Abstract
Fertilization is comprised of two sequential fusion processes; plasmogamy and karyogamy. Karyogamy completes with migration and fusion of the male and female nuclei in the fused cell. In animals, microtubules organized by the centrosome control female/male pronuclei migration. In contrast, the nuclear migration in fused gametes of angiosperms is controlled by actin filaments, but the mechanism that regulates actin filament-dependent nuclear migration is not clear. In this study, we prepared fused rice (Oryza sativa L.) gametes/zygotes using in vitro fertilization and observed the spatial and temporal movements of actin filaments and sperm nuclei. Our results show that actin filaments in egg cells form a meshwork structure surrounding the nuclei. Quantitative analysis of the actin meshwork dynamics suggests that actin meshwork converges toward the egg nucleus. In egg cells fused with sperm cells, actin filaments appeared to interact with a portion of the sperm nuclear membrane. The velocity of the actin filaments was positively correlated with the velocity of the sperm nucleus during karyogamy. These results suggest that sperm nuclear membrane and actin filaments physically interact with each other during karyogamy, and that the sperm nucleus migrates toward the egg nucleus through the convergence of the actin meshwork. Interestingly, actin filament velocity increased promptly after gamete fusion and was further elevated during nuclear fusion. In addition to the migration of gamete nuclei, convergence of actin meshwork may also be critical during early zygotic developments.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In fused eggs/zygotes of most animals, the movement of male and female pronuclei towards each other is essential for zygote formation (Wilson 1928). The pronuclear congression depends on the function of a microtubule aster (Chambers 1939; Schatten and Schatten 1981). The male pronucleus is associated with a centrosome, which produces microtubules to form a sperm aster (Reinsch and Gönczy 1998; Schatten 1994). Depending on the growth of the sperm aster, the centrosome and the associated male pronucleus migrate from the cell cortex toward the center of the fused egg. This microtubule-dependent migration of the male pronuclei is known to be driven by the activities of dynein and dynactin (Payne et al. 2003; Wühr et al. 2009, 2010).
The fusion of two haploid nuclei during yeast mating is an intensively investigated karyogamic event (Gibeaux et al. 2013; Kurihara et al. 1994). In budding yeast, kinesin motor Kar3, which exhibits microtubule minus end-directed non-processive motor activity (Endow et al. 1994), is required to bring the two nuclei together prior to nuclear fusion (Rose 1996).
In angiosperms, two sperm cells are delivered into an embryo sac through a pollen tube and fuse with the two female gametes, an egg cell and a central cell. Through such fusion processes called double fertilization, one sperm cell fuses with the egg cell, and the resultant zygote develops into an embryo. The second sperm cell fuses with the central cell to form a triploid primary endosperm cell, which develops into the endosperm that nourishes the developing embryo (Guignard 1899; Nawaschin 1898; Russell 1992). The mechanisms behind karyogamy and zygotic development, such as the involvement of heat shock protein70 chaperon in nuclear fusion (Maruyama et al. 2010), the synthesis of paternal mRNA in fused nucleus (Scholten et al. 2002) and the resetting of Histone3 variants in the zygotic nucleus (Ingouff et al. 2010), have gradually become evident. Recently, the dynamics of male and female nuclei during karyogamy in zygotes produced by electro-fusion of isolated rice gametes was reported (Ohnishi et al. 2014). In this case, the sperm nucleus migrated adjacently to the egg nucleus and the egg chromatin moved unidirectionally into the sperm nucleus through a possible nuclear connection. Thereafter, the sperm chromatin began to decondense with progression of nuclear fusion. Interestingly, the migration of the sperm nucleus in fused egg cells was inhibited by treatment with latrunculin B, an inhibitor of actin polymerization, although oryzalin, an inhibitor for tubulin assembly, had no effect on the progression of karyogamy. These results suggested that actin filaments play a role in sperm nucleus migration in rice. In addition to rice zygotes, Kawashima et al. (2014) demonstrated the involvement of actin filaments in sperm nuclear migration in transformed Arabidopsis zygotes in which the actin filaments were disrupted by the expression of semi-dominant negative actin in egg cells. Moreover, they reported that the sperm nucleus in fertilized central cells was surrounded by actin filaments and migrated toward the central nucleus via a constant movement of actin filaments. Taken together, these studies suggested that the actin filament-dependent nuclear migration system is a common feature of female gametes in angiosperms and represents an essential cytoskeletal mechanism in plant reproduction. However, the spatial distribution of actin filaments in egg cells and how these filaments are related to sperm nucleus migration during karyogamy in zygotes remain unclear. Furthermore, there is no study to our knowledge that addresses whether the function of actin filament movements is limited to nuclear migration in fused gametes or if such movement continues functioning after the completion of karyogamy. To elucidate these gametic and zygotic events, the dynamics of actin filaments in rice gametes and zygotes were investigated using an in vitro fertilization (IVF) system where the actin filaments, chromatins and/or the nuclear membrane of gametes were fluorescently labeled.
