Abstract
Polycyclic aromatic hydrocarbon (PAH) quinone reductase (PQR) and catechol-O-methyltransferase (COMT), from the PAH-degrading Mycobacterium vanbaalenii PYR-1, were demonstrated to be constitutive enzymes located in the soluble fraction of cell extracts. PQR activities for the reduction of 9,10-phenanthrenequinone and 4,5-pyrene- quinone were 1.40±0.13 and 0.12±0.01 μmol min−1 mg-protein−1, respectively. The exogenous catechols alizarin, anthrarobin, 2,3-dihydroxynaphthalene and esculetin inhibited PQR activity. Anthrarobin (100 μM) and esculetin (100 μM) inhibited 4,5-pyrenequinone reduction by 64–92%. COMT was involved in the O-methylation of 1,2-dihydroxyphenanthrene to form 1-methoxy-2-hydroxyphenanthrene and 1,2-dimethoxyphenanthrene. Both pyrene and 1-hydroxypyrene were metabolized by M. vanbaalenii PYR-1 to form 1-methoxypyrene, 1-methoxy-2-hydroxypyrene, 1-hydroxy-2-methoxypyrene and 1,2-dimethoxypyrene. Among the catechols tested, anthrarobin showed the highest COMT activity (1.06±0.04 nmol/30 min−1 mg-protein−1). These results suggest that the PQR and COMT activities of M. vanbaalenii PYR-1 may play an important role in the detoxification of PAH catechols.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
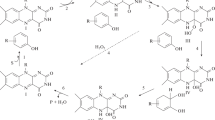
Recently, members of the genus Mycobacterium have been reported to degrade a wide variety of environmentally hazardous chemicals, including high molecular weight polycyclic aromatic hydrocarbons (PAHs), in soils and sediments [4, 13, 14, 23, 24, 27, 28]. Research in our laboratory has shown that Mycobacterium vanbaalenii PYR-1 [10] can extensively degrade PAHs with up to five fused aromatic rings [3, 5, 6, 8, 9, 15–17]. In contrast to Pseudomonas and Sphingomonas species, the initial oxidation reaction of PAHs by M. vanbaalenii PYR-1 seems to be highly stereo- and regio-specific, forming PAH dihydrodiols with cis and trans configurations [15–17]. This may be due to the involvement of a unique oxygenase system in the initial reactions. An NADH-linked dioxygenase produces cis-dihydrodiols [2, 10], and an NAD(P)H-linked cytochrome P450 and epoxide hydrolase system produce trans-dihydrodiols [6, 15–17] (Fig. 1). The initial enzyme system oxidizes a broad range of PAHs at different positions on the aromatic rings [15–17, 26, 27].
Dihydrodiol dehydrogenase catalyzes the conversion of PAH dihydrodiols to PAH catechols, which rapidly autoxidize to PAH o-quinones or serve as substrates for ring-fission enzymes [12]. PAH catechols and o-quinones facilitate one-electron redox cycling between quinones and semiquinones, which generate reactive oxygen species (ROS) with prominent cytotoxicity [22]. PAH o-quinones are genotoxic, producing in vitro and in vivo DNA strand scission, DNA adduct formation, and malondialdehyde generation [1]. The indigenous toxicity of PAH o-quinones can be reduced by an NAD(P)H-quinone reductase to form PAH catechols, which can either be metabolized by ring-cleavage enzymes or form glucuronide, reduced glutathione, sulfate or methyl conjugates. Recently, we observed that one of the two NADH-phenanthrenequinone reductases, PQR2, isolated from another pyrene-degrading Mycobacterium sp., strain PYR-100, reduced a broad range of PAH o-quinones and was strongly inhibited by exogenous catechols, such as alizarin, esculetin, and quercetin [11]. This implies that PAH catechols derived directly from PAH oxidation by enzymatic reduction of PAH o-quinones can adversely affect PAH degradation by inhibiting PAH quinone reductase (PQR) activity.
Mammalian catechol-O-methyltransferase (COMT) catalyzes the O-methylation of endogenous catechols (e.g., estrogen catechols, epinephrine, norepinephrine and dopamine) to protect cells from ROS-mediated cytotoxicity and genotoxicity, and to form O-methylated antioxidants, which act as competitive hydroxyl radical scavengers [18, 19, 30, 31]. COMT appears to play a role in deactivating non-metabolizable and biologically active PAH catechols, since 1-methoxy-2-hydroxyanthracene and 7-methoxy-8-hydroxyfluoranthene are produced as dead-end products from the degradation of anthracene and fluoranthene, respectively, by M. vanbaalenii PYR-1 [9, 15]. However, the COMT activity of M. vanbaalenii PYR-1 has not been determined with an exogenous catechol in the presence of the universal methyl donor, S-adenosyl-l-methionine.
