Abstract
Classification of malignant and benign pulmonary nodules is important for further treatment plan. The present work focuses on the classification of benign and malignant pulmonary nodules using support vector machine. The pulmonary nodules are segmented using a semi-automated technique, which requires only a seed point from the end user. Several shape-based, margin-based, and texture-based features are computed to represent the pulmonary nodules. A set of relevant features is determined for the efficient representation of nodules in the feature space. The proposed classification scheme is validated on a data set of 891 nodules of Lung Image Database Consortium and Image Database Resource Initiative public database. The proposed classification scheme is evaluated for three configurations such as configuration 1 (composite rank of malignancy “1” and “2” as benign and “4” and “5” as malignant), configuration 2 (composite rank of malignancy “1”,“2”, and “3” as benign and “4” and “5” as malignant), and configuration 3 (composite rank of malignancy “1” and “2” as benign and “3”,“4” and “5” as malignant). The performance of the classification is evaluated in terms of area (A z) under the receiver operating characteristic curve. The A z achieved by the proposed method for configuration-1, configuration-2, and configuration-3 are 0.9505, 0.8822, and 0.8488, respectively. The proposed method outperforms the most recent technique, which depends on the manual segmentation of pulmonary nodules by a trained radiologist.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Lung cancer accounts for the highest number of cancer-related deaths compared to any other cancer [1]. In the USA, an estimated number of new lung cancer cases is 2, 21, and 200 in 2015, accounting for about 13 % of all cancer’s diagnosed [1]. The estimated number of deaths due to lung cancer in the USA is 1, 58, and 040 in 2015, accounting for about 27 % of all cancer deaths [1]. Lung cancer is frequent for smokers as well as for non-smokers, and its causes remain unknown. At present, there is no effective way to prevent lung cancer except campaigning against smoking. Early diagnosis of lung cancer could improve 5-year survival rate from 15 to 80 % [9]. Several studies proved that screening program can substantially reduce the mortality rate due to lung cancer [9, 29]. The pulmonary nodules are potential manifestation of lung cancer [4]. The pulmonary nodules are blob-like structures with a diameter of 3 to 30 mm. Small nodules (diameter <1 cm) are difficult to access by standard needle biopsy. The biopsy procedure is more difficult for small nodules, located away from the chest wall. A classification scheme with minimal user intervention is required for the diagnosis of lung cancer.
Several features such as 2D shape-based [14], 3D shape-based [10], 3D texture-based [12], and combinations of 2D shape-based and texture-based [2, 11] features have been used for the classification of pulmonary nodules. No classification work is reported in the literature using the combination of 3D shape-based and 3D texture-based features. The aim of the present work is to improve the performance of classification of nodules using a combination of 2D shape-based, 3D shape-based, 3D margin-based, 2D texture-based, and 3D texture-based features. A semi-automated technique is used for segmentation of pulmonary nodules [7]. The proposed method provides the best classification accuracy using a set of 49 features. Support vector machine (SVM) is used for classification of benign and malignant nodules. The proposed method is compared with the most recent classification work of Han et al. [12]. The proposed classification scheme is also compared with the automated Han et al., where the pulmonary nodules are segmented using the method of Dhara et al. [7].
The paper is arranged as follows: “Prior Works on Classification of Pulmonary Nodules” section reviews the reported works in the literature, “Classification of Benign and Malignant Pulmonary Nodules” section describes the proposed classification framework, “Feature Selection” section describes the method of feature selection, and “Database” section describes the database. In “Results and Discussion” section, results of classification are stated.
Prior Works on Classification of Pulmonary Nodules
Several classifiers such as linear discriminant analysis (LDA) [2, 15, 20], artificial neural network (ANN) [19, 28], and SVM [12] have been used for the classification of benign and malignant pulmonary nodules. The reported classification works based on LDA, ANN, and SVM are reviewed in subsequent paragraphs.