Materials and methods
Plant materials, isolation of gametes and the electro-fusion of gametes
Rice (Oryza sativa L. cv Nipponbare) was grown in environmental chambers (K30-7248; Koito Industries) at 26 °C with a 13 h/11 h light–dark cycle. Transformed rice plants expressing the Lifeact-TagRFP, the histone H2B-GFP or the SUN2-GFP fusion protein were prepared as previously described (Abiko et al. 2013; Ohnishi et al. 2014). Isolation of egg and sperm cells from rice flowers, and the electro-fusion of isolated gametes for zygote production, were conducted as described by Uchiumi et al. (2006, 2007).
Microscopic observation
Egg cells and fused egg cells were transferred into droplets of mannitol adjusted to 370 and 450 mOsmol kg−1 of water, respectively. Cells were observed using a confocal laser scanning microscope (CLSM) (Carl Zeiss, LSM 710). H2B-GFP, SUN2-GFP, Acti-stain 488 phalloidin, and Calcium Green-1 acetyloxymethyl were observed by 488 nm excitation and 493–556 nm emission wavelengths. Lifeact-TagRFP was observed by 543 nm excitation and 568–712 nm emission wavelengths.
Phalloidin staining of egg cells
m-Maleimidobenzoic acid N-hydroxysuccinimide ester (MBS) was dissolved with ethyl acetate into 9.94 mM stock solutions and diluted to 400 µM with 370 mOsmol kg−1 mannitol solution containing 20 mM piperazine-N,N′-bis (2-ethane-sulfonic acid), 5 mM ethyleneglycol-bis-(β-aminoethyleter)-N,N,N,N′-tetraacetic acid, 5 mM MgCl2, 2% dimethylsulfoxide (DMSO) and 0.5 mM phenylmethylsulfonyl fluoride. Egg cells were fixed in a droplet of 400 µM MBS solution for 30 min, washed with phosphate-buffered saline (PBS: 20 mM NaH2PO4, 150 µM NaCl) for 5 min, and stained with 112 nM Acti-stain 488 phalloidin (Cytoskeleton) in PBS containing 1.5% DMSO for 30 min. After washing by transfer into fresh PBS droplets three times, cells were observed by CLSM.
Quantitative analysis for movement of actin filaments on set radial lines
Actin filaments were visualized in rice egg cells by the expression of Lifeact-TagRFP. After capturing time-lapse images of the actin meshwork, cell positions in the frame of all captured images were adjusted to the same position using GIMP2 (GNU General Public License). Fluorescent signals in all images were bandpass-filtered and binary-processed using Image J (NIH). Eight standard lines, each of which passed through the cell center, were fixed on the processed time-lapse images. The fluorescence intensity of actin filaments labeled with Lifeact-TagRFP was determined and presented as a graph showing the peak of fluorescent intensity along each standard line. Actin filaments were observed in at least three serial time-lapse images. The distance between the position of the cell center point and the actin filaments was measured. For the migration of actin filaments in egg cells ended near the nucleus, the actin filaments around the egg nucleus were excluded from this measurement. The distance of the movement after 30 s (between two serial images) was calculated as follows (1);
where l n is the distance of movement of the peaks; A n is the distance between the cell center point and the peak at n seconds; and A n+30 is the distance between the cell center point and the peak at n + 30 s. Then, velocities of the actin filament movements were calculated. The details of measurement are given in Supplementary Fig. S1.
