Abstract
Microbial genome mining is a rapidly developing approach to discover new and novel secondary metabolites for drug discovery. Many advances have been made in the past decade to facilitate genome mining, and these are reviewed in this Special Issue of the Journal of Industrial Microbiology and Biotechnology. In this Introductory Review, we discuss the concept of genome mining and why it is important for the revitalization of natural product discovery; what microbes show the most promise for focused genome mining; how microbial genomes can be mined; how genome mining can be leveraged with other technologies; how progress on genome mining can be accelerated; and who should fund future progress in this promising field. We direct interested readers to more focused reviews on the individual topics in this Special Issue for more detailed summaries on the current state-of-the-art.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Microbial genome mining is an alternative approach to more traditional methods for the discovery of novel secondary metabolites, which continue to serve as scaffolds for further embellishment by medicinal chemistry and combinatorial biosynthesis for the development of products for human medicine, animal health, crop protection, and numerous biotechnological applications. Historically, natural products have made a very large impact in these areas [25, 33, 59], and the large fraction of current investigational new drug applications based on natural products strongly suggests that they will continue to impact human therapeutic discovery and development in the future. The concept of genome mining has been maturing as a discipline since the discovery by David Hopwood and coworkers by whole-genome sequencing that Streptomyces coelicolor encodes many more secondary metabolites than had been anticipated from decades of study [15, 20]. In recent years, this observation has been generalized in many reports [1, 8, 40, 42, 58, 74]. It is fitting that we dedicate this Special Issue of the Journal of Industrial Microbiology and Biotechnology on Microbial Genome Mining to Sir David Hopwood on the occasion of his 80th birthday, August 19, 2013.
What is genome mining and why is it important?
For much of its history, secondary metabolite discovery has been a process driven in large part by chance. In most cases, discovery of new natural products has been driven either by bioactivity-guided fractionation of crude fermentation broth extracts, or via chemical screening (isolation of chromatographically resolvable metabolites with ‘interesting’ spectroscopic properties). As the natural pharmacopeia has grown and the preponderance of readily identified compounds has been catalogued, the long-term success of secondary metabolite discovery campaigns has generally been determined by the degree to which this dependence on blind chance can be minimized. Historically, this has been accomplished via a number of strategies including the exploration of new ecologies, judicious selection of genera [14, 43, 69], and by the development of new analytical methodologies with improved analytical separation and sensitivity [55].
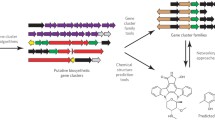
Genome mining is a radical re-envisioning of the process of secondary metabolite discovery, which has the theoretical potential to eliminate all chance from secondary metabolite discovery. In the context of this special issue, genome mining may be defined as the process of technically translating secondary metabolite-encoding gene sequence data into purified molecules in tubes. In comparison to the historical ‘grind and find’ mode of natural product discovery, the success of genome mining methods will be defined by the degree to which they unleash secondary metabolic gene clusters within a given system (Fig. 1a, b) and identify encoded metabolites. In recent years, the easy and inexpensive access to genomic sequence data, resulting from the advent of next-generation sequencing technologies [27], has created a potential embarrassment of riches regarding the starting point of genome mining. Indeed, most sequenced microorganisms with relatively large genomes, and plants contain dozens or more blueprints for the biosynthesis of secondary metabolites. Moreover, automated bioinformatics platforms now facilitate the semi-automated prediction of natural products encoded by secondary metabolic blueprints [16, 17]. However, the identification of genome-encoded secondary metabolism is only the first step in the process of genome mining. Indeed, genome mining now spans the full spectrum of the updated central dogma of molecular biology (Fig. 1c) including bioinformatic prediction of gene and pathway function, the control of gene expression and translation, and the identification and structural elucidation of new metabolites from within the metabolome of the producing organisms. As a consequence, genome-mining studies often become more than solely natural product discovery as they entail comprehensively understanding and manipulating cellular molecular systems. This issue contains articles that seek to address this navigation of the central dogma of genome to metabolites.
Strategies for natural product discovery. a The historical ‘grind and find’ mechanism of secondary metabolite discovery. b Post-genomic discovery now seeks to leverage prescience of gene sequence data to improve the yield of discovery. c The extended central dogma as it relates to genome mining for secondary metabolism
In the Sanger sequencing era (pre ~2005), genome mining efforts were primarily enabled by the genome of only two model Streptomyces and by gene clusters discovered using oligonucleotide gene sequence probes gene sequence tags (GSTs) based on known secondary metabolism. In the former case, S. coelicolor and Streptomyces avermitilis, revealed the apparently untapped potential of secondary metabolism and resulted, over the course of a decade, in the discovery of many new metabolites from these organisms that had previously been considered to be mined to exhaustion [20, 40]. The efforts of Ecopia Biosciences [30] generated thousands of high-quality gene clusters encoding the biosynthesis of secondary metabolites identified via a twofold process of (1) low-resolution shotgun genome scanning of potential microbial secondary metabolite producers (1 read/5–20 kpb) to identify short sequences with homology to sequences in annotated secondary metabolism databases and (2) follow-up sequencing of cosmids hybridizing to secondary metabolic GSTs. Regardless of the source of sequence data, the early days of genome mining efforts generally capitalized on the prescience of secondary metabolic potential to effectively ‘look harder’ in the producing organisms for the predicted metabolites. For instance, the prediction of a siderophore in S. coelicolor [21] prompted growth in low-iron media and application of siderophore assay for isolation and structural elucidation [48]. The prediction of an antifungal polyene in Streptomyces aizunensus prompted the use of antifungal screens using a range of growth conditions for the producing organism [4, 53]. Similarly, the observation of enediyne-encoding gene clusters prompted producing-organism growth-condition screening in combination with a DNA damage assay screen for detection of putative enediyne natural products [73].