McNitt-Gray et al. [20] used shape and texture features of a nodule slice and reported the classification accuracy of 90.3 % on a data set of 31 nodules using LDA. Armato et al. [2] also used several shape and texture features and classified the nodules using LDA. The method was validated on a data set of 470 nodules and reported A z is 0.79. Way et al. [31] developed a computer-aided diagnosis (CAD) system using morphological and gray-level features. A data set of 96 nodules was used and the reported A z is 0.83±0.04.
Matsuki et al. [19] considered seven clinical parameters and 16 radiological findings to categorize the pulmonary nodules into benign and malignant. The method was validated on a data set of 155 nodules using a three-layer, feed-forward ANN with a back-propagation algorithm. The performance of the ANN classifier was evaluated using the round-robin method of training and testing. The reported A z is 0.95. Suzuki et al. [28] applied massive training artificial neural network for the classification of benign and malignant nodules. Three gray-level-based features, two edge-based features, a morphological feature, and the clinical information were used to represent the pulmonary nodules. The data set consists of 76 malignant nodules and 413 benign nodules. They also evaluated the classifier performance using the round-robin method of training and testing, and the reported the A z of 0.88.
Han et al. [12] studied the role of three well-known texture features (Haralick, Gabor, and local binary patterns) in the classification of pulmonary nodules. They established the efficacy of 3D Haralick features as compared to the Gabor and local binary pattern-based features of a representative of the nodule. The method was validated on a data set of 1012 nodules of LIDC/IDRI. The highest classification accuracy was obtained by considering the nodules with composite rank of malignancy “1” and “2” as benign and “4” and “5” as malignant. They used SVM classifier and reported an A z of 0.94. This technique depends on radiologists for the segmentation of nodules.
Classification of Benign and Malignant Pulmonary Nodules
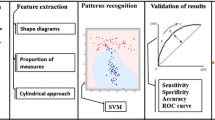
A cubic volume of interest (VOI) of size (40×40×40 mm) is selected considering the seed point as the centroid of the VOI. Pulmonary nodules are segmented using an established segmentation technique [7]. Several shape-based, margin-based, and texture-based features are computed from the segmented nodule. The selected features are used for the classification of nodules. The block diagram of the proposed classification scheme is provided in Fig. 1.
Segmentation of Pulmonary Nodules
Several works on segmentation based on morphological filtering [17, 18, 21] and active contour model [25] are reported in the literature. In this paper, pulmonary nodules are segmented using the method of Dhara et al. [7]. The segmentation technique is applicable for different types of pulmonary nodules based on their internal texture (viz, solid, part-solid, and non-solid) and external attachment (viz, juxta-pleural and juxta-vascular). In this segmentation technique, pulmonary nodules are classified into solid/part-solid or non-solid based on the analysis of intensity distribution in the core of nodule. Depending on the class level of nodules (solid/partsolid or non-solid), a particular algorithm is set for nodule segmentation.
The performance of the segmentation technique of Dhara et al. [7] was compared with the techniques of Kuhnigh et al. [17], Moltz et al. [21], and Kubota et al. [18] in terms of four contour-based metrics (mean distance, Pratt function, Hausdorff distance, and modified Hausdorff distance) and six region-based metrics (accuracy, overlap, sensitivity, specificity, similarity angle, and similarity region) [26] by mean of comparing the binary mask of the segmented nodule with the reference ground truth mask. The experimental results prove that the segmentation technique of Dhara et al. [7] is reliable for the segmentation of various types of pulmonary nodules with an improved accuracy.
Representation of Pulmonary Nodules
Malignant nodules have uneven surface due to the presence of spiculation and lobulation, whereas benign nodules have smooth surface (Fig. 2). The margin-sharpness of nodules and the gray scale distribution of nodules play an important role in discrimination of benign and malignant nodules. The imaging characteristics of nodules are represented in terms of several shape-based, margin-based, and texture-based features. The procedures of computing these features are described in subsequent sections.