Quantitative analysis for movement of branch/cross points of actin meshwork
Time-lapse images of the actin meshwork were obtained and processed as described. Skeletonization was then conducted for the time-lapse images, and the branch/cross points of actin filaments were identified. The coordinates of each branch/cross point and egg nucleus center were determined by time-lapse imaging when the points were detected in at least four serial images. The direction and the distance of movement during 30 s (between two serial images) were calculated according to Eqs. (2, 3) presented below;
where l n is the distance of movement of the branch/cross points after 30 s; θ n is the movement direction of the points after 30 s; (x a , y a ) are the coordinates of the point at n seconds; (x b , y b ) are the coordinates of the same point at n + 30 s; (x c , y c ) are the coordinates of the egg nucleus center point at n seconds; and p is the rate of change in length divided by the number of pixels. Upon calculation, the direction of the egg nucleus center point was assigned as 0°. The details of measurement are given in Supplementary Fig. S2.
Quantitative analysis for movement of sperm nucleus in fused egg cells
Egg cells expressing Lifeact-TagRFP were fused with sperm cells expressing H2B-GFP. Time-lapse images were captured every 30 s and sperm nucleus movement was measured along with actin filament movement. Central coordinates of the sperm nucleus and the egg nucleus in fused egg cells were obtained from the serial images. The distance between the sperm and egg nucleus center points was measured for all images. Subsequently, the distance of sperm nuclear migration after 30 s was calculated by Eq. (1) where l n is the length of movement of sperm nucleus; A n is the length between the sperm and the egg nucleus center points at n seconds; and A n+30 is the length between the sperm and egg nucleus center points at n + 30 s.
Inhibitor treatment of egg cells and fused egg cells
To inhibit the myosin-motor, egg cells were incubated in mannitol solution adjusted to 370 mOsmol kg−1 of water containing 50 mM 2,3-butanedione monoxime, BDM (Sigma) and/or 20 μM N-ethylmaleimide, NEM (Wako) for 30 min. These egg cells were then used for IVF with sperm cells, and the resulting zygotes were transferred into droplets of mannitol adjusted to 450 mOsmol kg−1 of water containing the inhibitors at the same concentrations. Observations of cells were conducted as above.
Data presentation in boxplot
To compare the velocities of actin filaments in an egg cell, fused egg cell and zygote during karyogamy, these velocities were presented as a boxplot in which the first quartile, the median, and the third quartile are represented by the lower edge, the middle line and the upper edge of each box, respectively. The lower and upper fences are defined as the 10–25th and the 75–90th percentiles, respectively. The average velocity is indicated as a square within the box. The lowest and highest values are shown as whiskers.
Monitoring the Ca2+ level of egg cells and fused egg cells
Calcium Green-1 acetyloxymethyl (AM) (Invitrogen) was dissolved with DMSO into 5 mM stock solutions. Egg cells and fused egg cells were incubated in droplets of mannitol solution adjusted to 370 mOsmol kg−1 of water containing 10 µM Calcium Green-1 AM and 1% DMSO for 20 min. After washing twice, the cells were observed by CLSM. To calculate fluorescence intensity vacuole regions were excluded from measurements of the fluorescent signal.
