Abstract
The presence of toxic compounds derived from biomass pre-treatment in fermentation media represents an important drawback in second-generation bio-ethanol production technology and overcoming this inhibitory effect is one of the fundamental challenges to its industrial production. The aim of this study was to systematically identify, in industrial medium and at a genomic scale, the Saccharomyces cerevisiae genes required for simultaneous and maximal tolerance to key inhibitors of lignocellulosic fermentations. Based on the screening of EUROSCARF haploid mutant collection, 242 and 216 determinants of tolerance to inhibitory compounds present in industrial wheat straw hydrolysate (WSH) and in inhibitor-supplemented synthetic hydrolysate were identified, respectively. Genes associated to vitamin metabolism, mitochondrial and peroxisomal functions, ribosome biogenesis and microtubule biogenesis and dynamics are among the newly found determinants of WSH resistance. Moreover, PRS3, VMA8, ERG2, RAV1 and RPB4 were confirmed as key genes on yeast tolerance and fermentation of industrial WSH.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Technology for conventional ethanol production from crops rich in starch or sugar with Saccharomyces cerevisiae strains is well established [28, 29], while bio-ethanol production from agricultural and agro-industrial residues is receiving growing scientific interest despite posing greater technical, engineering and biological challenges [19]. It is widely acknowledged that the main challenge when using microorganisms to produce bulk chemicals is the accumulation of toxic compounds inside the producing cells, which affects the regular activity of yeast metabolic machinery. Then, tolerance engineering is essential for the improvement of production of next-generation biofuels [7, 15, 33].
To make the sugars present in biomass residues available for fermentation, raw materials have to be subjected to pre-treatment and hydrolysis steps [26, 31]. Under the extreme conditions observed during the pre-treatment step some toxic compounds are released together with sugars. These can be grouped around three main classes: weak acids, furans and phenolics. While acetic acid, the most common weak acid derived from lignocellulosic hydrolysates, is formed by deacetylation of hemicelluloses, furan compounds, 2-furaldehyde (furfural) and 5-hydroxymethyl-2-furaldehyde (HMF), are formed by dehydration of pentoses and hexoses, respectively. Phenolic compounds are generated due to lignin breakdown and carbohydrate degradation during acid hydrolysis (recently reviewed by Almeida et al. [2]). During yeast cultivation and/or fermentation step, these inhibitors induce a harsh effect on yeast metabolic machinery reducing the ethanol yield and productivity [16, 23]. The yeast has inherent mechanisms to counteract the negative impact of this multiple effects, being the furan and weak acids inhibition patterns well studied in the last years. Weak acids have been described to induce a strong intracellular acidification, with negative consequences for the activity of metabolic enzymes, and to cause the dissipation of the plasma membrane potential which is an essential feature for secondary transport [20]. This can be partly compensated by plasma membrane ATPase activity, which pumps protons out of the cell at the expense of ATP hydrolysis [20]. On the other hand, furan compounds have to be reduced to their corresponding less toxic alcohols by yeast cells decreasing the fermentation process productivity (and increasing the lag phase). Modig et al. [24] showed that the extended lag phase results from a reduction in available cellular energy caused by the inhibition of several enzymes (alcohol dehydrogenase, aldehyde dehydrogenase and pyruvate dehydrogenase), which coupled with the deficiency of the cofactor nicotinamide adenine dinucleotide phosphate (NADPH), are directly involved in oxidative damages to yeast cells. Moreover, furfural and HMF are known to cause RNA, DNA, protein and membrane damage at low concentrations [3, 14].
To overcome the problem of toxicity, several strategies have been considered such as biological or chemical detoxification step prior to fermentation (reviewed by Palmqvist and Hahn-Hagerdal [27]), optimization of fermentation environment to minimize the toxic effects of inhibitors [11] or the improvement of resistance of the organism itself [10]. Results emerging from genome-wide screenings have the potential to identify phenotype-specific genes under selective conditions that could be targets for subsequent genetic engineering aiming to obtain more robust industrial yeast strains [30]. Chemogenomic analysis has been successfully applied to identify genes, at a genomic scale, required for maximal tolerance to ethanol [18, 34] or high concentrations of glucose [35], single stress relevant for very high gravity (VHG) fermentations, or to inhibitory concentrations of furfural [9], acetic acid [21], and vanillin [8], and single stress relevant for lignocellulosic biomass fermentations. However, genes found to be specifically required to confer resistance to individual stresses may not be relevant in a multi-stress environment. Since during the bio-detoxification step yeast cells are subjected to a wide range of toxic compounds, where their combined effect can have a large impact on yeast metabolic machinery, it would be interesting to find key genes able to increase yeast tolerance to multiple inhibitory compounds presented in lignocellulosic hydrolysates produced for industrial applications. Although some efforts have been put into the study of the genome-wide expression response to cultivation in hydrolysate [5], a chemogenomic analysis under stresses induced by industrial lignocellulosic hydrolysates provides a straightforward and more realistic approach to better understand molecular and biological mechanisms during biomass in situ detoxification by yeast cells.