3D Shape-Based Features
Several 3D shape-based features are computed from the binary mask of the segmented nodules as described below.
-
Sphericity: The irregularity of the shape of a nodule is represented by sphericity [27]
$$ \mathit{Sphericity} = 6 \sqrt{\pi}V{A}^{-3/2}, $$(1)where V is the volume of the nodule and A is its surface area. The volume of a nodule is computed by multiplying the total number of voxel with voxel resolution. The surface area of a nodule is computed by summing the area of all triangular faces of the nodule mesh. In this article, the nodule mesh is generated using marching cubes algorithm [16] whenever needed. The range of sphericity is [0,1].
-
Spiculation: The spiculation is computed using the method of Dhara et al. [8]. The height and base area of each spicule are determined. The net spiculation of a nodule is computed in two different ways
$$ \mathit{Spiculation}_{1} = {\Sigma}_{i = 1}^{N}{e^{- {\omega_{i}}}}{h_{\text{i}}}, $$(2)and,
$$ \mathit{Spiculation}_{2} = \frac{\sum\limits_{i=1}^{N}{h_{\text{i}}\cos{\omega_{i}}}}{\sum\limits_{i=1}^{N}{h_{\text{i}}}}, $$(3)where ω i is the solid angle subtended at peak point of i th spicule, h i is the height of a spicule, and N is the total number of spicules of a nodule under consideration. The nonlinear weighting factor is introduced to emphasize the contribution of sharp spicules.
-
Lobulation: Lobulated surface is associated with the uneven growth rate of nodules. Dhara et al. [8] represented the lobulation in terms of the ratio of total concave surface area and total convex surface area of the nodule mesh. Region growing is applied to get the concave and convex part of the nodule mesh. The lobulation of a nodule is defined as
$$ Lobulation = \frac{{{S_{\text{concave}}}}}{{{S_{\text{convex}}}}}, $$(4)where S convex is the total convex surface area and S concave is the total concave surface area of the nodule.
-
Volume: Volume of a nodule is computed by multiplying the total number of voxels with voxel resolution [8].
-
Surface area: Surface area of a nodule is computed by summing the area of all triangular faces of the nodule mesh.
-
Equivalent diameter 3D: It is defined as [27]
$$ D = 2\sqrt[3]{{\frac{{3V}}{{4\pi }}}}, $$(5)where V is the volume of the nodule.
-
Major axis length 3D: The major axis length of the bounding ellipsoid of a nodule [21].
-
Minor axis length 3D: The minor axis length of the bounding ellipsoid of a nodule [21].
-
Convex surface area: It is computed by summing the area of all triangular faces in the equivalent triangular mesh (generated by marching cubes algorithm [16]) of the convex hull of a nodule.
2D Shape-Based Features
Several 2D shape-based features are computed from the biggest axial slice (S biggest) of each nodule [24]. The procedures of computing these features are described below.
-
Area: The number of pixel in S biggest multiplied by pixel resolution.
-
Perimeter: The perimeter of S biggest multiplied by pixel resolution.
-
Equivalent diameter 2D: It is the equivalent diameter of S biggest and represented as \(\frac {2}{\pi }\sqrt {Area}\)
-
Convex area: The area of convex hull of S biggest
-
Convex perimeter: The perimeter of the convex hull of S biggest
-
Compactness: It is defined as \(\frac {{Perimeter{^{2}}}}{{4\pi Area}}\)
-
Major axis length: The major axis length of the bounding ellipse of S biggest
-
Minor axis length: The minor axis length of the bounding ellipse of S biggest
-
Circularity: The circularity is defined as
$$ Circularity = \frac{{4\pi A}}{{{q^{2}}}}, $$(6)where A is the area of S biggest and q is the convex perimeter of S biggest.
-
Standard deviation of radial distance 2D: The standard deviation of radial distances of all boundary pixels S biggest.