Results
Actin meshwork in rice egg cells continuously changes structure
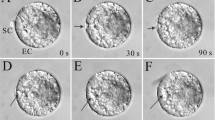
In egg cells expressing Lifeact-TagRFP under the control of the egg cell specific Arabidopsis DD45 promoter, actin filaments were fluorescently labeled (Ohnishi et al. 2014). In the present study, egg cells were observed using a CLSM to gain detailed structural information of the actin filaments. The results showed that actin filaments were bundled together to form a mesh-like structure (Fig. 1a). The egg nucleus was surrounded by actin filaments, as were other organelles such as vacuoles, mitochondria and amyloplasts. Additionally, when the structures of actin filaments in wild-type egg cells were observed by staining with fluorescent phalloidin, the actin meshwork structure was similar to that in egg cells expressing Lifeact-TagRFP (Fig. 1b). These results suggested that the structure of the actin meshwork visualized by Lifeact-TagRFP represents in situ actin organization in egg cells of wild-type rice plants. To monitor the dynamics of actin filaments in egg cells expressing Lifeact-TagRFP, time-lapse images were obtained every 30 s for 60 min and represented as serial images (Fig. 1c, d) and a serial movie (Movie 1). In these images, the structure of the actin meshwork in egg cells actively changed, and the actin filaments appeared to migrate toward the egg nucleus from the cell cortex. In addition, time-lapse and three-dimensional observations of actin filaments at the cell cortex suggested that actin filaments emerged from the cell cortex and moved toward the inside of the egg cell (Fig. S3; Movie 2).
Spatial and temporal organizations of actin meshwork in rice egg cells. a Structure of actin filaments in an isolated egg cell expressing Lifeact-TagRFP. Upper and lower panels show bright field and fluorescence images, respectively. The arrowhead indicates the egg nucleus. b Phalloidin staining of actin filaments in egg cells. Egg cells expressing Lifeact-TagRFP (upper panels) and the wild-type egg cells (lower panels) were stained with Acti-stain 488 phalloidin, and observed by CLSM. Three-dimensional images were constructed from z-stack images for Acti-stain 488 phalloidin. c, d Representative time-lapse images of actin filaments in egg cells expressing Lifeact-TagRFP. To show the actin filament movements toward the egg nucleus from the cell cortex, five serial images were captured every 30 s and presented as examples of the movements at the cortex region (c) and at the nuclear region (d). The blue and red dotted line indicates approximate plasma membrane location and nuclear membrane location, respectively. Arrowheads indicate actin filaments that migrate toward the egg nucleus. Observation times were indicated in panels. The time-lapse images are also presented as Movie 1. Bars 10 µm in (a, b), 5 µm in (c, d)
Quantitative measurements of the dynamics of the actin meshwork in egg cells indicate continuous convergence toward the egg nucleus
To understand the spatial and temporal changes in the structure of the actin meshwork, two types of quantitative measurements for movement were conducted. Egg cells expressing Lifeact-TagRFP were observed every 30 s for 15 min and time-lapse images were used to determine the movement distance of the actin filaments. A standard line passing through the cell center was fixed on the time-lapse images (Fig. 2a). Fluorescent signals of actin filaments on the standard line in all 31 time-lapse images were obtained and the temporal changes were presented in Fig. 2b. The length and the direction of actin filament movement on the standard line were calculated, and are presented in Supplementary Fig. S1a–e. In the study, eight standard lines were set as radial line passing through the cell center point (Fig. S1f) and the measuring was done for these eight lines. The movement of 125 actin filaments in three independent egg cells were traced and it was shown that most actin filaments migrated in the direction toward the cell center (Fig. 2c). The average velocity of the actin filament movements was 0.27 ± 0.03 µm min−1 (n = 125, mean ± SE).