In this context, the aim of this study was to systematically identify, at a genomic scale, the genes required for simultaneous and maximal tolerance to inhibitors derived from lignocellulosic biomass pre-treatment, by screening the EUROSCARF haploid mutant collection for susceptibility in industrial wheat straw hydrolysate (WSH) and in an inhibitor-supplemented synthetic hydrolysate (SH).
Methods
Strains and growth media
S. cerevisiae BY4741 strain (MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and the EUROSCARF collection of BY4741-derived haploid mutant strains, with all non-essential open reading frames (ORFs) individually deleted (http://web.uni-frankfurt.de/fb15/mikro/euroscarf/), were used for the chemogenomics analysis carried out during this study. Batch cultures of yeast were pre-grown in liquid minimal medium (MM4) that contains, per liter, 1.7 g yeast nitrogen base without amino acids or NH4+ and 20 g glucose, 2.65 g (NH4)2SO4, 20 mg methionine, 20 mg histidine, 60 mg leucine and 20 mg uracil. A yeast peptone dextrose (YPD) medium that contains, per liter, 20 g glucose, 20 g bactopeptone and 10 g yeast extract was also used for yeast growth in standard laboratory conditions. Solid YPD and MM4 growth media were obtained by supplementing the liquid medium with 2 % (w/v) agar.
Preparation of wheat straw and synthetic hydrolysates
A lignocellulosic wheat straw hydrolysate (WSH) was prepared following the method described by Ruiz et al. [31]. Briefly, the milled wheat straw (with particle size distribution of: >1 mm, 10 %; between 1 and 0.5 mm, 40 %; between 0.5 and 0.3 mm, 40 %; <0.3 mm, 10 %) and water were mixed to obtain a ratio 10:1 liquid/solid and treated for 30 min in a 3.75 L stainless steel reactor, at 180 °C for autohydrolysis. After hydrolysis, the liquid phase (hemicellulosic liquor) was collected by filtration and stored at −20 °C. Prior to its use for yeast growth, the hemicellulosic liquor was centrifuged for 10 min at 9,000 rpm (4 °C) to remove the solid fraction, supplemented with 2 % (w/v) agar, the pH was adjusted to 4.5 (NaOH 1 M) and then sterilized at 121 °C during 20 min. Afterward, the hemicellulosic liquor at 55 °C was supplemented with glucose and aminoacids to a final concentration of 20 g/L glucose, 20 mg/L methionine, 20 mg/L histidine, 60 mg/L leucine and 20 mg/L uracil to account for the auxotrophies of the BY4741 parental strain and its derived single deletion strains tested. All strains were also cultured in YPD solid medium supplemented with the same aminoacids mix to compare and evaluate their growth phenotype in standard laboratory conditions. A synthetic hydrolysate was also used to test the susceptibility of yeast cells to the inhibitors found in industrial WSH containing the same nutritional base of MM4 medium (see composition above) and supplemented or not with 30 mM acetic acid, 4.5 mM furfural and 0.67 mM HMF. The plates of both solid hydrolysates were prepared a day prior to use. The concentrations of glucose, acetic acid, furfural and HMF in the WSH and SH hydrolysates prepared as described above were quantified by high-performance liquid chromatography (HPLC). Glucose and acetic acid were quantified upon separation of an aliquot of the hydrolysate in a Varian MetaCarb 87H column, eluted at 60 °C with 0.005 M sulfuric acid, at a flow rate of 0.7 mL/min. The peaks corresponding to glucose were detected using a refractive index detector, whereas acetic acid was detected using an UV detector set at 210 nm. Furfural and HMF were quantified upon separation of an aliquot of the hydrolysate in a Macherey-Nagel C18 column, eluted with 20 % acetonitrile to 80 % water at a flow rate of 0.9 mL/min. Peak detection was performed using an UV detector set at 276 nm.
Screening for lignocellulosic inhibitor-sensitive deletion mutants
To screen the entire EUROSCARF deletion mutant collection for susceptibility response to lignocellulosic inhibitors all strains were inoculated from stock cultures (96-well plates at −80 °C) to batch cultures at pH 4.5, 30 °C with orbital agitation (250 rpm) in MM4 medium under aerobic conditions. After 12 h of growth, a 96-pin replica platter was used to spot these cells onto the surface of WSH, YPD and SH with either 0 or the cocktail inhibitor described above. Susceptibility phenotypes were registered after incubation at 30 °C for 2 and 3 days, depending on the severity of growth inhibition. At least two independent replicates were conducted for each set of mutants. The inhibition phenotype in WSH and SH was scored as (-), if the strain showed a residual growth after 72 h; and (--), if the strain showed no growth after 72 h (see Fig. 1). Strains exhibiting no growth or growth that was difficult to score were rescreened to confirm the results. Only the mutants that present normal growth in control media (YPD) comparatively to parental strain have been considered susceptible to inhibitory WSH or SH (Fig. 1). Groups of genes that are overrepresented in our dataset, compared to yeast genome, were assigned to determined functional groups using the GOToolBox tool and the enrichment was considered for P values below 0.01. Moreover, the description of gene function was complemented using the information available in Saccharomyces Genome Database, SGD (http://www.yeastgenome.org/).