The importance of genome mining extends well beyond its potential to completely circumvent the chance component of the process of secondary metabolite discovery. For instance, understanding the connection between metabolites, which represent one of the end points of the central dogma, and the gene sequences that encode them, can provide insight into the basic biology of producing organisms as discrete individuals, and as members of the microbiota of their environment. It is becoming increasingly clear that many if not most secondary metabolites play roles in interspecies, intergeneric, and/or interkingdom chemical ecological associations. In this special issue, Crawford summarizes exciting developments in microbial ecology as they relate to genome mining of Photorhabdis and Xenorhabdis species [67]. It is becoming increasingly apparent that understanding the roles of secondary metabolites in their endogenous contexts has the potential to reveal new strategies for controlling undesirable interkingdom relationships [57]. For instance, bacterial infections in humans may be addressed through the discovery of new antibiotic substances or bioactive metabolite antibiotic combinations discovered via gene mining methods. Beyond antibiosis, applications for interrogating interkingdom cell signaling provide inroads to new therapeutics for cancer and other human diseases. The discovery of the antifungal compound rapamycin from a strain of Streptomyces hygroscopicus resulted in the revelation of a whole area of cell signaling in the mammalian target of rapamycin (mTOR, for which over 11,000 PubMed entries are available) [18] and identification of new therapeutic targets cascading from this central signaling kinase [64, 71].
What microbes should be mined?
There has been an ongoing debate as to which microorganisms are the best sources for current and future discovery of natural products. Some scientists have suggested that unculturable microorganisms might serve as “untapped” sources for novel secondary metabolites [41]. With the advent of inexpensive microbial DNA sequencing, it became possible to explore the genetic capacity of different groups of microorganisms, and to ask: (1) which microbial taxa have the highest potential to produce large numbers of complex secondary metabolites with drug-like properties; (2) which taxa have moderate potential; and (3) which have the lowest potential. It stands to reason that effort should be focused on the microorganisms with the highest potential, and those with the lowest potential should not be heavily emphasized. If we use the numbers of type I polyketide synthase (PKS) and non-ribosomal peptide synthetase (NRPS) genes per microbe as yardsticks to measure the potential to produce secondary metabolites (i.e., pathways that contain type I PKS, NRPS, or mixed PKS-NRPS account for well over 60 % of important secondary metabolites discovered over the past 50 years [14], then it is clear that microbes with large genomes are generally more productive sources of secondary metabolites, and that among these the actinomycetes are the most productive [12, 28, 75]. It has been shown that the number of functional NRPS pathways can be estimated by counting the number of mbtH homologs in a microbial genome. MbtH homologs are generally small chaperone proteins (65–75 amino acids) that enhance adenylation reactions of some adenylation (A) domains during peptide assembly by NRPS proteins [12, 38, 54]. Because of the diversity of A domains encountered in NRPS genes, MbtH homologs can be orthologous for related pathways and paralogous for unrelated pathways. This dichotomy renders MbtH homologs ideal surrogates or “beacons” to count the numbers of NRPS pathways, and to triage known and unknown pathways by using low pass sequencing [12, 14]. Twenty-four internal segments of diverse MbtH homologs were concatenated to generate a probe for BLASTp analyses of MbtH-like proteins. The relative homologies to the individual 24 MbtH homolog probes were converted into numerical MbtH codes, which can facilitate the triage process. The MbtH code analysis confirmed that among the actinomycetes with large genomes, there are gifted, average, and not-so-gifted species. The MbtH codes for several gifted actinomycetes suggested that there are many new and novel NRPS pathways to be unraveled and possibly exploited for natural product discovery and development. Some other microbial groups are essentially devoid of mbtH (and NRPS) genes [12]. However, within the Proteobacteria, mbtH homologs are observed in Burkholderia, Photorhabdus, and Xenorhabdus species, and the full extent of secondary metabolite biosynthetic capabilities of these genera are being revealed by genome mining [49, 67].
While prioritization using NRPS and PKS potential is likely to enrich efforts in new molecule discovery in the near future, caution is warranted in focusing exclusively on large modular biosynthetic systems deriving from highly characterized systems. If we look beyond NRPS and type I PKS pathways, which require substantial coding capacity, it is becoming apparent from recent genome mining successes that many microbes with smaller genomes encode ribosomally synthesized and post-translationally modified peptides (RiPPs) or phosphonates [24, 44, 52]. Perhaps these discoveries will contribute to a more robust drug discovery process in the coming years. Moreover, more traditional microbial natural product discovery efforts continuously reveal new molecular diversity that is only associable with gene clusters after the fact, due to a lack of biosynthetic precedence. For instance, considering the structures of platensimycin, an oxidatively modified cyclic terpenoid [68], and merochlorin, a mixed polyketide-terpenoid natural product [45], both would unlikely be prioritized by comparison to well-characterized biosynthetic systems. These discoveries underline the continued importance of biosynthetic studies to understand newly discovered natural products.