Quantitative analysis of the actin filament movement on set radial lines in egg cells. a A CLSM image of an egg cell expressing Lifeact-TagRFP. The yellow line indicates a standard line passing through the center of the cell. b Kymograph for temporal changes of actin filament position on the standard line. The minus (−) and plus (+) signs below the panel indicate cell cortex and center, respectively. c Movement distance of the actin filaments on eight standard radial lines. Movements of 125 actin filaments in three independent egg cells were calculated as described in “Materials and methods”. The movement toward the cell center is indicated as a positive value. Bars 10 µm
The actin meshwork in egg cells is composed of actin filaments and their branch or cross points (Fig. 1a). In addition to the quantitative analysis of actin filament movement on the radial lines, the movement of the branch/cross points of actin filaments was measured to further elucidate how the actin meshwork moves in egg cells. In the same time-lapse images, all points at which actin filaments branched or crossed were marked with dots (Fig. 3a), and the direction and distance of the serial movements of these dots during observation were measured. These measurements are presented in Supplementary Fig. S2. In this analysis, the most direct path toward the center point of the egg nucleus was assigned as 0°. Data obtained from three independent egg cells were plotted according to the direction and distance of the movement (Fig. 3b). Plotted points were divided into two groups: branch/cross points moving toward the egg nucleus (forward; red dots in Fig. 3b) or branch/cross points moving toward the cell cortex (backward; blue dots in Fig. 3b). A comparison of their summed movements suggested that the branch/cross points also tended to move toward the egg nucleus (Fig. 3c, d). Z-stack images of the half-hemispheric region of an egg cell were obtained every 2 min for 20 min and the movement of the branch/cross point of actin filaments was quantitatively analyzed at the three-dimensional level (Fig. S4a–c), and showed movement toward the egg nucleus (Fig. S4d). In the study, the movements of actin filaments on set standard lines (Fig. 2) and the movement of the branch/cross points of actin filaments (Fig. 3) each independently supported the putative migration of actin filaments in egg cells from the cell cortex to the egg nucleus.
Quantitative analysis of the movement of actin filament branch/cross points in egg cells. a Branch/cross points of the actin meshwork in a time-lapse image. Yellow dots indicate the branch/cross points. b Distribution of the direction and the distance of branch/cross point movement. The direction toward the egg nucleus center point is set as 0°. Coordinates from 227 branch/cross points in three independent egg cells were determined, and the resulting 1965 movements were plotted. Moving distances are indicated as the distance from the center point of the panel to each point. Plotted dots are red when their direction of travel (θ) is −90° < θ < 90° (forward; toward the egg nucleus) and blue when their direction of travel is θ < −90°, 90° < θ (backward; toward the cell cortex). c The ratio of the total distance between branch/cross points with forward motion (red dots in panel b) and backward motion (blue dots in panel b). d Directional angle-based distribution of the branch/cross points. The total movement distance (purple bars) and the number of points (green bars) are categorized every 4°. Bars 10 µm in (a) and 1 µm in (b). Error bars in c represent means ± SE (n = 3). *P < 0.001 by Student’s t test
Migration of the sperm nucleus is mediated by the continuous convergence of the actin meshwork toward the egg nucleus in fused egg cells
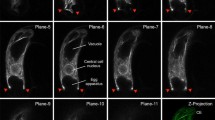
It has been suggested that sperm nuclei in the fused egg and the central cell are surrounded by actin filaments (Kawashima et al. 2014; Ohnishi and Okamoto 2015). To make clear this subcellular relationship between the sperm nucleus and actin filaments during karyogamy, egg cells expressing Lifeact-TagRFP were fused with sperm cells expressing GFP tagged with SUN2, a Sad1/UNC84 (SUN) domain protein localized on the inner nuclear membrane (Granumann et al. 2010). In fused egg cells, the sperm nucleus expressing SUN2-GFP was enmeshed by the actin meshwork and migrated with the meshwork toward the egg nucleus (Fig. 4), suggesting a physical interaction of actin filaments with the sperm nuclear membrane and the direct involvement of the actin meshwork in sperm nuclear migration.