Example of growth phenotypes of the parental BY4741 strain and single deletion mutant strains on WSH, SH and YPD (CTRL) media after 72 h. In this example, Vps16 was classified as a inhibitor-sensitive deletion mutant with “high phenotype” (--, no growth after 72 h); Gim3 and Alf1 were classified as inhibitor-sensitive deletion mutants with “low phenotype” (-, residual growth after 72 h); the growth phenotype of the other single gene deletion strains was considered “n.i”, no inhibition)
Results
Screening of genes conferring resistance to stress induced by cultivation in WSH
Genome-wide identification of genes implicated in S. cerevisiae resistance to stresses induced by cultivation in an industrial WSH was based on the comparison of the susceptibility to this growth medium of the EUROSCARF haploid mutant collection (approximately 5,100 mutants individually deleted for non-essential genes) with the corresponding parental strain BY4741. Two-hundred and forty-two mutant strains were found to be more susceptible to cultivation in WSH than the BY4741 parental strain, corresponding to approximately 5 % of the strain collection tested. Two levels of inhibition were considered [(-) and (--)] based on increasing levels of growth deficiency of the deletion mutants tested in WSH, compared to the parental strain (Table S1).
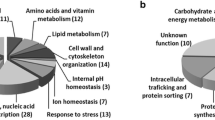
Clustering of the specific genes required for maximal tolerance to WSH-derived inhibitors, based on their biological process, was performed according to their associated GO terms, using the GOToolBox software (http://genome.crg.es/GOToolBox/). The frequency of each biological class in the dataset under analysis was compared to that in the genome and a statistical test was applied to correct the data. The 242 identified genes were then predominantly associated to six enriched GO terms (those having an associated p value below 0.01): “vacuolar transport”, “regulation of gene expression”, “response to nutrient levels”, “cellular ion homeostasis”, “vitamin metabolic process”, and “lipid metabolic process” (Fig. 2).
Clustering, based on biological function, of yeast determinants required for maximal tolerance to cultivation in wheat straw hydrolysate (WSH). Genes were clustered using GOToolBox, and only classes (#genes >10) found to be statistically overrepresented in our dataset are displayed (P value below 0.01). Black bars, gene frequency within each class in the WSH dataset; white bars, frequency registered for the whole genome
Based on this classification, and using the gene descriptions deposited in the Saccharomyces Genome Database (www.yeastgenome.org), Table S1 (Additional File 1, Supplementary Material) was put together, in which the 242 genes were grouped according to their biological functions. Among these biological functions, the most represented include intracellular trafficking (34 genes), transcriptional machinery and RNA processing (30 genes), protein synthesis (17 genes), lipid metabolism (14 genes), amino acid metabolism (12 genes), ion transport (10 genes), vitamin metabolism (8 genes), cell wall metabolism (8 genes), stress response (7 genes), vacuolar acidification (7 genes), degradation (6 genes) and folding (5 genes).
Biological functions, involving lipid metabolism, including ergosterol and phospholipid composition, had been previously found in relation to yeast resistance to furfural [9], vanillin [8] and acetic acid [22]. The same is also true for vacuolar function, which appears to be essential for the maintenance of intracellular and vacuolar pH levels and associated functions, affected by the presence of acetic acid [22] and other growth inhibitors of relevance in the fermentation industry [34, 35]. The importance of vacuolar function and intracellular trafficking may further relate to the requirement to target membrane transporters to overcome inhibitor stress imposed by acetic acid [22].
The observed diversity of biological functions found to be required for yeast resistance to cultivation in WSH is consistent with the complexity of this medium exhibiting multiple sources of stress.