How should microbial genomes be mined?
Genome-mining campaigns can range in complexity from simple trial-and-error approaches to breathtakingly ambitious programs in synthetic biology. Currently, these campaigns can be organized into one of two major categories: those involved in eliciting expression in the encoding producing organism (homologous expression) and those endeavoring to recapitulate pathways in non-producing hosts (heterologous expression) (Fig. 2). In the case of homologous expression, the power of secondary metabolic prescience alone should not be underestimated and foreknowledge of metabolic potential has forever changed the process of natural product discovery. Simply ‘looking harder’ via growth condition parameterization with structural guidance by (bio) analytical chemistry has unlocked a significant fraction of unknown metabolites [23, 32]. Indeed, this strategy saturated the discovery pipeline at Ecopia Biosciences and lead to farnesylated benzodiazepinone ECO4601, the first genome mining-derived natural product entered into human clinical trials in 2003 [5, 36]. However, there are limits to eliciting gene cluster expression and product detection by media formulation and analytical foreknowledge. Genetic regulation of secondary metabolism remains poorly understood across the diversity of secondary metabolite-producing organisms and identifying a low-abundance discrete predicted metabolite from within a crude metabolome is a non-trivial challenge. Consequently, more recently, a host of methods for activating regulated secondary metabolism have been developed with the ability to unlock tightly regulated clusters [11, 26, 51, 61, 76]. In combination with ‘looking harder’, and given the rate-limiting steps of isolation and structural elucidation, these approaches can likely occupy discovery efforts for some time. Heterologous expression can often aid in the production of adequate levels of compound for evaluation, particularly when expression hosts are derived from industrial production strains or highly engineered laboratory strains [9, 11, 34, 40]. Synthetic biology approaches are also being developed which focus on refactoring secondary metabolic gene clusters, either in producing hosts via genetic recombination, or in ‘clean’ heterologous hosts via gene synthesis [22, 62]. Heterologous expression approaches have many advantages in that they are not limited to gene clusters derived from cultivatable microbes, that the products of heterologous expression can be identified by comparatively straightforward differential metabolomic analysis of the clean and transformed host, and that heterologous systems, once established, can be readily genetically manipulated to diversify the encoded natural products. This special issue addresses both major categories of approaches. The work of Zhu et al. [76], Ochi et al. [61], and Yoon and Nodwell [72] focus specifically on creative new methods to activate secondary metabolism in actinomycetes and the work of Gomez-Escribo and Bibb [34], Ikeda et al. [40] and Cobb et al. [22] discuss cutting-edge approaches to heterologous expression.
Two major categories of approaches being investigated in genome-mining efforts. In heterologous expression, whole gene clusters are mobilized into expression strains and differentially analyzed to identify new compounds. In homologous expression approaches, endogenous transcriptional, translational, or metabolic elements are manipulated, either by mutation or (bio) chemical stimulants to activate secondary metabolite production. In either case, these approaches entail the labor-intensive process of identification of new peaks within a metabolome and isolation of these compounds for structural elucidation
Indeed, there are now no conceptual barriers towards the future unlocking of all previously cryptic and/or orphan secondary metabolic gene clusters. However, many significant technical and practical barriers must be addressed in order to realize the full potential of genome mining. Arguably, improvements in methods for isolation and structure elucidation of secondary metabolites from complex extracts have not kept pace with genomic advances, and it is likely that these will become the rate-limiting step in metabolite discovery [2, 56]. With the current state-of-the-art, isolation and elucidation of new natural products at moderate abundance (1 mg/l) require weeks to months or longer per compound for full characterization. Clearly this is an area in need of substantial innovation if the goals of genome mining are to be even partially realized. Remaining to be determined is the rate and cost per metabolite/gene cluster via genome-mining approaches. For instance, refactoring secondary metabolic gene clusters via a synthetic biology approach is highly resource intensive using standard technologies. In considering that most secondary metabolic gene clusters consist of dozens of genes (at a typical length of 20–150 kbp), and taking into account current gene synthesis costs and the time entailed in homologous recombination-based assembly of gene clusters and the likely requirement for generating multiple cluster variants, it seems probable that the cost per compound will be quite high ($50,000 USD or more per compound). Additionally, a method for universal heterologous protein expression has not yet been discovered, as indicated by the Protein Structure Initiative, in which only 18 % of human proteins could be expressed and purified in soluble form [19]. Correspondingly, in large secondary metabolite gene clusters bearing dozens of open reading frames, the potential for failure is quite high. For these reasons, homologous producer-cluster activation approaches, although they may unlock only a fraction of secondary metabolism, may prove in the short term to yield a substantially lower cost per compound and saturate isolation/elucidation pipelines in the immediate future. Ultimately however, it is expected that rapid and inexpensive gene synthesis and effective expression and more effective translation technologies will be developed to access not only the full potential of genome mining of the full natural pharmacopeia of culturable and non-culturable organisms, but also new natural chemical entities via mutasynthesis and recombineering.