Localization of actin filaments and sperm nuclear membrane in fused egg cells. a Three-dimensional distribution of the actin filaments (magenta) and the sperm nucleus (green) in fused egg cells after gamete fusion. The fused egg cell was produced by electro-fusion of an egg cell expressing Lifeact-TagRFP with a sperm cell expressing SUN2-GFP, and three-dimensional fluorescent signals were observed. Actin filaments labeled with Lifeact-TagRFP (magenta) and sperm nuclear membranes labeled with SUN2-GFP (green) are indicated. The times after fusion (min) are shown in each panel. b Close-up view of the sperm nucleus. Arrowheads indicate where actin filaments may have adhered to the sperm nuclear membrane. Bars 10 µm in (a) and 2 µm in (b)
To investigate this relationship further, egg cells expressing Lifeact-TagRFP were electro-fused with isolated sperm cells expressing histone H2B (H2B)-GFP fusion protein. In the fused egg cells, the sperm nucleus labeled with H2B-GFP migrated adjacently to the egg nucleus approximately 20–30 min after fusion (Fig. 5a). Thereafter, a possible bridge between gamete nuclei was formed, male chromatin began to decondense and the karyogamy process was completed (Movie 3). Karyogamy progressed normally in the fused egg cells in which Lifeact-TagRFP was expressed, but the migration of the sperm nucleus toward the egg nucleus appeared to be delayed (Fig. 5a). When fused egg cells were previously produced from wild-type egg cells and sperm cells expressing H2B-GFP, most sperm nuclei migrated to be adjacent to the egg nucleus within only 5–10 min after fusion (Ohnishi et al. 2014). To estimate the timetable from gamete fusion to the onset of sperm chromatin decondensation in the fused egg cells expressing Lifeact-TagRFP, 46 independent zygotes produced by the fusion of egg cells expressing Lifeact-TagRFP with sperm cells expressing H2B-GFP were observed every 15–20 min after fusion. Comparison of this timetable with that of wild-type egg cells fused with sperm cells expressing H2B-GFP shows karyogamy, including sperm nuclear migration and nuclear fusion, was delayed for approximately 20 min in fused egg cells prepared from egg cells expressing Lifeact-TagRFP (Fig. S5). It is suggested that the expression of Lifeact-TagRFP in rice egg cells results in the delay of the migration of sperm nucleus, but does not hinder the post-nuclear migration events during karyogamy. This delay may be caused by a reduction of the reorganization rate of actin filaments by the high expression of Lifeact (van der Honing et al. 2011). However, the sperm nucleus migrates directionally from cell peripheral to egg nucleus in the fused egg cells with or without expression of Lifeact-TagRFP (Ohnishi et al. 2014; Ohnishi and Okamoto 2015), and the structure of the actin meshwork in egg cells Lifeact-TagRFP showed in situ organization of actin filaments in the egg cells (Fig. 1b). Therefore, we further analyzed the dynamics of actin meshwork and sperm nucleus migration using fused egg cells expressing Lifeact-TagRFP.
Quantitative analyses of movements of the actin meshwork and the sperm nucleus in fused egg cells (a–d) and the effects of inhibitors for ATPase on the dynamics of actin meshwork and nuclear migration (e–h). a The actin meshwork and the sperm nucleus in fused egg cells after gamete fusion. The RFP and GFP signals are presented as white and cyan colors, respectively. b Direction and distance of the movement of actin filaments (n = 130) and the sperm nuclei (n = 3) in fused egg cells. White lines indicate movements of the sperm nuclei. c The ratio of the total distance between branch/cross points with forward motion and backward motion. d Directional angle-based distribution of the branch/cross points. Total movement distance (purple bars) and number of points (green bars) were categorized. e The actin meshwork and the sperm nucleus in fused egg cells after treatment with BDM and NEM. f Direction and distance of the movements of actin filaments (n = 109) and sperm nucleus (n = 3) in fused egg cells that were treated with inhibitors. g The ratio of total movement distance between branch/cross points with forward motion and backward motion. h Directional angle-based distribution of the branch/cross points in fused egg cells treated with inhibitors. Total movement distance (purple bars) and number of points (green bars) are categorized every 4°. Bars 10 µm. The error bars in panels c and f represent means ± SE (n = 3) *P < 0.001 by Student’s t test