The present study shows that a constitutive COMT enzyme exists in the cytosol of M. vanbaalenii PYR-1 and that this enzyme may be responsible for the production of methylated catechols from phenanthrene and pyrene. Evidence is also provided for the involvement of quinone reductase(s) in the detoxification of PAH catechols derived from PAH degradation pathways.
Materials and methods
Chemicals
The following chemicals were obtained from Sigma-Aldrich (St Louis, Mo.): alizarin (1,2-dihydroxyanthraquinone), anthrarobin (1,2,10-anthracenetriol), 2,3-dihydroxynaphthalene, esculetin (6,7-dihydroxycoumarin), 1-hydroxypyrene, phenanthrene, 9,10-phenanthrenequinone, pyrene, dl-dithiothreitol (DTT), NADH, NADPH, S-(5,adenosyl)-l-methionine (SAM) and [methyl-3H] S-5′-adenosyl-l-methionine ([methyl-3H]SAM). Synthetic 4,5-pyrenequinone was a gift from P.P. Fu, National Center for Toxicological Research (US FDA, Jefferson, Ark.) and M.D. Aitken of the University of North Carolina (Chapel Hill, N.C.). Other solvents and chemicals were of the highest grades available from Baker (Phillipsburg, N.J.).
Cultural conditions
M. vanbaalenii PYR-1, isolated from microcosm sediments of a petrogenic site [5, 10], was cultivated continuously using a Bioflo 3000 bioreactor and a 5 l glass vessel containing 2 l mannitol-yeast extract (MYE) medium consisting of 10 g d-mannitol and 2 g yeast extract in 1 l phosphate-basal minimal medium [11]. The pH of media varied from 6.5 to 9.5, adjusted with 5 M NaOH and 5 M phosphoric acid, and harvested cells were subsequently used for batch cultures to determine the apparent maximum growth rates (μmax′, d−1) with an initial cell density (optical density at 546 nm) of 0.05 under the same pH conditions. The bioreactor had an airflow of 2 l/min and the medium was agitated at 250 rpm with a marine blade impeller. The heat exchanger and the exhaust condenser were operated at 30 and 4°C, respectively. Cells were harvested by centrifugation for 15 min at 4°C and 14,000 g, and washed three times with equal volumes of sterile 50 mM sodium phosphate buffer (pH 7.4) chilled to 4°C.
Whole cell biotransformation of PAHs
Freshly harvested cells (5 g wet weight) were suspended in 100 ml sterile 50 mM sodium phosphate buffer (pH 7.4) in 500 ml flasks, and phenanthrene or pyrene was added to a final concentration of 0.5 mM. As a control, no PAH was added to the cell suspension in one flask. The flasks were incubated at 30°C with shaking at 250 rpm. Samples (250 μl) were taken at various timed intervals, immediately mixed with 750 μl cold acetonitrile, centrifuged at 14,000 g for 15 min and analyzed by HPLC. At the same time, a 100 μl sample was serially diluted with 900 μl sterile 50 mM sodium phosphate buffer (pH 7.4), and the dilution orders of magnitude from 10−6 to 10−8 were evenly plated on Remel (Lenexa, Kan.) tryptic soy agar plates. After incubation at 30°C for 1 week, plates containing 30–300 colonies were counted to determine the colony-forming units (cfu).
At the midpoint of PAH degradation, the reaction mixtures were centrifuged for 15 min at 4°C and 14,000 g, and the supernatants were extracted with three volumes of ethyl acetate at neutral and acidic (pH 2.5) conditions. The ethyl acetate was vacuum evaporated and the residue was dissolved in a small volume of acetone and filtered through a 0.2-μm Teflon filter. The samples were dried in vacuo and stored at −20°C.
Cell pastes were washed three times with equal volumes of 50 mM sodium phosphate (pH 7.4). Cells were disrupted by three passes through a pre-chilled French pressure cell at 1,400 psi and five 30 s cycles of ultrasonication at 200 W and 30 s cooling on ice. Cell debris was removed by centrifugation for 1 h at 120,000 g. To extract PAH metabolites formed in the cytosolic fractions, the cell extracts were filtered through Sep-Pak Plus tC18-cartridges (Waters, Milford, Mass.), which had been washed with 10 ml methanol and reconstituted with 50 mM sodium phosphate (pH 7.4). The cell extracts were stored at −20°C. After the tC18-cartridges were washed with water, the hydrophobic residues were eluted with 5 ml methanol. The methanol was removed using a Speed-Vac system (Savant Instruments, Holbrook, N.Y.), and the remaining solid was dissolved in a small volume of acetone. The insoluble particles were removed through a 0.2-μm Teflon filter, and the samples were dried in vacuo and stored at −20°C.