How can genome mining be leveraged?
Enrichment for gifted microbes
It has been estimated that ~1026 actinomycete colony-forming units (mostly spores?) exist in the top 10 cm of soil covering the Earth, but only ~107 actinomycetes have been screened for secondary metabolite production by the pharmaceutical industry over the past 50 years [6]. Brady and coworkers [62, 63] have demonstrated that genome sequencing can be applied to environmental DNA (eDNA) extracted from soils, and the diversity of PKS and NRPS sequences assessed. This approach could be extended to include “beacon” analysis with the MbtH multiprobe and other pathway-specific probes to identify soils that contain gifted actinomycetes for cultivation and whole-genome sequencing.
Many actinomycete genera have traits that are amenable to enrichment by antibiotic selection and other nutritional methods [35]. As the most “gifted” genera are identified by sequencing many different species already identified, then specific enrichments can be used to build substantially larger collections of gifted microbes for whole-genome sequencing.
Coupling genome mining with combinatorial biosynthesis for accelerated evolution
In the past three decades, progress has been made on developing methodologies and biochemical rules for combinatorial biosynthesis of complex PKS, NRPS, and other pathways [7, 13, 22, 70]. For NRPSs, rules have been developed for coupling of domains to maintain proper upstream and downstream protein–protein interactions [13]. For instance, there are three types of condensation (C) domain for coupling fatty acid to l-amino acid, l-amino acid to l-amino acid, and d-amino acid to l-amino acid. Likewise, there are three types of thiolation (T) or peptidyl carrier protein (PCP) domain, depending on downstream interactions with C domains, epimerase (E) and C domains, or thioesterase (Te) domains. Successful genetic engineering of NRPS pathways requires the correct assembly of the right types of C and T domains in the right context (e.g., by keeping homologous C and A domains together whenever possible). Although many different NRPS modules have been already discovered, there are not many combinations of highly specialized modules (e.g., the combinations of C for fatty acid coupling have a limited number of A domain partners for initiating lipopeptide biosynthesis). Genome mining should provide a wealth of new NRPS parts and devices to facilitate expanded synthetic biology approaches to combinatorial biosynthesis of novel NRPS pathways. The same should be true for PKS and mixed NRPS/PKS pathways, and tailoring reactions (e.g., sugar biosynthesis and glycosyl transfer, hydroxylations, and transfer of methyl groups).
Modified natural products can also be generated by mutation outside of the modular biosynthetic systems. For instance, pactamycin analogs have been generated via mutasynthetic approaches that have superior activities to the progenitor natural products [3, 50], glycovariants have been generated of a large number of natural products via enzymatic methods [31] and genetic knockouts [29].
How can progress on genome mining be accelerated?
Knowing which peak to isolate
The success of genome mining hinges on being able to correlate metabolites of interest within complex biological extracts following pathway activation or heterologous expression. This endeavor is not a trivial process for either homologous or heterologous expression categories. While in theory this process should be simplified by heterologous expression approaches, there is no guarantee that new metabolites will be easily observed (e.g., due to low abundance, lack of chromophore to simplify detection, chemical incompatibility with selected extraction conditions, etc.). In response to this limitation, more sophisticated statistical methods are being developed to identify new compounds generated by modified growth conditions or recombinant strains [26, 39, 46]. The development of genomisotopic approaches [37] is another methodology that has the potential for advancing efforts for identifying metabolites of interest.
Speeding up isolation of secondary metabolites from complex extracts
Extraction followed by chromatographic partitioning and separation remain the primary means of isolating compounds from cultures. In most cases, this process involves multiple steps with target compound losses at every step resulting in diminishing yields of compounds throughout the purification process. In nearly all discovery efforts, compound purity, which is a function of sample homogeneity and stability, is the threshold parameter for initiating structure elucidation. It is also the rate-limiting step, often requiring scale-up fermentation, extraction, and purification protocols. The isolation process alone can require weeks to months to perfect and the follow-up elucidation process can also require an equivalent amount of time, depending on the structural complexity and inherent properties of the sample. The ideal process would be small molecule ‘teleportation’ from crude extracts into tubes, a process that is surprisingly not beyond the reach of ion soft landing mass spectrometry, an analytical technique that separates ionized compounds from mixtures using a mass analyzer and lands them on a surface forming compound arrays [66]. This technology has already been successfully demonstrated in purifying and landing small molecules and even active enzymes in sufficient quantities for analysis, but has not been scaled to isolate compounds in sufficient quantities for cryogenic NMR. In the absence of the wide-scale availability of this technology, assuming isolation and elucidation workflows cannot be accelerated otherwise, genome mining efforts may grind to a snail’s pace.
Developing universal tools for gene manipulation and expression in producing organisms
Secondary metabolite-producing organisms are taxonomically diverse and, despite decades of research, generalizable tools for genetic manipulation (e.g., transformation, intergeneric conjugation, homologous recombination, gene expression) are sparse across phyla. Even within a genus or species, quirks in gene uptake, regulator and genetic marker compatibility can confound efforts at genetic manipulation required for many homologous and heterologous expression techniques. The development of reliable genus- and species-specific tools for genetic manipulation of secondary metabolic gene clusters will be essential for rapid progress in genome mining.