Genes required for maximal tolerance to cultivation in an SH
Acetic acid and furan compounds (furfural and HMF), frequently the most dominant inhibitor cocktail present in plant-biomass hydrolysates, are also the main stress factors found to be present in the industrial WSH used in this study. Indeed, using HPLC analysis it was possible to assess that the concentration of furfural, acetic acid and HMF was of 4.5, 30 and 0.67, respectively. Several studies, based on molecular biology and genome-wide approaches, aiming the elucidation of the mechanisms underlying yeast tolerance to each of these single-stress agents were previously conducted [15]. To look further into the possibility that these stress agents may, when in combination, exert a non-additive toxic effect in yeast cells, SH was devised, based on the supplementation of minimal YNB-derived medium with the exact same concentrations of furfural, HMF and acetic acid found in the used industrial WSH. The entire EUROSCARF collection was then screened for sensitivity mutants in this SH medium, and 200 and 216 mutant strains were identified as displaying increased susceptibility to the simultaneous presence of acetic acid, furfural and HMF. Again using the gene descriptions deposited in the Saccharomyces Genome Database (www.yeastgenome.org), Table S2 (Additional File 2, Supplementary Material) was put together, in which these 216 genes were grouped according to their biological functions. Most of the biological functions found to be required for WSH resistance were also found to be required for SH resistance, although with different levels of relative importance. Indeed, the majority of the genes identified as determinants of resistance to WSH are also determinants of SH resistance. However, 59 genes were found to be determinants of SH resistance alone, while 79 genes were identified as exclusively determining WSH resistance in yeast (Fig. S1). This difference highlights the importance of examining stress tolerance mechanisms beyond lab conditions, especially when aiming industrial applications, even when a combination of stresses is considered.
Among the groups of proteins found to be required for WSH resistance, but not for SH resistance, the following are highlighted: vitamin metabolic enzymes, including a number of those contributing to thiamine and pantothenate metabolism; proteins involved in iron limitation response, including the transcription factor controlling this response, Aft1, and the oxidoreductase required for high-affinity iron up-take, Fet3; subunits of the heteromeric cochaperone prefoldin complex, Gim3, Gim4 and Pac10, which plays a key role in the folding of actin, tubulin and other aggregation-sensitive polypeptides, thereby allowing their efficient folding [32]; microtubule dynamics-related proteins, including Kar3, a microtubule motor, the Nip100 subunit of the dynactin complex, and the alpha-tubulin Tub3.
On the other hand, some genes were found to be required for SH resistance but not for WSH resistance, including especially a number involved in phospholipid biosynthesis: the transcription factors INO2 and INO4, controlling the response to inositol depletion; IPK1, an inositol-1,3,4,5,6-pentakisphosphate 2-kinase, whose activity has been shown to be important for the process of mRNA export [37]; and PDR17, a multidrug resistance phosphatidylinositol transfer protein, which downregulates Plb1p-mediated turnover of phosphatidylcholine.
These differences are expected to result from the fact that the more complex WSH medium contains further stress sources than SH and is defective in some of the key nutrients that exist in the nutrient-balanced SH medium. However, the fact that WSH appears to exert a less stressing environment in terms of lipid homeostasis than the used SH medium is interesting, possibly relating to the fact that SH is derived from the minimal YNB-based growth medium.
Comparison of the genes required for S. cerevisiae resistance to SH or WSH with those required for furfural or acetic acid resistance
Given that one of the key goals of this study is to understand the differences between analyzing the effect of individual stress agents and analyzing the effect of their combination, a comparison between the genes identified in this study as conferring resistance to SH with those previously identified as involved in acetic acid [21] or furfural [9] tolerance was carried out. The intersection between the three datasets (Fig. S1B) reveals that there is little overlap between the identified furfural resistance determinants and those required for SH tolerance.
Indeed, only 5.6 % of the genes found to play a role in SH resistance were previously involved in furfural tolerance, including PRS3, RPB4, RPE1, STB5, VMA8, ZWF1, also involved in acetic acid tolerance, and BUD27, DCC1, EAF7, FYV6, VMA22 and YDR049w. On the other hand, around 43 % of the determinants of SH resistance had been previously identified as conferring acetic acid resistance, suggesting that this stress agent is a key factor in the toxicity exerted by the used SH. However, the observation that more than 50 % of the determinants of SH resistance were not found to be required for yeast tolerance to acetic acid or furfural is also significant. The finding of 117 genes that are required for the resistance to the SH formulation, but not to the individual stresses therein, reinforces the notion that studies carried out just looking at individual stress conditions do not provide a complete picture of what is going on in the complex real-life stress environments. It also points out to a possible combinatorial, or eventually, synergistic action of acetic acid, furfural and HMF.
This notion is also clear when comparing the identified determinants of WSH resistance with the furfural or acetic acid resistance determinant (Fig. S1C). Again, only a very small fraction of the WSH resistance determinants, 3.7 %, are required for furfural tolerance, including PRS3, RPE1, STB5, VMA8 and ZWF1, also involved in acetic acid tolerance, and BUD27, DCC1, FYV6 and VMA22. On the contrary, 45.7 % of the WSH resistance determinants are required for acetic acid tolerance. Despite this partial overlap, more than 50 % of the WSH resistance determinants are not necessary for yeast to tolerate stress induced by furfural or acetic acid alone.
