang Z et al (2020a) Microplastics provide new microbial niches in aquatic environments. Appl Microbiol Biotechnol 104:6501–6511
Yang Y, Wang J, Xia M (2020b) Biodegradation and mineralization of polystyrene by plastic-eating superworms Zophobas atratus. Sci Total Environ 708:135233. https://doi.org/10.1016/j.scitotenv.2019.135233
Yoshida S, Hiraga K, Takehana T et al (2016) A bacterium that degrades and assimilates poly(ethylene terephthalate). Science 351:1196–1199. https://doi.org/10.1126/science.aad6359
Yu H, Zhang X, Hu J et al (2020) Ecotoxicity of polystyrene microplastics to submerged carnivorous Utricularia vulgaris plants in freshwater ecosystems. Environ Pollut 265:114830. https://doi.org/10.1016/j.envpol.2020.114830
Zhang B, Du X, Jia S et al (2011) Rapid assessment of DNA damage induced by polystyrene nanosphere suspension using a photoelectrochemical DNA sensor. Sci China Chem 54:1260–1265. https://doi.org/10.1007/s11426-011-4302-2
Zhou B, Wang J, Zhang H et al (2020) Microplastics in agricultural soils on the coastal plain of Hangzhou Bay, east China: Multiple sources other than plastic mulching film. J Hazard Mater 388:121814. https://doi.org/10.1016/j.jhazmat.2019.121814
Zhu M, Chernick M, Rittschof D, Hinton DE (2020) Chronic dietary exposure to polystyrene microplastics in maturing Japanese medaka (Oryzias latipes). Aquat Toxicol 220:105396. https://doi.org/10.1016/j.aquatox.2019.105396
Zulkifley MA, Mustafa MM, Hussain A et al (2014) Robust identification of polyethylene terephthalate (PET) plastics through bayesian decision. PLoS ONE 9:e114518. https://doi.org/10.1371/journal.pone.0114518
Zurier HS, Goddard JM (2020) Biodegradation of microplastics in food and agriculture. Curr Opin Food Sci 37:37–44. https://doi.org/10.1016/j.cofs.2020.09.001
Funding
This research is funded by the Universiti Kebangsaan Malaysia research Grant (GUP-2019-026).
Author information
Authors and Affiliations
Contributions
ARO: was involved conceptualization, writing—original draft, writing—review & editing, HAH: helped in writing—review & editing, N'II: contributed to writing—original draft, MHM: was involved in writing—review & editing, and SRSA: helped in supervision.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Othman, A.R., Hasan, H.A., Muhamad, M.H. et al. Microbial degradation of microplastics by enzymatic processes: a review. Environ Chem Lett 19, 3057–3073 (2021). https://doi.org/10.1007/s10311-021-01197-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10311-021-01197-9