Along the same line, a universal strategy for up-regulating the desired genetic elements for production of new metabolites would have huge ramifications for genome mining efforts. Evidence is increasing that a significant fraction of secondary metabolism in microorganisms can be activated to isolable levels by chemical and biochemical cues [48, 51, 61, 67, 72, 76]. A molecular understanding of the connection of these cues to specific transcriptional, translational, or other metabolic elements across genera would no doubt be very valuable in this context. Ideally, a comprehensive signaling network will be mapped in response to a potential elicitor that can be extended beyond a single metabolic pathway or species, thus helping to streamline the genome-mining process.
Synthetic biology tools
Heterologous expression of synthetic or cloned gene clusters is also in need of robust methods for the synthesis and assembly of small to large gene clusters in a reliable, inexpensive, and a high-throughput format. The aforementioned problems of functional gene expression will likely require the generation of multiple variants of targeted gene clusters that often consist of dozens of large biosynthetic genes such as those found in modular PKS and NRPS systems. De novo production of these genetic variants poses technological challenges in gene assembly and potential financial issues until costs per base decline. Operationally, refactoring polycistronic clusters also requires multiple orthogonal tools for selection, promoting, or otherwise marking, reassembled gene clusters, the feasibility of which has recently been described by refactoring a 20-gene, seven-operon nitrogen fixation cluster from Klebsiella oxytoca and functional expression in Escherichia coli [65].
Merge with the high-throughput model
The dominant paradigm in drug discovery, for better or worse, is via high-throughput screening (HTS) of large chemical libraries against biochemical and/or phenotypic assays. Notwithstanding the modest track record of this approach, the associated technologies are immensely powerful tools for efforts in drug discovery. Natural product discovery, which is becoming strongly associated with genome mining, would benefit greatly if natural products can be assembled in sufficient numbers, or if technology existed to assay them in sufficient numbers, to be complementary and compatible with current HTS methods and paradigms.
Investment in fundamental biosynthetic research
Bioinformatic approaches for the estimation of the secondary metabolic products of sequenced gene clusters [16, 17] and future engineering studies to generate chemical diversity are entirely dependent upon biosynthetic precedent established by basic research into the biochemistry of secondary metabolism. Indeed, decades of unraveling the molecular logic of NRPS and PKS systems has provided a sound foundation for searching genomes and predicting the chemical output (i.e., metabolite identity). As a relatively recent example, progress in understanding the biosynthesis of RiPPs has unleashed a torrent of identification of gene clusters encoding this previously poorly understood class of compounds, and created an entire new category of genome mining and synthetic biology efforts [52]. There are undoubtedly many such uninvestigated systems for currently known secondary metabolites that could create new domains for genome mining. Thus, a continued investment into unraveling the underlying biosynthetic mechanisms of structurally diverse metabolites will foreseeably refine what is meant by a “gifted” organism.
Who should fund future progress in genome mining?
In the past, natural product discovery and development has been mainly funded by large pharmaceutical companies or chemical companies with animal health or plant sciences subsidiaries. This worked well when discoveries came easily, and returns on investments were sufficient to drive the process, but most pharmaceutical companies have abandoned natural products discovery during the past two decades. More recently, biotechnology companies have been carrying much of the load, but no individual company has the resources to fully exploit the rapidly developing field of genome mining, and develop it into a robust discipline commensurate with its sizable potential. It would seem that this is an opportune time for the NIH, NSF, and DOE in the US and other funding agencies in Europe and Asia to put sizable resources into bringing this important new discipline to a technological level commensurate with its potential to generate new molecules for drug discovery not obtainable by medicinal or combinatorial chemistry. As an example, the DOE has funded a Microbial Genome Project that focuses on mission areas of alternative fuels, global carbon cycling, and biogeochemistry (http://www.jgi.doe.gov/CSP/user_guide/). This approach has generated important fundamental information on a few actinomycetes; in particular, the finished quality of the genome sequences assures a high level of confidence in the assembly of complex PKS and NRPS pathways. This in turn can serve as part of an important expanding baseline for current and future genome mining. It is intriguing that among the small number of actinomycetes sequenced so far in this program, Actinosynnema mirum [47] and Streptosporangium roseum [60] can be classified as gifted by the MbtH counting method [14], and Saccharomonospora viridis has yielded a cryptic daptomycin biosynthetic gene cluster [10], even though none of these strains were sequenced with secondary metabolite discovery in mind. To further exploit this approach of finishing subsets of microbial genomes, it would be highly valuable to develop a program to generate finished genome sequences of many actinomycetes that produce interesting secondary metabolites, and to fully annotate all known secondary metabolite clusters. Having a baseline of all known secondary metabolite pathways will accelerate the discovery of novel secondary metabolite pathways, while streamlining the de-replication of known pathways, the bane of natural products discovery in industry that has impeded progress for the past three decades.