Margin-Based Features
Margin sharpness is a measure of the difference in the intensity of the boundary region of a nodule and its surroundings. Margin sharpness is represented using the combination of acutance and histogram spread of averaged gradient of the nodule. The averaged gradient for K th boundary normal at a nodule slice is represented as [23]
where f(k) and b(k) are part of normal boundary, located in the foreground and background of the nodule. The acutance of the nodule is defined as
where d max is the maximum value of averaged gradient computed at all boundary pixel of the nodule. The average gradient of all boundary pixels for all the slices containing the nodule is denoted by a vector named N_gradient. The dimension of N_gradient is (P×1), where P is the total number of boundary pixel in all image slices containing the nodule. Histogram spread of average gradient (HSAG) of nodule is defined as
The histogram spread was introduced by Tripathi et al. [30]. The range of histogram spread is (0,1].
2D Texture-Based Features
Six Haralick features [13] (viz, entropy, energy, inverse difference moment, sum entropy, difference entropy, and contrast) are computed from the biggest representative slice of the segmented nodule. Each combinations of directions (0,45,90,135 degree) and distances (1,2,3,4 pixels) generate a separate gray-level co-occurrence matrix (GLCM). Eleven Haralick features are calculated from each GLCM [24]. The mean of each Haralick feature computed from several GLCMs is used in the proposed work. Histogram of oriented gradient (HOG) [5] features is computed from the biggest representative slice of the segmented nodule. The mean, variance, and standard deviation of HOG features are used in the proposed classification scheme.
3D Texture-Based Features
Han et al. [12] introduced 3D Haralick features. They reported that the Haralick features considering nine directions (Fig. 3) provide better classification performance than five directions. In the proposed method, GLCM was computed for each nodule considering nine directions as described Fig. 3. Fourteen Haralick features [13] were computed from each GLCM matrix. The maximum correlation coefficient is not considered in our experiment, as it is computationally expensive [12]. The mean and the range of each Haralick feature over the nine directions is computed as
where H ik is the k th Haralick feature considering the i th direction. Total number of 3D texture features is 26.
Feature Selection
The performance of the proposed classification scheme can be improved by selecting a set of relevant features. The relevant features are selected by considering A z and p values. The A z is computed using ROCKIT [22], and p values is computed by performing 2-tailed Student’s t test. The list of selected features is provided in “Results of Feature Selection”.
Database
The proposed classification scheme and the competing technique are evaluated on a data set of 891 nodules of Lung Image Database Consortium and Image Database Resource Initiative (LIDC/IDRI) public database [3]. Each slice of lung CT images consists of a matrix of 512×512 pixels. The pixel resolution range from 0.5 to 0.8 mm. The mean ± standard deviation of volume of nodules is 855.48±1545.10 mm3. The meam ± standard deviation of number of slices comprise nodules is 6.44±4.84. In LIDC/IDRI, the nodules with texture index 1 are considered as non-solid, texture index 2 or 3 are considered as part-solid and texture index 4 or 5 are considered as solid. The result of biopsy is not provided for pulmonary nodules. A group of four radiologists have provided the rank of malignancy of each nodule on a scale of 1:5, where rank 1 indicates the least chance of malignancy and rank 5 indicates the highest chance for malignancy. The composite rank of malignancy is the mode of malignancy rank of four radiologists [6]. The composition of nodules based on the composite rank of malignancy is provided in Table 1. Han et al. [12] categorized the pulmonary nodules into benign and malignant, based on the composite rank of malignancy. They considered three different configurations (see Table 2) for the evaluation of their CAD system. The same configurations are used for the evaluation of proposed classification scheme.