For 1-hydroxypyrene biotransformation experiments, cultures of M. vanbaalenii PYR-1 were grown in 250-ml Erlenmeyer flasks containing 50 ml minimal basal salts medium supplemented with 0.38 g/ml each of peptone, yeast extract, and soluble starch. A 100 μl aliquot of phenanthrene in N,N-dimethylformamide (DMF; 12 mg/ml) was added to each flask for enzyme induction. The cultures were grown for 4 days in the dark at 28°C with shaking at 110 rpm. 1-Hydroxypyrene (6 mg) was dissolved in DMF and added to the cultures. After 48 h of incubation, the flasks were extracted with three equal volumes of ethyl acetate and dried. The residue was dissolved in 3 ml methanol and concentrated to approximately 100 μl, using a model SS21 Savant Speed-Vac system (Savant Instruments) for analysis by reversed-phase HPLC.
PQR assay
PQR activity was measured by monitoring the oxidation of NADH at 340 nm (ɛ=6,220 M−1 cm−1), and 1 unit (U) PQR activity was defined as the oxidation of 1 μmol NADH per minute. The reaction mixtures (1 ml) contained 5 μl 4,5-pyrenequinone (final concentration, 10 μM) or 9,10-phenanthrenequinone (final concentration, 50 μM) dissolved in dimethylsulfoxide, 5 μl 20 mM NADH (final concentration, 100 μM) and 10–50 μl cell extract (100 μg/ml) in 50 mM sodium phosphate (pH 7.4). The reaction was started by addition of the enzyme solution and the initial rate was measured for 30 s at 25°C using a Shimadzu UV-2101PC spectrophotometer equipped with a thermostat (Shimadzu, Kyoto, Japan). To examine the inhibitory effects of exogenous catechols, PQR activity on 9,10-phenanthrenequinone or 4,5-pyrenequinone was measured in the presence or absence of an exogenous catechol (100 μM) as described above. All enzyme assays were performed in triplicate. Protein concentrations were determined by the Bio-Rad protein assay (Bio-Rad, Hercules, Calif., using bovine serum albumin as a standard.
Southern hybridization screening for pqr1 and pqr2
For Southern hybridization, the degenerate probes PQR1 (AAYCCNMGNGAYGTNGCNGTNYTNGTNGGNWSNYTNMGNAARGARWSNYTNAAYYTNAARYTNGCNAARGC) and PQR2 (GCNAARGTNYTNTAYATHACNGCNCAYCCNCAYGAYGARACNGTNWSNTAYWSNATGGCNACNGCNAARGCNTT) were used, which correspond with the published N-terminal amino acid sequences of the quinone reductases PQR1 and PQR2 from Mycobacterium sp. PYR-100 [11]. The oligonucleotide probes were labeled with a DIG oligonucleotide tailing kit, 2nd generation, according to the manufacturer’s instructions, procedure 3.2.1 (Roche, Indianapolis, Ind.).
XbaI-digested DNA from Mycobacterium sp. PYR-100 and M. vanbaalenii PYR-1 was prepared, separated by pulsed field gel electrophoresis and blotted onto nylon membranes [2]. Southern hybridization was performed repeatedly under various stringency conditions [25].
PCR screening for PQR1 and PQR2 genes
To detect the pqr1 and pqr2 genes of M. vanbaalenii PYR-1 by PCR, primers were designed using the conserved domains of the homologous genes identified in the GenBank database (Table 1). The translated protein sequences showed the closest homology to the determined N-terminal amino acid sequences of PQR1 and PQR2 of Mycobacterium sp. PYR100 [11]. Total genomic DNA extracted from M. vanbaalenii PYR-1 using a DNA Easy Tissue Kit (Qiagen, Valencia, Calif.) was used as a PCR template. PCR was performed with Taq DNA polymerase and the supplied PCR solutions according to the manufacturer’s instructions (Qiagen). The PCR regime consisted of 3 min preincubation at 95°C, 30 cycles of 30 s denaturation at 94°C, 30 s annealing at 55°C and 1 min extension at 72°C, followed by a final hold at 72°C for 7 min. PCR products of the expected sizes were sequenced using amplification primers that contained no degenerate bases.
COMT assay
COMT activity was assayed by a modification of the method of Zhu et al. [30–32]. The reaction mixture (total volume, 100 μl) consisted of 0.1 M sodium phosphate (pH 7.4), 1.2 mM MgCl2, 1 μCi [methyl−3H]SAM, 50 μM S-adenosyl-l-methionine (SAM), 1 mM DTT, 0.5 mM NADPH, 0.5 mM NADH, 50 μM catechol substrate dissolved in dimethylsulfoxide, and an appropriate amount of cell extract containing 100 μg protein. Reactions were initiated by addition of cell extract and were stopped at 30 min by addition of 1 ml hexane. The solvent extract (0.5 ml) was mixed with 7.5 ml Ultima Gold LSC-cocktail (Packard, Meriden, Conn.) and the radioactivity (dpm) was counted using a Packard 2000CA Tri-Carb liquid scintillation counter (Packard, Downers Grove, Ill.). The specific activity (U/mg) of COMT was determined as the consumption of 1 μmol SAM mg-protein−1 30 min−1. As controls, either the catechol substrate or the cell extract was omitted.