The functional groups exclusively represented in the WSH or SH datasets include vitamin metabolism, mitochondrial function, peroxisomal function, ribosome biogenesis and response to reactive oxygen species. The specific genes and gene functions are indicated in Table 1.
Discussion
In this study, the determinants of yeast resistance to cultivation in industrial wheat straw hydrolysate (WSH) were identified. The understanding of the mechanisms of resistance to the individual stress agents that are present in WSH has been looked into in some detail in studies focused not only on specific mechanisms [2, 15] but also on a genome-wide perspective [8, 9, 21, 34–36].
However, the effect of the combination of these stresses in yeast cell ability to thrive and ferment in high yield has been mostly neglected. Although it is true that some studies have focused on the genome-wide response to lignocellulosic hydrolysates and related stresses [5, 22], or to the characterization of the genome-wide expression patterns in strains evolved to thrive in industrial hydrolysates [1], the fact is that the best way to characterize the mechanisms of resistance to a given stress is through the screening of systematic mutant libraries [6]. Using such an approach, 242 genes were found to be required for cultivation of S. cerevisiae cells in industrial WSH, half of them being linked to this phenomenon for the first time.
Looking in more detail into the new insights provided by this work, our analysis was focused on the determinants of resistance to WSH cultivation that had not been previously identified as conferring resistance to stresses caused upon exposure to single-stress agents present in WSH such as furfural [9] and acetic acid [21]. Among the 128 genes matching this criterion, a few functional categories were highlighted, including response to reactive oxygen species (ROS), mitochondrial function, ribosome biogenesis, peroxisomal function and vitamin metabolism.
Two reactive oxygen species-responsive genes, namely those encoding the cytosolic, Sod1, and mitochondrial, Sod2, superoxide dismutases, were found to confer WSH resistance, while only Sod2 was found to confer SH resistance and none was implicated in acetic acid or furfural resistance. HMF and furfural have been shown to affect redox metabolism, draining the cells of reductive power [17] and inducing the expression of genes involved in the redox balance of the cell [5]. Consistently, among the proteins found to be required for furfural resistance are NADPH-regenerating enzymes, such as those of the pentose phosphate pathway, but not ROS-responsive genes [9]. The reason for this appears to relate to the ability of S. cerevisiae to convert furfural to furan methanol and HMF to furan di-methanol through multiple NADPH-dependent aldehyde reduction, such as the reductases ALD4 and GRE3 [32].
The involvement of mitochondrial function in yeast resistance to WSH is in agreement with the increased transcript levels of some mitochondria-associated genes in response to acetic acid and furfural [12]. Mitochondrial genes had also been previously linked to acetic acid resistance [22], suggesting that, even in the presence of glucose, mitochondrial function is essential for tolerance to cultivation in WSH. Ribosome biogenesis genes were also found to increase yeast ability to grow in WSH. The beneficial effect of the expression of these genes is in agreement with the dramatic increase of the degradation rate of ribosomal RNA in acetic acid-stressed cells [25]. However, it is interesting to point out that the ribosome- or mitochondria-related genes found to confer acetic acid resistance do not coincide with those conferring WSH resistance. Surprisingly, the expression of ribosomal genes, as well of genes functioning in the synthesis and transport of proteins, metabolism of carbohydrates, lipids, vitamins and vacuolar proteins, was found to decrease significantly in yeast T2 cells exposed to hardwood-spent sulfite liquor [5]. The huge difference between the genes found to be determinants of WSH and those up-regulated upon exposure to this stress environment reinforces the notion that gene expression analysis is not the single way to study resistance mechanisms.
Peroxisomal function was also a new biological function associated to WSH resistance within this study. Although its precise role in stress resistance has been elusive, a recent study showed that under stress the peroxisome was the most severely affected compartment in terms of redox state and pH recovery [4]. Since both oxidative and acidic stress appear to be a clear consequence of the joint action of furfural, HMF and acetic acid, it is reasonable to assume that the deletion of peroxisomal genes would increase the susceptibility of yeast cells to this combination of stress agents. It would be interesting to further inspect the role of peroxisomes in the maintenance of pH and redox balance under WSH stress. These results further suggest that conditions leading to peroxisomal proliferation might be beneficial for yeast cells to thrive in industrial WSH.
Another finding of this study is the association of vitamin metabolism and WSH resistance. More specifically, most of the key enzymes and some regulators leading to the synthesis of pantothenate and thiamine were found to be determinants of WSH tolerance, but, in most cases, not required for SH resistance. Thiamine is an essential cofactor for enzymes that decarboxylate α-keto acids, including α-ketoglutarate dehydrogenase, branched-chain α-ketoacid dehydrogenase, and transketolase, during amino acid and carbohydrate metabolism, and has also been linked to the maintenance of NAD + homeostasis [13]. Pantothenate, on the other hand, is a metabolic precursor to coenzyme A (CoA) and acyl carrier protein, which are also cofactors required by a large number of metabolic enzymes.