Results and Discussion
Results of Feature Selection
Total number of features computed from the segmented nodule is 56. The A z and p values of these features are provided in Table 3. Out of 56 features, A z is ≥0.60 and p value is less than 0.05 for 49 features (rank 1 to rank 50 excluding feature of rank 8 in Table 3). These 49 features are used in the proposed classification scheme. The spiculation of nodule is an important feature as it provides A z of 0.78 using Eq. 2. However, the computation of spiculation using Eq. 2 is very sensitive to segmentation error. Hence, spiculation is computed using Eq. 3, as the value of spiculation is more reproducible using Eq. 3.
Results of Classification
SVM is used for the classification of benign and malignant nodules. Separate training and test data sets are constructed using five-fold cross-validation approach. The training and testing are performed over 100 times for each configuration. The mean A z for a particular configuration is obtained from the set of 100 classification outcomes. The proposed method is compared with the method of Han et al., which depends on the manual segmentation of pulmonary nodules by a trained radiologist. The method of Han et al. is automated using the segmentation technique of Dhara et al. and named as automated Han et al. The average value of A z for the proposed method, the method of Han et al., and automated Han et al. are provided in Table 4 for different configurations. The ROC plots for different configurations are provided in Fig. 4. The results in Table 4 depict that the proposed method outperforms automated Han et al. as well as the method of Han et al.
The method of Han et al. makes use of the segmented nodule provided by the experienced radiologists. On the other hand, the proposed technique and automated Han et al. use an established segmentation technique and they are affected by the segmentation error. Hence, the method of Han et al. outperforms automated Han et al. though both of them use the same set of features. The proposed classification scheme considers several 2D shape-based, 3D shape-based, margin-based, and texture-based features of nodules, whereas the method of Han et al. uses only texture-based features. Due to the inclusion of more relevant features, the proposed classification scheme outperforms automated Han et al. and the method of Han et al.
The results in Table 4 depict that the classification accuracy is higher in configuration 1 as compared to configuration 2 and configuration 3 due to the absence of uncertain nodules in configuration 1. The classification accuracy in configuration 2 is higher than configuration 3, and the result is consistent with the work of Han et al. The data suggests that nodules with composite rank of malignancy “3” have much similarity towards benign category. The sensitivity and specificity at the best operating point (a point on the ROC curve nearest to the point [0,1]) are provided in Table 5. The results depict that the sensitivity and specificity of the proposed method are slightly better compared to automated Han et al. and the method of Han et al. for three configurations.
Conclusion
We have developed a classification scheme for differentiation of benign and malignant pulmonary nodules. The classification scheme could be operated with minimal user intervention and could assists radiologists in the diagnosis of lung cancer. While the most recent technique depends on the user for nodule segmentation, the proposed system would be fast and easy to use as the radiologists need to provide only a seed point for segmentation of nodules. Several shape-based, margin-based, and texture-based features are analyzed to improve the accuracy of classification. An optimal feature set is determined for efficient representation of nodules in multidimensional feature space. The proposed classification scheme outperforms the competing techniques for all configurations. Further work is required to improve the performance of classification. To improve the performance, more research work is required on segmentation of pulmonary nodules and improvement of feature set for proper representation of nodules in the feature space.