Physical and chemical analysis
PAHs and their metabolites were separated by reversed-phase HPLC with UV detection at 254 nm. The chromatographic system consisted of an Inertsil ODS3 column (5 μm, 4.6×250 mm, MetaChem, Torrance, Calif.), a guard column packed with the same stationary phase, a Waters 600 pump and controller, a Waters 2487 dual wavelength absorbance detector, and a Waters 717 Plus autosampler with a 200-μl injection loop. The system control and data analysis used a Waters Millennium32 Chromatography Manager. The mobile phase, delivered at 1 ml/min, was a linear gradient of 30% (v/v) to 90% (v/v) acetonitrile in water for 20 min, then held at 90% acetonitrile for 10 min.
Gas chromatography/mass spectrometry (GC/MS) and direct exposure probe mass spectrometry (DEP/MS) analyses were performed on a TSQ 700 triple quadrupole mass spectrometer (ThermoFinnigan, San Jose, Calif.) operated in the single quadrupole analyzer mode. Electron ionization (EI) was employed at 70 eV with a 150°C ion-source temperature. For the DEP/EI-MS analyses, the DEP current was increased linearly to 800 mA at 5 mA/s. A DB-5ms capillary column (J&W Scientific, Folsom, Calif.) (30 m ×0.25 mm ×0.25 μm) was used for chromatographic separation (GC/MS).
Proton NMR spectra were recorded at 500.13 MHz on a Bruker AM500 spectrometer (Bruker, Billerica, Mass.). The metabolites were dissolved in 0.5 ml deuterated acetone (99.96 atom% D); 1H chemical shifts were reported on the δ scale (ppm) by assigning the residual solvent peak to 2.04 ppm. Typical 1H data acquisition conditions were as follows: data size 32,000; sweep width 7,042 Hz; filter width 8,900 Hz; acquisition time 2.33 s; flip angle 90°; relaxation delay 0 s; temperature 301 K. For measurements of coupling constants, the free induction decay was zero-filled to 64 K, resulting in a final data point resolution of 0.215 Hz per point. Coupling constants reported were first-order. Assignments were made from homonuclear decoupling experiments, nuclear Overhauser effect (NOE) experiments, integration, analysis of substituent effects, and comparison with spectra of authentic compounds.
Results
Degradation of phenanthrene and pyrene by M. vanbaalenii PYR-1
By varying the pH of MYE media in chemostat cultures, it was determined that M. vanbaalenii PYR-1 grew optimally at pH 7.5, and the apparent maximum specific growth rate (μ′max) was 1.42 day−1 (Fig. 2). At that pH, approximately 15 g wet weight of cells were harvested from 1 l culture medium.
Because the dissolution rate of solid phenanthrene and pyrene into the aqueous phase limits the rate of degradation, phenanthrene and pyrene began to be metabolized only after 6–12 h of incubation (Fig. 3). After these lag times, both degradation curves appeared to be linear (r2>0.95). The specific degradation rate of phenanthrene (0.42±0.04 μmol min−1 mg-protein−1) was much greater than that of pyrene (0.17±0.01 μmol min−1 mg-protein−1).
Degradation curves of phenanthrene (filled triangles) and pyrene (filled circles) by whole cells (5 g wet weight) of M. vanbaalenii PYR-1 in 100 ml 50 mM sodium phosphate buffer (pH 7.4). Colony-forming units (cfu) for the degradation of phenanthrene (open triangles) and pyrene (open circles) are shown on a logarithmic scale
PQR activity of M. vanbaalenii PYR-1
When M. vanbaalenii PYR-1 was continuously cultivated in MYE medium, the cell extract contained PQR activity for the reduction of 9,10-phenanthrenequinone (PQ) and 4,5-pyrenequinone (PyQ) with the respective specific activities (μmol mg-protein−1 min−1) of 1.40±0.13 and 0.12±0.01. There was little difference in PQR activity when M. vanbaalenii PYR-1 was grown in the absence or presence of PAHs.
To further examine whether this strain has a constitutive PQR enzyme specific for the reduction of both PQ and PyQ, or different PQR isozymes showing different specificities towards PQ and PyQ, the PQR activity was inhibited by exogenous catechols (Table 2). Alizarin and 2,3-dihydroxynaphthalene produced no significant difference in the inhibitory effects on either PQR activity for reduction of PQ or PyQ, whereas anthrarobin and esculetin inhibited PQR activity for PyQ reduction more strongly than that for PQ reduction. Similarly, we recently found two constitutive PQR isozymes (PQR1 and PQR2) from another pyrene-degrading Mycobacterium sp., PYR100. PQR2 was specific for PyQ reduction and was more strongly inhibited by the exogenous catechol, quercetin [11]. This indicated that each of the PAH-degrading Mycobacterium spp. strains had at least two different PQR isozymes.