The observation that the genes required for the biosynthesis of these vitamins are essential for WSH, and less significantly for SH, resistance, may reflect the fact that not only WSH contains a pool of synergistic stress agents, but might also be deficient in key nutrients, whose absence would impair S. cerevisiae ability to grown in inhibitory WSH.
Interestingly, a small group of determinants of yeast resistance to cultivation in WSH were previously identified as conferring resistance to furfural during a chemogenomic analysis using standard laboratory conditions [9]. This reinforces that these genes including PRS3, RPE1, STB5, VMA8 and ZWF1 play an important role on WSH resistance. In particular, Gorsich et al. 2006 showed that ZWF1 overexpression in S. cerevisiae allowed growth at furfural concentrations that are normally toxic confirming a strong relationship between these genes and furfural tolerance complex [9]. Altogether, results from this study suggest that the combination of acetic acid, furfural and the environment found in industrial WSH appears to exacerbate oxidative stress upon yeast cells, in a way that goes beyond the individual action of each of these stress agents.
Based on the direct intersection of the genes previously identified as conferring resistance to lignocellulose biomass fermentation-related stresses, namely furfural, vanillin and acetic acid, ERG2, PRS3, RAV1, RPB4 and VMA8 were also reported to be required for cell viability maintenance and fermentation in wheat straw hydrolysate [30]. Since our focus was the identification of genes whose expression confers simultaneous resistance growth and fermentation of inhibitory hydrolysates, the previous results from Pereira et al. [30] obtained in fermentation tests mimicking industrial relevant conditions (limiting oxygen conditions) allow us to conclude about the feasibility of the screening strategy in solid growth media and confirm the importance of the identified genes in this screening study—PRS3, VMA8, ERG2, RAV1, RPB4—for maximal tolerance to key inhibitors present in WSH. Interestingly, in this study ERG2 and PRS3 genes were again found to be required for simultaneous SH and WSH resistance, which highlight the importance of these genes in yeast global adaptation to lignocellulose-derived inhibitors.
Knowledge-based medium manipulation or/and genetic engineering, after genome-wide identification of target genes, for improved S. cerevisiae tolerance appears as a promising approach that can be used, in combination with strain adaptation and fermentation control for the development of efficient lignocellulose-based ethanol processes.
Conclusion
Understanding fermentation inhibitor tolerance in yeasts at the genetic level is of considerable importance to the fermentation industry. The focused and more realistic screening strategy presented in this study proved to be useful in identifying genes involved in inhibitor tolerance. Specifically, the present genome-wide survey conducted in industrial WSH and SH uncovered 242 determinants of resistance. The results obtained highlight the relevance of the vacuolar acidification, ribosomal, mitochondrial and peroxisomal functions, microtubule biogenesis and dynamics and oxidative stress in WSH resistance in yeast. Finally, PRS3, VMA8, ERG2, RAV1 and RPB4 were highlighted as key genes on yeast tolerance and fermentation of industrial WSH.
References
Almario MP, Reyes LH, Kao KC (2013) Evolutionary engineering of Saccharomyces cerevisiae for enhanced tolerance to hydrolysates of lignocellulosic biomass. Biotechnol Bioeng. doi:10.1002/bit.24938
Almeida JR, Modig T, Petersson A, Hähn-Hägerdal B, Lidén G, Gorwa-Grauslund MF (2007) Increased tolerance and conversion of inhibitors in lignocellulosic hydrolysates by Saccharomyces cerevisiae. J Chem Technol Biotechnol 82:340–349. doi:10.1002/jctb.1676
Ask M, Bettiga M, Mapelli V, Olsson L (2013) The influence of HMF and furfural on redox-balance and energy-state of xylose-utilizing Saccharomyces cerevisiae. Biotechnol Biofuels 6:22. doi:10.1186/1754-6834-6-22
Ayer A, Sanwald J, Pillay BA, Meyer AJ, Perrone GG, Dawes IW (2013) Distinct Redox Regulation in Sub-Cellular Compartments in Response to Various Stress Conditions in Saccharomyces cerevisiae. PLoS ONE 8:e65240. doi:10.1371/journal.pone.0065240