References
Aigle B, Lautra S, Spiteller D, Dickschat JS, Challis GL, Leblond P, Pernodet J-L (2013) Genome mining of Streptomyces ambofaciens. J Ind Microbiol Biotechnol. doi:10.1007/s10295-013-1379-y
Albright JC, Goering AW, Doroghazi JR, Metcalf WW, Kelleher NL (2013) Strain-specific proteogenomics accelerates discovery of natural products via their biosynthetic pathways. J Ind Microbiol Biotechnol. doi:10.1007/s10295-013-1373-4
Almabruk KH, Lu W, Li Y, Abugreen M, Kelly JX, Mahmud T (2013) Mutasynthesis of fluorinated pactamycin analogues and their antimalarial activity. Org Lett 15:1678–1681
Bachmann BO, McAlpine JB, Zazopoulos E, Farnet CM (2003) Polyene polyketides, process for their production and their use as a pharmaceutical. US Patent 7,375,088
Bachmann BO, McAlpine JB, Zazopoulos E, Farnet CM, Piraee M (2006) Farnesyl dibenzodiazepinone, and processes for its production. US Patent 7,101,872
Baltz RH (2005) Antibiotic discovery from actinomycetes: will a renaissance follow the decline and fall? SIM News 55:186–196
Baltz RH (2006) Molecular engineering approaches to peptide, polyketide and other antibiotics. Nat Biotechnol 24:1533–1540
Baltz RH (2008) Renaissance in antibacterial discovery from actinomycetes. Curr Opin Pharmacol 8:557–563
Baltz RH (2010) Streptomyces and Saccharopolyspora hosts for heterologous expression of secondary metabolite gene clusters. J Ind Microbiol Biotechnol 37:759–772
Baltz RH (2010) Genomics and the ancient origins of the daptomycin biosynthetic gene cluster. J Antibiot 63:506–511
Baltz RH (2011) Strain improvement in actinomycetes in the postgenomic era. J Ind Microbiol Biotechnol 38:657–666
Baltz RH (2011) Function of MbtH homologs in non-ribosomal peptide biosynthesis and applications in secondary metabolite discovery. J Ind Microbiol Biotechnol 38:1747–1760
Baltz RH (2012) Combinatorial biosynthesis of cyclic lipopeptide antibiotics: a model for synthetic biology to accelerate the evolution of secondary metabolite biosynthetic pathways. ACS Synth Biol. doi:10.1021/sb3000673
Baltz RH (2013) MbtH homology codes to identify gifted microbes for genome mining. J Ind Microbiol Biotechnol. doi:10.1007/s10295-013-1360-9
Bentley SD, Chater KF, Cerdeño-Tárraga AM et al (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147
Blin K, Medema MH, Kazempour D, Fischbach MA, Breitling R, Takano E, Weber T (2013) antiSMASH 2.0—a versatile platform for genome mining of secondary metabolite producers. Nucl Acids Res 41:W204–W212
Boddy CN (2013) Bioinformatics tools for genome mining of polyketide and non-ribosomal peptides. J Ind Microbiol Biotechnol. doi:10.1007/s10295-013-1368-1
Brown EJ, Albers MW, Shin TB, Ichikawa K, Keith CT, Lane WS, Schreiber SL (1994) A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature 369:756–758
Büssow K, Scheich C, Sievert V, Harttig U, Schultz J, Simon B, Bork P, Lehrach H, Heinemann U (2005) Structural genomics of human proteins—target selection and generation of a public catalogue of expression clones. Microb Cell Fact 4:21
Challis G (2013) Exploitation of the Streptomyces coelicolor A3(2) genome sequence for discovery of new natural products and biosynthetic pathways. J Ind Microbiol Biotechnol. doi:10.1007/s10295-013-1383-2
Challis GL, Ravel J (2000) Coelichelin, a new peptide siderophore encoded by the S. coelicolor genome: structure prediction from the sequence of its non-ribosomal peptide synthetase. FEMS Microbiol Lett 187:111–114
Cobb RE, Ning JC, Zhao H (2013) DNA assembly techniques for next generation combinatorial biosynthesis of natural products. J Ind Microbiol Biotechnol. doi:10.1007/s10295-013-1358-3
Corre C, Challis GL (2009) New natural product biosynthetic chemistry discovered by genome mining. Nat Prod Rep 26:977–986
Deane CD, Mitchell DA (2013) Lessons learned from the transformation of natural product discovery to a genome-driven endeavor. J Ind Microbiol Biotechnol. doi:10.1007/s10295-013-1361-8
Demain AL (2013) Importance of microbial natural products and the need to revitalize their discovery. J Ind Microbiol Biotechnol. doi:10.1007/s10295-013-1325-z
Derewacz DK, Goodwin CR, McNees CR, McLean JA, Bachmann BO (2013) Antimicrobial drug resistance affects broad changes in metabolomic phenotype in addition to secondary metabolism. Proc Natl Acad Sci USA 110:2336–2341
Didelot X, Bowden R, Wilson DJ, Peto TE, Crook DW (2012) Transforming clinical microbiology with bacterial genome sequencing. Nat Rev Genet 13:601612