The actin meshwork dynamics in fused egg cells during nuclear migration were measured using the same analytical procedures as those used in egg cells, and the movement of the sperm nucleus in fused egg cells was quantitatively investigated as described previously. Most actin filaments and sperm nuclei appeared to migrate toward the egg nucleus in fused egg cells (Fig. 5b). The average velocity of actin filaments in the three fused egg cells was 0.66 ± 0.06 µm min−1 (n = 130, mean ± SE). The branch/cross points of actin filaments were also shown to move toward the center of the egg nucleus (Fig. 5c, d). In addition, to address whether nuclear migration in fused egg cells is dependent on the continuous convergence of the actin meshwork, the function of myosin, ATP-dependent motor proteins responsible for actin-based motility, was inhibited, and sperm nuclear migration was observed. The egg cells were treated with 50 mM 2,3-butanedione monoxime (BDM), 20 μM N-ethylmaleimide (NEM) or both, which are inhibitors of the ATPase activity of myosin, and then fused with sperm cells expressing H2B-GFP. When cells were treated by BDM or NEM, convergence of the actin meshwork and sperm nuclear migration were detectable, although the average velocities of actin filament movement were reduced (Fig. S6). On the other hand, when cells were treated with both BDM and NEM, the convergence of the actin meshwork was almost entirely inhibited but there was no effect on the structure of the actin meshwork (Fig. 5e). Moreover, sperm nuclear migration was also arrested. Quantitative analysis of the movement of the actin meshwork and the sperm nuclei in fused egg cells treated with these inhibitors supported the arrest of both motions (Fig. 5f). The average velocity of the actin filaments was dramatically reduced to 0.01 ± 0.01 µm min−1 (n = 109, mean ± SE), and the branch/cross points migrated randomly even if only slightly from their original positions (Fig. 5g, h).
To estimate the correlation between the velocity of sperm nucleus migration and the velocity of actin filament movements around the sperm nucleus, these velocities were measured and the correlation coefficient was calculated (n = 21). There was a positive correlation between these two velocities (Fig. 6, r = 0.795), suggesting that the velocity of the sperm nucleus migration is correlated with the velocity of the actin filament movement. Taken together, these results suggest that the sperm nucleus in fused egg cells is enmeshed by the actin filaments in the cell cytoplasm and migrates toward the egg nucleus through the use of a convergence of the actin meshwork as the motivity.
Correlation between velocities of sperm nucleus migration and actin filament movement. a A CLSM image of a fused egg cell produced by fusion of an egg cell expressing Lifeact-TagRFP with a sperm cell expressing H2B-GFP. Yellow lines indicate the five standard lines used to quantitative analyses of actin filaments and sperm nucleus movement. The arrow indicates the cell center. b Correlation between velocities of actin filaments and sperm nucleus movements (n = 21). Pearson’s correlation coefficient (r), the contribution rate (r2), Spearman’s rank correlation coefficient (rs) and P values of the coefficients (P r and P rs ) are shown in the graph. Bar 10 µm
Velocity of actin filaments changes after gamete fusion and during zygotic development
The convergence of the actin meshwork occurred continuously in both egg cells and fused egg cells. Interestingly, the velocity of the actin filaments seemed to increase after fusion with sperm cells (Figs. 2c, 5b). In addition, the migration distance of the branch/cross points of actin filaments per 30 s also appeared to increase after fusion (Figs. 3d, 5d). To provide more detail of this increase in actin filament velocity, further time-lapse observations were conducted with egg cells (n = 12) and fused egg cells (n = 11) every 30 s for 5 min, and the migration velocity of actin filaments toward the egg nucleus was measured as described above. For fused egg cells, each observation started 20 min after gamete fusion. Actin filament velocity increased approximately 1.4 fold after fusion (Fig. 7a). Moreover, to monitor the changes in the actin meshwork velocity during zygote development, quantitative analyses with zygotes at 4, 12 and 22 h after fusion were conducted. After 4 h and the completion of nuclear fusion, the velocity of actin filament movement in zygotes, was twofold higher than that of egg cells. After this point, the velocity of actin filaments in zygotes slowed gradually until just before the first division at 22 h (Fig. 7b). After continuous observation of the actin meshwork dynamics in fused egg cells (early zygotes) during 5–165 min post-fusion, there was an initial increase of velocity at 20 min after fusion, and the velocity of actin filaments was further increased 85–95 min after fusion, in which nuclear fusion and decondensation of sperm chromatin were in progress (Fig. 7c; Movie 3).