Bajwa PK, Ho CY, Chan CK, Martin VJ, Trevors JT, Lee H (2013) Transcriptional profiling of Saccharomyces cerevisiae T2 cells upon exposure to hardwood spent sulphite liquor: comparison to acetic acid, furfural and hydroxymethylfurfural. Antonie Van Leeuwenhoek 103:1281–1295. doi:10.1007/s10482-013-9909-1
Dos Santos SC, Teixeira MC, Cabrito TR, Sa-Correia I (2013) Yeast toxicogenomics: genome-wide responses to chemical stresses with impact in environmental health, pharmacology, and biotechnology. Front Genet 3:63. doi:10.3389/fgene.2012.00063
Dunlop MJ (2011) Engineering microbes for tolerance to next-generation biofuels. Biotechnol Biofuels 4:32. doi:10.1186/1754-6834-4-32
Endo A, Nakamura T, Ando A, Tokuyasu K, Shima J (2008) Genome-wide screening of the genes required for tolerance to vanillin, which is a potential inhibitor of bioethanol fermentation, in Saccharomyces cerevisiae. Biotechnol Biofuels. doi:10.1186/1754-6834-1-3
Gorsich SW, Dien BS, Nichols NN, Slininger PJ, Liu ZL, Skory CD (2006) Tolerance to furfural-induced stress is associated with pentose phosphate pathway genes ZWF1, GND1, RPE1, and TKL1 in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 71:339–349. doi:10.1007/s00253-005-0142-3
Heer D, Sauer U (2008) Identification of furfural as a key toxin in lignocellulosic hydrolysates and evolution of a tolerant yeast strain. Microbial Biotechnol 1:497–506. doi:10.1111/j.1751-7915.2008.00050.x
Huang RL, Su RX, Qi W, He ZM (2011) Bioconversion of lignocellulose into bioethanol: process intensification and mechanism research. Bioenergy Res 4:225–245. doi:10.1007/s12155-011-9125-7
Li BZ, Yuan YJ (2010) Transcriptome shifts in response to furfural and acetic acid in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 86:1915–1924. doi:10.1007/s00253-010-2518-2
Li M, Petteys BJ, McClure JM, Valsakumar V, Bekiranov S, Frank EL, Smith JS (2010) Thiamine biosynthesis in Saccharomyces cerevisiae is regulated by the NAD + -dependent histone deacetylase Hst1. Mol Cell Biol 30:3329–3341. doi:10.1128/MCB.01590-09
Lin FM, Qiao B, Yuan YJ (2009) Comparative proteomic analysis of tolerance and adaptation of ethanologenic Saccharomyces cerevisiae to furfural, a lignocellulosic inhibitory compound. Appl Environ Microbiol 75:3765–3776. doi:10.1128/AEM.02594-08
Liu ZL (2011) Molecular mechanisms of yeast tolerance and in situ detoxification of lignocellulose hydrolysates. Appl Microbiol Biotechnol 90:809–825. doi:10.1007/s00253-011-3167-9
Liu ZL (2006) Genomic adaptation of ethanologenic yeast to biomass conversion inhibitors. Appl Microbiol Biotechnol 73:27–36. doi:10.1007/s00253-006-0567-3
Liu ZL, Ma M, Song M (2009) Evolutionarily engineered ethanologenic yeast detoxifies lignocellulosic biomass conversion inhibitors by reprogrammed pathways. Mol Genet Genomics 282:233–244. doi:10.1007/s00438-009-0461-7
Ma M, Liu LZ (2010) Quantitative transcription dynamic analysis reveals candidate genes and key regulators for ethanol tolerance in Saccharomyces cerevisiae. BMC Microbiol 10:169. doi:10.1186/1471-2180-10-169
Madhavan A, Srivastava A, Kondo A, Bisaria VS (2012) Bioconversion of lignocellulose-derived sugars to ethanol by engineered Saccharomyces cerevisiae. Crit Rev Biotechnol 32:2–48. doi:10.3109/07388551.2010.539551
Mira NP, Becker JD, Sa-Correia I (2010) Genomic expression program involving the Haa1p-regulon in Saccharomyces cerevisiae response to acetic acid. OMICS 14:587–601. doi:10.1089/omi.2010.0048
Mira NP, Palma M, Guerreiro JF, Sa-Correia I (2010) Genome-wide identification of Saccharomyces cerevisiae genes required for tolerance to acetic acid. Microb Cell Fact 9:79. doi:10.1186/1475-2859-9-79
Mira NP, Teixeira MC, Sá-Correia I (2010) Adaptative response and tolerance to weak acid stress in Saccharomyces cerevisiae: a genome-wide view. OMICS 14:525–540. doi:10.1089/omi.2010.0072
Modig T, Almeida JR, Gorwa-Grauslund MF, Liden G (2008) Variability of the response of Saccharomyces cerevisiae strains to lignocellulose hydrolysate. Biotechnol Bioeng 100:423–429. doi:10.1002/bit.21789
Modig T, Liden G, Taherzadeh MJ (2002) Inhibition effects of furfural on alcohol dehydrogenase, aldehyde dehydrogenase and pyruvate dehydrogenase. Biochem J 363:769–776