Southern hybridization and PCR screening for pqr1 and pqr2
A hypothesis that two quinone reductases are present in M. vanbaalenii PYR-1, homologous to proteins PQR1 and PQR2 found in Mycobacterium sp. PYR-100 [11], was tested using Southern hybridization. Under several stringency conditions, multiple nonspecific hybridization bands were present in XbaI-digested total genomic DNA of M. vanbaalenii PYR-1. The same situation was observed in Mycobacterium sp. PYR-100. This is understandable, since probes PQR1 and PQR2 were degenerate. Thus, the presence of PQR1 or PQR2 homologues could be neither confirmed nor excluded by this experiment. However, when screening for a PQR1 homologue in M. vanbaalenii PYR-1, the combination of primers PQR1f and PQR1r2 yielded a PCR product with the expected size of 300 bp. The DNA sequence of this PCR product was obtained (GenBank accession number AY426337). Comparison with other entries in the GenBank database using the program BlastX revealed that, on the translated peptide level, this sequence is 67% identical to the conserved hypothetical protein Rv3054c from M. tuberculosis and 62% identical to a putative secreted protein from Streptomyces avermitilis. Some of the lower homology matches included 42% identity to a chromate reductase (ChrR) from Pseudomonas putida, 43% identity to an NADPH:quinone oxidoreductase (mll4710) from Mesorhizobium loti, and 44% identity to a predicted flavoprotein gene (Reut2486) from Ralstonia metallidurans. An RPS-Blast search of a conserved domains database shows that the sequence AY426337 partially covers the conserved domain of NADPH-dependent FMN reductases [GenBank Consolidated Domain Database (CDD) 7037]. Although some of the combinations of primers for PQR2 yielded PCR products of expected size, these bands were always minor. Thus, no unambiguous DNA sequence was obtained for the PQR2-like gene.
COMT activity of M. vanbaalenii PYR-1
The use of a PAH as sole carbon and energy source did not significantly change the expression level of COMT. As seen in Table 3, M. vanbaalenii PYR-1 appeared to have a constitutive COMT activity. Among the exogenous catechol substrates tested, anthrarobin (1,2,10-anthracenetriol) showed the highest COMT activity.
In contrast, the COMT activity towards alizarin (1,2-dihydroxyanthraquinone), 2,3-dihydroxynaphthalene, and esculetin (6,7-dihydroxycoumarin) was less than the activity for the O-methylation of anthrarobin. Although alizarin seems to be structurally analogous to anthrarobin and 1,2-dihydroxyanthracene, the O-methylation of alizarin occurred more slowly. This may be because the hydroxyl groups of alizarin can be highly delocalized at the C1, C2, C9 and C10 positions.
Identification of O-methylated phenanthrene and pyrene metabolites
The HPLC profile of a tC18-solid phase extract of the cell extract from phenanthrene degradation showed two major products. The DEP/EI mass spectrum of the compound eluting at 19.2 min with UV absorption peaks (λmax) of 211, 246 (shoulder), 254 nm and a valley of 230 nm, had a base peak molecular ion [M+.] at m/z 224. Significant fragment ions included m/z 209 [M–CH3]+ (78), 181 [M–CH3-CO]+ (38), and 152 [M–CH3OCCOH]+ (32). The mass spectrum was consistent with a hydroxymethoxy-phenanthrene. The NMR spectrum (Fig. 4a) consisted of six doublets, two triplets and one singlet. Homonuclear decoupling and NOE experiments, used to make resonance assignments, showed a methoxyl group at C1 and a hydroxyl group at C2. The NMR assignments, with the proton chemical shifts (δ, ppm) and coupling constants (JH,H, Hz), are 3.96 (H1), 7.33 (H3; J3,4=9.2), 8.45 (H4), 8.67 (H5; J5,6=8.6), 7.62 (H6; J6,7=7.7, J6,8=1.5), 7.53 (H7; J7,8=7.7), 7.91 (H8), 7.79 (H9; J9,10=9.2) and 8.00 (H10). This compound was identified as 1-methoxy-2-hydroxyphenanthrene.