References
American Cancer Society, Cancer facts and figure 2015
Armato III SG, Altman MB, Wilkie J, Sone S, Li F, Doi K, Roy AS: Automated lung nodule classification following automated nodule detection on CT: a serial approach. Med Phys 30(6):1188–1197, 2003
Armato III SG, McLennan G, Bidaut L, McNitt-Gray MF, Meyer CR, Reeves AP, Zhao B, Aberle DR, Henschke CI, Hoffman EA, Kazerooni EA, MacMahon H, Beek EJR, Yankelevitz D, Biancardi AM, Bland PH, Brown MS, Engelmann RM, Laderach GE, Max D, Pais RC, Qing DPY, Roberts RY, Smith AR, Starkey A, Batra P, Caligiuri P, Farooqi A, Gladish GW, Jude CM, Munden RF, Petkovska I, Quint LE, Schwartz LH, Sundaram B, Dodd LE, Fenimore C, Gur D, Petrick N, Freymann J, Kirby J, Hughes B, Casteele AV, Gupte S, Sallam M, Heath MD, Kuhn MH, Dharaiya E, Burns R, Fryd DS, Salganicoff M, Anand V, Shreter U, Vastagh S, Croft BY, Clarke LP: The lung image database consortium (LIDC) and image database resource initiative (IDRI): a completed reference database of lung nodules on CT scans. Med Phys 38(2):915–931, 2011
Austin J, Müller N, Friedman PJ, Hansell DM, Naidich DP, Remy-Jardin M, Webb WR, Zerhouni EA: Glossary of terms for CT of the lungs: recommendations of the nomenclature committee of the fleischner society. Radiology 200(2):327–331, 1996
Dalal N, Triggs B, Schmid C: Human detection using oriented histograms of flow and appearance. Computer Vision–ECCV 2006. Springer, 2006, pp 428–441
Dasovich GM, Kim R, Raicu DS, Furst JD: A model for the relationship between semantic and content based similarity using LIDC. Proceedings of SPIE Medical Imaging, 2010, pp 762, 431–762, 431–10
Dhara AK, Mukhopadhyay S, Das Gupta R, Garg M, Khandelwal N: A segmentation framework of pulmonary nodules in lung CT images. J Digit Imaging, 2015. doi:10.1007/s10278-015-9812-6
Dhara AK, Mukhopadhyay S, Saha P, Garg M, Khandelwal N: Differential geometry-based techniques for characterization of boundary roughness of pulmonary nodules in CT images. Int J Comput Assist Radiol Surg, 2015. doi:10.1007/s11548-015-1284-0
Diederich S, Wormanns D, Semik M, Thomas M, Lenzen H, Roos N, Heindel W: Screening for early lung cancer with low-dose spiral CT: Prevalence in 817 asymptomatic smokers. Radiology 222(3):773–781, 2002
El-Baz A, Nitzken M, Vanbogaert E, Gimel’farb G, Falk R, El-Ghar MA: A novel shape-based diagnostic approach for early diagnosis of lung nodules. Proceedings of IEEE International Symposium on Biomedical Imaging: From Nano to Macro, 2011, pp 137–140
Elizabeth D, Nehemiah H, Raj CR, Kannan A: Computer-aided diagnosis of lung cancer based on analysis of the significant slice of chest computed tomography image. IET Image Process 6(6):697–705, 2012
Han F, Wang H, Zhang G, Han H, Song B, Li L, Moore W, Lu H, Zhao H, Liang Z: Texture feature analysis for computer-aided diagnosis on pulmonary nodules. J Digit Imaging 28(1):99–115, 2014
Haralick RM, Shanmugam K, Dinstein IH: Textural features for image classification. IEEE Trans Syst Man Cybern 6:610–621, 1973
Iwano S, Nakamura T, Kamioka Y, Ishigaki T: Computer-aided diagnosis: a shape classification of pulmonary nodules imaged by high-resolution CT. Comput Med Imaging Graph 29(7):565–570, 2005
Li F, Aoyama M, Shiraishi J, Abe H, Li Q, Suzuki K, Engelmann R, Sone S, MacMahon H, Doi K: Radiologists’ performance for differentiating benign from malignant lung nodules on high-resolution CT using computer-estimated likelihood of malignancy. Am J Roentgenol 183(5):1209– 1215, 2004
Lorensen WE, Cline HE: Marching cubes: a high resolution 3D surface construction algorithm. ACM Siggraph Comput Graph 21(4):163–169, 1987