Mroczek S, Kufel J (2008) Apoptotic signals induce specific degradation of ribosomal RNA in yeast. Nucleic Acids Res 36:2874–2888. doi:10.1093/nar/gkm1100
Mussatto SI, Dragone G, Guimaraes PM, Silva JP, Carneiro LM, Roberto IC, Vicente A, Domingues L, Teixeira JA (2000) Technological trends, global market, and challenges of bio-ethanol production. Biotechnol Adv 28:817–830. doi:10.1016/j.biotechadv.2010.07.001
Palmqvist E, Hahn-Hagerdal B (2000) Fermentation of lignocellulosic hydrolysates. I: inhibition and detoxification. Bioress Technol 74:17–24. doi:10.1016/S0960-8524(99)00160-1
Pereira FB, Gomes DG, Guimaraes PM, Teixeira JA, Domingues L (2011) Cell recycling during repeated very high gravity bio-ethanol fermentations using the industrial Saccharomyces cerevisiae strain PE-2. Biotechnol Lett 34:45–53. doi:10.1007/s10529-011-0735-0
Pereira FB, Guimaraes PM, Teixeira JA, Domingues L (2011) Robust industrial Saccharomyces cerevisiae strains for very high gravity bio-ethanol fermentations. J Biosci Bioeng 112:130–136. doi:10.1016/j.jbiosc.2011.03.022
Pereira FB, Guimaraes PM, Gomes DG, Mira NP, Teixeira MC, Sa-Correia I, Domingues L (2011) Identification of candidate genes for yeast engineering to improve bioethanol production in very high gravity and lignocellulosic biomass industrial fermentations. Biotechnol Biofuels 4:57. doi:10.1186/1754-6834-4-57
Ruiz HA, Ruzene DS, Silva DP, da Silva FF, Vicente AA, Teixeira JA (2011) Development and characterization of an environmentally friendly process sequence (autohydrolysis and organosolv) for wheat straw delignification. Appl Biochem Biotechnol 164:629–641. doi:10.1007/s12010-011-9163-9
Siegers K, Waldmann T, Leroux MR, Grein K, Shevchenko A, Schiebel E, Hartl FU (1999) Compartmentation of protein folding in vivo: sequestration of non-native polypeptide by the chaperonin–GimC system. EMBO J 18:75–84. doi:10.1093/emboj/18.1.75
Taylor MP, Mulako I, Tuffin M, Cowan D (2012) Understanding physiological responses to pre-treatment inhibitors in ethanologenic fermentations. Biotechnol J 7:1169–1181. doi:10.1002/biot.201100335
Teixeira MC, Raposo LR, Mira NP, Lourenco AB, Sa-Correia I (2009) Genome-wide identification of Saccharomyces cerevisiae genes required for maximal tolerance to ethanol. Appl Environ Microbiol 75:5761–5772. doi:10.1128/AEM.00845-09
Teixeira MC, Raposo LR, Palma M, Sa-Correia I (2010) Identification of genes required for maximal tolerance to high-glucose concentrations, as those present in industrial alcoholic fermentation media, through a chemogenomics approach. OMICS 14:201–210. doi:10.1089/omi.2009.0149
Teixeira MC, Mira NP, Sa-Correia I (2010) A genome-wide perspective on the response and tolerance to food-relevant stresses in Saccharomyces cerevisiae. Curr Opin Biotechnol 22:150–156. doi:10.1016/j.copbio.2010.10.011
York JD, Odom AR, Murphy R, Ives EB, Wente SR (1999) A phospholipase C-dependent inositol polyphosphate kinase pathway required for efficient messenger RNA export. Science 285:96–100. doi:10.1126/science.285.5424.96
Acknowledgments
The authors thank Juan Carlos Parajó and Héctor Ruíz for assistance in the pre-treatment of lignocellulose biomass. Research described in this article was financially supported by FEDER and “Fundação para a Ciência e a Tecnologia” (FCT) (Contracts PEst-OE/EQB/LA0023/2011, PTDC/BIO/66151/2006, PTDC/AGR-ALI/102608/2008 and ERA-IB/0002/2010 and PhD grant (SFRH/BD/64776/2009) to FP).
Conflict of interests
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
10295_2014_1519_MOESM3_ESM.pdf
Supplementary material 3 (PDF 17 kb) Figure S1. Venn diagram representing the intersection of yeast determinants of a wheat straw hydrolysate (WSH) and synthetic hydrolysate (SH) resistance; b Synthetic hydrolysate (SH) and acetic acid [21] or furfural resistance [9]; c wheat straw hydrolysate (WSH) and acetic acid [21] or furfural resistance [9]
Rights and permissions
About this article
Cite this article
Pereira, F.B., Teixeira, M.C., Mira, N.P. et al. Genome-wide screening of Saccharomyces cerevisiae genes required to foster tolerance towards industrial wheat straw hydrolysates. J Ind Microbiol Biotechnol 41, 1753–1761 (2014). https://doi.org/10.1007/s10295-014-1519-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-014-1519-z