A compound eluting at 23 min had UV absorption peaks of 215 and 259 nm and a valley at 235 nm. The DEP/EI mass spectrum had a base peak, [M+.], at m/z 238. Significant fragment ions included m/z 223 [M–CH3]+ (39), 195 [M–CH3–CO] + (23), 180 (22), 177 [C14H9]+ (21), and 152 [C12H18] + (29). The mass spectrum was consistent with a dimethoxyphenanthrene. The NMR spectrum (Fig. 4b) was similar to that of the 19.2-min peak but had one additional singlet. The sharp singlet peaks at 3.98 ppm and 4.02 ppm were characteristic of two methoxyl groups. NOE and homonuclear decoupling experiments indicated that the methoxyl groups were at C1 and C2. NMR assignments with proton chemical shifts (δ, ppm) and coupling constants (JH,H, Hz) are: 3.98 (H1), 4.02 (H2), 7.52 (H3; J3,4=9.0), 8.53 (H4), 8.69 (H5; J5,6=8.2, J5,7=1.3), 7.63 (H6; J6,7=7.7, J6,8=1.5), 7.56 (H7; J7,8=8.4), 7.91 (H8), 7.78 (H9; J9,10=9.5) and 8.06 (H10). Therefore, this compound was identified as 1,2-dimethoxyphenanthrene.
M. vanbaalenii PYR-1 also produced O-methylated products from pyrene and 1-hydroxypyrene. The extracted ion chromatograms of an extract from the incubation of 1-hydroxypyrene are shown in Fig. 5.
The compound eluting at 12.7 min had a molecular ion at m/z 232 [M+.] and fragment ions at m/z 217 [M–CH3]+ and m/z 189 [M–CH3–CO]+, which are consistent with 1-methoxypyrene (Fig. 6a).
The EI mass spectra of compounds eluting at 14.4 and 15.7 min both had molecular ions [M+.] at m/z 248. Significant fragment ions at m/z 233 [M–CH3]+, 205 [M–CH3-CO]+ and 176 [M–HOCCOCH3]+ were observed. These ions are consistent with either 1-hydroxy-2-methoxypyrene or 2-hydroxy-1-methoxypyrene (Fig. 6b,c).
The molecular ion for the compound eluting at 14.8 min was at m/z 262 with a relative intensity of 96%. Significant fragment ions included m/z 247 [M–CH3]+, 232 [M-CH3–CO]+ and 176. These ions are consistent with 1,2-dimethoxypyrene (Fig. 6d). The peak at tR=13.1 min is diisooctyl phthalate.
Discussion
M. vanbaalenii PYR-1 uses two possible pathways to produce PAH-dihydrodiols [6]. An NADH-linked dioxygenase is stereo- and regio-specific for PAH-cis-dihydrodiol production. In contrast, an NAD(P)H-linked cytochrome P450 monooxygenase seems to have less stereo- and regio-specificity than the dioxygenase. P450s can be involved in the production of various PAH epoxides and PAH phenols from PAH degradation. PAH epoxides are hydrolyzed to PAH-trans-dihydrodiols by an epoxide hydrolase, and the corresponding PAH catechols are produced by dihydrodiol dehydrogenase activity. The proportions of the resulting PAH catechols vary with different hydrocarbons [21]. K-Region PAH catechols of phenanthrene and pyrene, including the 9,10-dihydroxyphenanthrene and 4,5-dihydroxypyrene that are produced by M. vanbaalenii PYR-1, are oxidized by ortho-cleavage, resulting in formation of dicarboxylic acids [6, 15]. If non-K- and non-bay-region PAH catechols remain in the cell for a prolonged time, cytotoxicity and genotoxicity may be increased by the one-electron redox recycling generating ROS, DNA strand scission, DNA adduct formation and malondialdehyde generation [1, 22]. In particular, PAH catechols are likely to inhibit NADH-linked PQR. PQR is specific for the reduction of various PAH o-quinones, including PQ and PyQ. We found a constitutive PQR enzyme specific for the reduction of PQ and PyQ in M. vanbaalenii PYR-1. This enzyme activity towards PyQ was strongly inhibited by exogenous catechol inhibitors such as anthrarobin and esculetin.
With a PCR approach, we found the DNA sequence AY426337, which could be a part of the gene, or of one of the genes, coding for PQR activity. This sequence is homologous to a gene coding for the quinone reductase PQR1 in Mycobacterium sp. PYR-100. When translated, sequence AY426337 contains an FMN-reductase domain, consistent with the observation that PQR1 in strain PYR-100 contains an FAD cofactor. Although strain PYR-100 also contains a second quinone reductase, PQR2, DNA evidence for its counterpart in M. vanbaalenii PYR-1 was limited and requires future investigation.
O-Methylation, catalyzed by mammalian COMT, plays an important role in deactivating recalcitrant PAH catechols into less reactive forms. The O-methylated products then act as hydroxyl radical (·OH) scavengers to stop metal ion (e.g., Fe2+/Fe3+ and Cu+/Cu2+)-mediated Fenton chemistry [18, 19]. Wunder et al. [29] found that the fungus Penicillium glabrum strain TW 9424 metabolized pyrene and 1-hydroxypyrene to 1-methoxypyrene and 1,6-dimethoxypyrene by COMT activity. Narro et al. [20] reported that the O-methylation of hydroxyphenanthrene derived from phenanthrene degradation by a marine cyanobacterium, Agmenellum quadruplicatum PR-6, significantly reduced the toxicity.