Donadio S, Monciardini P, Sosio M (2007) Polyketide synthases and non-ribosomal peptide synthetases: the emerging view from bacterial genomics. Nat Prod Rep 24:1073–1109
Du Y, Derewacz DK, Deguire SM, Teske J, Ravel J, Sulikowski GA, Bachmann BO (2011) Biosynthesis of the apoptolidins in Nocardiopsis sp. FU 40. Tetrahedron 67:6568–6575
Farnet CM, Zazopoulos E (2005) Improving drug discovery from microorganisms. In: Zhang L, Demain AL (eds) Natural products: drug discovery and therapeutic medicine. Humana Press Inc, Totowa, pp 95–106
Gantt RW, Peltier-Pain P, Thorson JS (2011) Enzymatic methods for glyco (diversification/randomization) of drugs and small molecules. Nat Prod Rep 28:1811–1853
Genilloud O, González I, Salazar O, Martín J, Tormo JR, Vicente F (2011) Current approaches to exploit actinomycetes as a source of novel natural products. J Ind Microbiol Biotechnol 38:375–389
Giddings LA, Newman DJ (2013) Microbial natural products: molecular blueprints for antitumor drugs. J Ind Microbiol Biotechnol 40:1181–1210
Gomez-Escribano JP, Bibb MJ (2013) Heterologous expression of natural product biosynthetic gene clusters in S. coelicolor: from genome mining to manipulation of biosynthetic pathways. J Ind Microbiol Biotechnol. doi:10.1007/s10295-013-1348-5
Goodfellow M (2010) Selective isolation of Actinobacteria. In: Baltz RH, Davies JE, Demain AL (eds) Manual of industrial microbiology and biotechnology. American Society for Microbiology, Washington, pp 13–27
Gourdeau H, McAlpine JB, Ranger M, Simard B, Berger F, Beaudry F, Farnet CM, Falardeau P (2008) Identification, characterization and potent antitumor activity of ECO-4601, a novel peripheral benzodiazepine receptor ligand. Cancer Chemother Pharmacol 61:911–921
Gross H, Stockwell VO, Henkels MD, Nowak-Thompson B, Loper JE, Gerwick WH (2007) The genomisotopic approach: a systematic method to isolate products of orphan biosynthetic gene clusters. Chem Biol 14:53–63
Herbst DA, Boll B, Zocher G, Stehle T, Heide L (2013) Structural basis of the interaction of MbtH-like proteins, putative regulators of non-ribosomal peptide biosynthesis, with adenylating enzymes. J Biol Chem 288:1991–2003
Hou Y, Braun DR, Michel CR, Klassen JL, Adnani N, Wyche TP, Bugni TS (2012) Microbial strain prioritization using metabolomics tools for the discovery of natural products. Anal Chem 84:4277–4283
Ikeda H, Shin-Ya K, Ōmura S (2013) Genome mining of the Streptomyces avermitilis genome and development of genome-minimized hosts for heterologous expression of biosynthetic gene clusters. J Ind Microbiol Biotechnol. doi:10.1007/s10295-013-1327-x
Iqbal HA, Feng Z, Brady SF (2012) Biocatalysts and small molecule products from metagenomic studies. Curr Opin Chem Biol 16:109–116
Jensen PR, Chavarria K, Fenical W, Moore BS, Ziemert N (2013) Challenges and triumphs to genomics-based natural product discovery. J Ind Microbiol Biotechnol. doi:10.1007/s10295-013-1353-8
Jensen PR, Mincer TJ, Williams PG, Fenical W (2005) Marine actinomycete diversity and natural product discovery. Antonie Van Leeuwenhoek 87:43–48
Ju K-S, Doraghazi JR, Metcalf WW (2013) Genomics enabled discovery of phosphonate natural products and their biosynthetic pathways. J Ind Microbiol Biotechnol. doi:10.1007/s10295-013-1375-2
Kaysser L, Bernhardt P, Nam SJ, Loesgen S, Ruby JG, Skewes-Cox P, Jensen PR, Fenical W, Moore BS (2012) Merochlorins A–D, cyclic meroterpenoid antibiotics biosynthesized in divergent pathways with vanadium-dependent chloroperoxidases. J Am Chem Soc 134:11988–11991
Krug D, Zurek G, Schneider B, Garcia R, Müller R (2008) Efficient mining of myxobacterial metabolite profiles enabled by liquid chromatography-electrospray ionization-time-of-flight mass spectrometry and compound-based principal component analysis. Anal Chim Act 624:97–106
Land M, Lapidus A, Mayilraj S (2009) Complete genome sequence of Actinosynnema mirum type strain (101). Stand Genom Sci 1:46–53
Lautru S, Deeth RJ, Bailey LM, Challis GL (2005) Discovery of a new peptide natural product by S. coelicolor genome mining. Nat Chem Biol 1:265–269
Liu X, Cheng Y-Q (2013) Genome-guided discovery of diverse natural products from Burkholderia sp. J Ind Microbiol Biotechnol. doi:10.1007/s10295-013-1376-1
Lu W, Roongsawang N, Mahmud T (2011) Biosynthetic studies and genetic engineering of pactamycin analogs with improved selectivity toward malarial parasites. Chem Biol 18:425–431