Kuhnigk JM, Dicken V, Bornemann L, Bakai A, Wormanns D, Krass S, Peitgen HO: Morphological segmentation and partial volume analysis for volumetry of solid pulmonary lesions in thoracic CT scans. IEEE Trans Med Imaging 25(4):417–434, 2006
Kubota T, Jerebko AK, Dewan M, Salganicoff M, Krishnan A: Segmentation of pulmonary nodules of various densities with morphological approaches and convexity models. Med Image Anal 15(1):133–154, 2011
Matsuki Y, Nakamura K, Watanabe H, Aoki T, Nakata H, Katsuragawa S, Doi K: Usefulness of an artificial neural network for differentiating benign from malignant pulmonary nodules on high-resolution CT: evaluation with receiver operating characteristic analysis. Am J Roentgenol 178(3):657–663, 2002
McNitt-Gray MF, Hart EM, Wyckoff N, Sayre JW, Goldin JG, Aberle DR: A pattern classification approach to characterizing solitary pulmonary nodules imaged on high resolution ct: preliminary results. Med Phys 26(6):880–888, 1999
Moltz JH, Kuhnigk JM, Bornemann L, Peitgen H: Segmentation of juxtapleural lung nodules in ct scan based on ellipsoid approximation. Proceedings of First International Workshop on Pulmonary Image Processing. New York, 2008, pp 25–32
Noessner J, Niepert M, Stuckenschmidt H: Rockit: Exploiting parallelism and symmetry for map inference in statistical relational models. arXiv:1304.4379, 2013
Rangayyan RM, El-Faramawy NM, Desautels JL, Alim OA: Measures of acutance and shape for classification of breast tumors. IEEE Trans Med Imaging 16(6):799–810, 1997
Seitz KA, Giuca AM, Furst J, Raicu D: Learning lung nodule similarity using a genetic algorithm. Proceedings of SPIE Medical Imaging 2012. San Deigo, USA, 2012, pp 831,537–831,537–7
Saini K, Dewal ML, Manojkumar R: A fast region-based active contour model for boundary detection of echocardiographic images. J Digit Imaging 25(2):271–278, 2012
Silva JS, Santos JB, Roxo D, Martins P, Castela E, Martins R: Algorithm versus physicians variability evaluation in the cardiac chambers extraction. IEEE Trans Inf Technol Biomed 16(5):835–841, 2012
Sladoje N, Nyström I, Saha PK: Measurements of digitized objects with fuzzy borders in 2D and 3D. Image Vis Comput 23(2):123–132, 2005
Suzuki K, Li F, Sone S, et al.: Computer-aided diagnostic scheme for distinction between benign and malignant nodules in thoracic low-dose CT by use of massive training artificial neural network. IEEE Trans Med Imaging 24(9):1138– 1150, 2005
Swensen SJ, Jett JR, Sloan JA, Midthun DE, Hartman TE, Sykes AM, Aughenbaugh GL, Zink FE, Hillman SL, Noetzel GR, et al.: Screening for lung cancer with low-dose spiral computed tomography. Am J Respir Crit Care Med 165(4):508–513, 2002
Tripathi AK, Mukhopadhyay S, Dhara AK: Performance metrics for image contrast. Proceedings of IEEE International Conference on Image Information Processing. India: Simla, 2011, pp 1–4
Way TW, Hadjiiski LM, Sahiner B, Chan HP, Cascade PN, Kazerooni EA, Bogot N, Zhou C: Computer-aided diagnosis of pulmonary nodules on ct scans: segmentation and classification using 3d active contours. Med Phys 33(7):2323–2337, 2006
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
This work is done using a public lung CT image data set and for this type of study formal consent is not required. This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interests
This study was funded by Department of Electronics Technology, Govt. of India, Grant number 1(3)2009-ME&TMD and 1(2)/2013-ME &TMD/ESDA, respectively. The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Dhara, A.K., Mukhopadhyay, S., Dutta, A. et al. A Combination of Shape and Texture Features for Classification of Pulmonary Nodules in Lung CT Images. J Digit Imaging 29, 466–475 (2016). https://doi.org/10.1007/s10278-015-9857-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-015-9857-6