M. vanbaalenii PYR-1 has a COMT that catalyzes the conversion of potentially toxic catechol compounds to less active O-methylated forms. The COMT activity showed higher activity against anthrarobin than the other exogenous catechol inhibitors tested. A structurally similar compound, 1,2-dihydroxyanthracene, is a transient intermediate from anthracene degradation that is converted to 1-methoxy-2-hydroxyanthracene by the same strain [15]. This result is similar to those of previous studies in that this strain can produce 1-methoxy-2-hydroxyanthracene and 7-methoxy-8-hydroxyfluoranthene as dead-end products from the degradation of anthracene and fluoranthene [9, 15].
Even though phenanthrene and pyrene served as sole carbon and energy sources for degradation by M. vanbaalenii PYR-1, a variety of O-methylated products were found in the cytosol. The corresponding PAH catechols formed via non-K-region PAH- dihydrodiols served as precursors for the O-methylation reactions mediated by COMT activity.
In contrast, K-region catechols PAH (e.g., 9,10-dihydroxyphenanthrene) and bay-region PAH catechols (e.g., 3,4-dihydroxyphenanthrene) can be further metabolized by the intradiol catechol dioxygenase activity of M. vanbaalenii PYR-1 [3, 15], although high concentrations allow rapid accumulation of the corresponding PAH o-quinones that can produce acute toxic effects [7].
In this paper, we provide evidence that M. vanbaalenii PYR-1 has constitutive COMT and PQR enzymes that inactivate PAH catechols derived from PAH degradation pathways. The COMT catalyzes two O-methylation reactions with the PAH catechols to subsequently form monomethoxyhydroxy-PAHs and dimethoxy-PAHs. The COMT activity is a detoxification process for non-K- and non-bay-region PAH catechols, which do not serve as carbon and energy sources. Quinone reduction by M. vanbaalenii PYR-1 PQRs may play an eminent role to protect the cells against cytotoxicity of PAH o-quinones and to supply PAH catechols for further degradation.
References
Bolton JL, Michael MA, Penning TM, Dryhurst G, Monks TJ (2000) Role of quinones in toxicity. Chem Res Toxicol 13:135–160
Brezna B, Khan AA, Cerniglia CE (2003) Molecular characterization of dioxygenases from polycyclic aromatic hydrocarbon-degrading Mycobacterium spp. FEMS Microbiol Lett 223:177–183
Cerniglia CE (1993) Biodegradation of polycyclic aromatic hydrocarbons. In: Rosenberg E (ed) Microorganisms to combat pollution. Kluwer, Dordrecht, pp 227–244
Cheung P, Kinkle B (2001) Mycobacterium diversity and pyrene mineralization in petroleum-contaminated soils. Appl Environ Microbiol 67:2222–2229
Heitkamp MA, Franklin W, Cerniglia CE (1988) Microbial metabolism of polycyclic aromatic hydrocarbons: isolation and characterization of a pyrene-degrading bacterium. Appl Environ Microbiol 54:2549–2555
Heitkamp MA, JP Freeman, DW Miller, Cerniglia CE (1988) Pyrene degradation by a Mycobacterium sp.: identification of ring oxidation and ring fission products. Appl Environ Microbiol 54:2556–2565
Kazunga C, Aitken MD (2000) Products from the incomplete metabolism of pyrene by polycyclic aromatic hydrocarbon-degrading bacteria. Appl Environ Microbiol 66:1917–1922
Kelley I, Cerniglia CE (1995) Degradation of a mixture of high-molecular-weight polycyclic aromatic hydrocarbons by a Mycobacterium strain PYR-1. J Soil Contam 4:44–91
Kelley I, Freeman JP, Evans FE, Cerniglia CE (1993) Identification of metabolites from the degradation of fluoranthene by Mycobacterium sp. strain PYR-1. Appl Environ Microbiol 59:800–806
Khan AA, Kim S-J, Paine DD, Cerniglia CE (2002) Classification of a polycyclic aromatic hydrocarbon-metabolizing bacterium, Mycobacterium sp. strain PYR-1, as Mycobacterium vanbaalenii sp. nov. Int J Syst Evol Microbiol 52:1997–2002
Kim Y-H, Engesser K-H, Cerniglia CE (2003) Two polycyclic aromatic hydrocarbon o-quinone reductases from a pyrene-degrading Mycobacterium. Arch Biochem Biophys 416:209–217
Kulakov L, Allen CCR, Lipscomb DA, Larkin MJ (2000) Cloning and characterization of a novel cis-napthalene dihydrodiol dehydrogenase gene (narB) from Rhodococcus sp. NCIMB12038. FEMS Microbiol Lett 182:327–331