Luzhetskyy A, Rebets Y, Brötz E, Tokovenko B (2013) Actinomycetes biosynthetic potential: how to bridge in silico and in vivo. J Ind Microbiol Biotechnol. doi:10.1007/s10295-013-1352-9
Maksimov MO, Link AJ (2013) Prospecting genomes for lasso peptides. J Ind Microbiol Biotechnol. doi:10.1007/s10295-013-1357-4
McAlpine JB, Bachmann BO, Piraee M, Tremblay S, Alarco AM, Zazopoulos E, Farnet CM (2005) Microbial genomics as a guide to drug discovery and structural elucidation: ECO-02301, a novel antifungal agent, as an example. J Nat Prod 68:493–496
McMahon MD, Rush JS, Thomas MG (2012) Analyses of MbtB, MbtE, and MbtF suggest revisions to the mycobactin biosynthesis pathway in Mycobacterium tuberculosis. 194:2809–2818
Molinski TF (2010) Microscale methodology for structure elucidation of natural products. Curr Opin Biotechnol 21:819–826
Molinski TF (2010) NMR of natural products at the nanomole-scale. Nat Prod Rep 27:321–329
Moree WJ, Phelan VV, Wu CH, Bandeira N, Cornett DS, Duggan BM, Dorrestein PC (2012) Interkingdom metabolic transformations captured by microbial imaging mass spectrometry. Proc Natl Acad Sci USA 109:13811–13816
Nett M, Ikeda H, Moore BS (2009) Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat Prod Rep 26:1362–1384
Newman DJ, Cragg GM (2012) Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod 75:311–335
Nolan M, Sikorski J, Jando M et al (2010) Complete genome sequence of Streptosporangium roseum type strain (NI 9100). Stand Genom Sci 2:29–37
Ochi K, Tanaka Y, Tojo S (2013) Activating the expression of bacterial cryptic genes by rpoB mutations in RNA polymerase or by rare earth elements. J Ind Microbiol Biotechnol. doi:10.1007/s10295-013-1349-4
Owen JG, Reddy BV, Ternel MA, Charlop-Powers Z, Calle PY, Kim JH, Brady SF (2013) Mapping gene clusters within arrayed metagenomic libraries to expand the structural diversity of bio-medically relevant natural products. Proc Nat Acad Sci USA 110:11797–11802
Reddy BV, Kallifidas D, Kim JH, Charlop-Powers Z, Feng Z, Brady SF (2012) Natural product biosynthetic gene diversity in geographically distinct soil microbiomes. Appl Environ Microbiol 78:3744–3755
Roden J, Dienstmann R, Serra V, Tabernero J (2013) Development of PI3 K inhibitors: lessons learned from early clinical trials. Nat Rev Clin Oncol 10:143–153
Temme K, Zhao D, Voigt CA (2012) Refactoring the nitrogen fixation gene cluster from Klebsiella oxytoca. Proc Nat Acad Sci USA 109:7085–7090
Ouyang Z, Takats Z, Blake TA, Gologan B, Guymon AJ, Wiseman JM, Oliver JC, Davisson VJ, Cooks RG (2003) Preparing protein microarrays by soft-landing of mass-selected ions. Science 301:1351–1354
Vizcaino MI, Guo X, Crawford JM (2013) Merging chemical ecology with bacterial genome mining for secondary metabolite discovery. J Ind Microbiol Biotechnol. doi:10.1007/s10295-013-1356-5
Wang J, Soisson SM, Young K et al (2006) Platensimycin is a selective FabF inhibitor with potent antibiotic properties. Nature 441:358–361
Weissman KJ, Müller R (2010) Myxobacterial secondary metabolites: bioactivities and modes-of-action. Nat Prod Rep 27:1276–1295
Wong FT, Khosla C (2012) Combinatorial biosynthesis of polyketides––a perspective. Curr Opin Chem Biol 16:117–123
Yea SS, Fruman DA (2013) Achieving cancer cell death with PI3 K/mTOR-targeted therapies. Ann NY Acad Sci 1280:15–18
Yoon V, Nodwell JR (2013) Activating secondary metabolism with stress and chemicals. J Ind Microbiol Biotechnol. doi:10.1007/s10295-013-1387-y
Zazopoulos E, Huang K, Staffa A, Liu W, Bachmann BO, Nonaka K, Ahlert J, Thorson JS, Shen B, Farnet CM (2003) A genomics-guided approach for discovering and expressing cryptic metabolic pathways. Nat Biotechnol 21:187–190
Zerikly M, Challis GL (2009) Strategies for the discovery of new natural products by genome mining. ChemBioChem 10:625–633
Zhu F, Qin C, Tao L et al (2011) Clustered patterns of species origins of nature-derived drugs and clues for future bio-prospecting. Proc Nat Acad Sci USA 31:12943–12948
Zhu H, Sandiford SK, van Wezel GP (2013) Triggers and cues that activate antibiotic production by actinomycetes. J Ind Microbiol Biotechnol. doi:10.1007/s10295-013-1309-z
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bachmann, B.O., Van Lanen, S.G. & Baltz, R.H. Microbial genome mining for accelerated natural products discovery: is a renaissance in the making?. J Ind Microbiol Biotechnol 41, 175–184 (2014). https://doi.org/10.1007/s10295-013-1389-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-013-1389-9