McLellan S, Warshawsky D, Shann J (2002) The effect of polycyclic aromatic hydrocarbons on the degradation of benzo[a]pyrene by Mycobacterium sp. strain RJGII-135. Environ Toxicol Chem 21:253–259
Molina M, Araujo R, Hodson R (1999) Cross-induction of pyrene and phenanthrene in a Mycobacterium sp. isolated from polycyclic aromatic hydrocarbon contaminated river sediments. Can J Microbiol 45:520–529
Moody JD, Freeman JP, Doerge DR, Cerniglia CE (2001) Degradation of phenanthrene and anthracene by cell suspensions of Mycobacterium sp. strain PYR-1. Appl Environ Microbiol 67:1476–1483
Moody JD, Fu PP, Freeman JP, Cerniglia CE (2003) Regio- and stereoselective metabolism of 7,12-dimethylbenz[a]anthracene by Mycobacterium vanbaalenii PYR-1. Appl Environ Microbiol 69:3924–3931
Moody JD, Freeman JP, Fu PP, Cerniglia CE (2004) Degradation of benzo[a]pyrene by Mycobacterium vanbaalenii PYR-1. Appl Environ Microbiol 70:340–345
Nappi AJ, Vass E (1998) Hydroxyl radical formation via iron-mediated Fenton chemistry is inhibited by methylated catechols. Biochim Biophys Acta 1425:159–167
Nappi AJ, Vass E, Collins MA (1999) Contrasting effects of catecholic and O-methylated tetrahydroisoquinolines on hydroxyl radical production. Biochim Biophys Acta 1434:64–73
Narro ML, Cerniglia CE, Van Baalen C, Gibson DT (1992) Metabolism of phenanthrene by the marine cyanobacterium Agmenellum quadruplicatum PR-6. Appl Environ Microbiol 58:1351–1359
Pangrekar J, Kandaswami C, Kole P, Kumar S, Sikka HC (1995) Comparative metabolism of benzo[a]pyrene, chrysene and phenanthrene by brown bullhead liver microsomes. Mar Environ Res 39:51–55
Penning TM (1993) Dihydrodiol dehydrogenase and its role in polycyclic aromatic hydrocarbon metabolism. Chem Biol Interact 89:1–34
Rehmann K, Noll H, Steinberg C, Kettrup A (1998) Pyrene degradation by Mycobacterium sp. strain KR2. Chemosphere 36:2977–2992
Rehmann K, Hertkorn N, Kettrup A (2001) Fluoranthene metabolism in Mycobacterium sp. strain KR20: identification of pathway intermediates during degradation and growth. Microbiology 147:2783–2794
Sambrook J, Russell DW (2001) Molecular cloning, a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
Schneider J, Grosser R, Jayasimhulu K, Xue W, Warshawsky D (1996) Degradation of pyrene, benz[a]anthracene, and benzo[a]pyrene by Mycobacterium sp. strain RJGII-135, isolated from a former coal gasification site. Appl Environ Microbiol 62:13–19
Vila J, López Z, Sabaté J, Minguillón C, Solanas AM, Grifoll M (2001) Identification of a novel metabolite in the degradation of pyrene by Mycobacterium sp. strain AP1: actions of the isolate on two- and three-ring polycyclic aromatic hydrocarbons. Appl Environ Microbiol 67:5497–5505
Willumsen P, Karlson U, Stackebrandt E, Kroppenstedt R (2001) Mycobacterium frederiksbergense sp. nov., a novel polycyclic aromatic hydrocarbon-degrading Mycobacterium species. Int J Syst Evol Microbiol 51:1715–1722
Wunder T, Marr J, Kremer S, Sterner O, Anke H (1997) 1-Methoxypyrene and 1,6-methoxypyrene: two novel metabolites in fungal metabolism of polycyclic aromatic hydrocarbons. Arch Microbiol 167:310–316
Zhu BT, Conney AH (1998) Is 2-methoxyestradiol an endogenous estrogen metabolite that inhibits mammary carcinogenesis? Cancer Res 58:2269–2277
Zhu BT, Conney AH (1998) Functional roles of estrogen metabolism in target cells: Review and perspectives. Carcinogenesis 19:1–27
Zhu BT, Patel UK, Cai MX, Conney AH (2000) O-Methylation of tea polyphenols catalyzed by human placental cytosolic catechol-O-methyltransferase. Drug Metab Dispos 28:1024–1030
Acknowledgements
The authors thank John B. Sutherland for critical review of the manuscript, and Diana Mathews for clerical assistance. This research was supported by the Postgraduate Research Participation Program at the National Center for Toxicological Research administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the United States Department of Energy and the Food and Drug Administration.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kim, YH., Moody, J.D., Freeman, J.P. et al. Evidence for the existence of PAH-quinone reductase and catechol-O-methyltransferase in Mycobacterium vanbaalenii PYR-1. J IND MICROBIOL BIOTECHNOL 31, 507–516 (2004). https://doi.org/10.1007/s10295-004-0178-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-004-0178-x