The velocity of actin filament movement toward the egg/zygote nucleus during karyogamy and early zygotic development. a Velocities of actin filament movement in egg cells (n = 12) and fused egg cells (n = 11) at 20 min after electro-fusion. The average velocities in egg cells and fused egg cells were 0.47 ± 0.03 µm min−1 (n = 336) and 0.65 ± 0.03 µm min−1 (n = 302), respectively. b Velocities of actin filament movement in zygotes at 4, 12 and 22 h after fusion. The average velocities in zygotes at 4, 12 and 22 h after fusion were 1.00 ± 0.12 µm min−1 (n = 48), 0.46 ± 0.10 µm min−1 (n = 98), 0.33 ± 0.03 µm min−1 (n = 103), respectively. c Changes over time in the velocity of actin filament movement in fused egg cells during karyogamy. Observation was conducted every 60 s during the period 5–165 min after gamete fusion, and velocity of actin filaments was measured every 10 min. The upper illustration indicates the progression of karyogamy at the time indicated. Cyan and magenta circles indicate sperm and egg nuclei/chromatin, respectively. Yellow indicates the merged sperm and egg chromatin. The serial images are presented in Movie 3. Average velocities represent means ± SE. *P < 0.001 by Student’s t test
Upon gamete fusion, Ca2+ levels rapidly increase in plant zygotes produced in vitro (Antoine et al. 2000; Digonnet et al. 1997) and in vivo (Denninger et al. 2014; Hamamura et al. 2014), as is the case for animal zygotes (Stricker 1999). Moreover, it is also known that Ca2+ levels in animals regulate muscle contraction via a calmodulin-mediated change of interaction between actin filaments and myosin (Ohtsuki et al. 1986). When Ca2+ levels were observed in fused egg cells/zygotes produced by IVF, the fertilization-induced increases in cytosolic Ca2+ levels were detected (Fig. S7).
Discussion
In egg cells, actin filaments form a meshwork structure that surrounds the egg nucleus and other organelles (Fig. 1a). The actin meshwork consists of multiple actin filaments and emerges at the cell cortex (Fig. S3). Quantitative analysis of the movements of the actin meshwork and the sperm nucleus in this study clearly suggest that the actin meshwork continuously migrates toward the egg nucleus and is involved in sperm nucleus migration (Figs. 2, 3, 4, 5). After the arrival of the actin filaments around the egg nucleus, these filaments seemed to remain around the nuclear membrane, but accumulation of an excessive amount of the actin filaments around the nucleus was not observed. It is unclear how the turnover and depolymerization of actin filaments around the egg nuclear membrane occurs. It has been reported that the constant movement of actin filaments toward the nucleus occurs in the central cell of Arabidopsis (Kawashima et al. 2014), suggesting that there may be a common mechanism for the convergence of actin meshwork functions in both egg cells and central cells. On the other hand, actin filament dynamics such as continuous convergence in plant female gametes for the cellular trafficking of organelles has not been reported in plant somatic cells or in animal cells (Gundersen and Worman 2013; Higa et al. 2014). This suggests that female gametes in angiosperms may possess a characteristic cytoskeleton system for organelle trafficking. However, an aster-like structure of actin filaments, which was detected around the sperm nucleus in fertilized Arabidopsis central cells (Kawashima et al. 2014), was not observed in the fused egg cells of rice (Fig. 4). This suggests the possibility that the factors governing actin meshwork convergence are different in egg and central cells.
When compared to egg cells, the average velocity of actin filament movement in fused egg cells was approximately 1.4 fold faster. This velocity change was detected shortly after gamete fusion (Fig. 7a), and is likely to be related to the increase of Ca2+ levels in the fused gamete. The fertilization-induced increase of Ca2+ levels was detected in fused egg cells/zygotes in rice (Fig. S7). Muscle contraction in animals is known to be regulated with Ca2+ levels through a calmodulin-mediated change of interaction between actin filaments and myosin (Ohtsuki et al. 1986). In this study, inhibitors for the activity of myosin blocked the dynamics of the actin meshwork in rice egg cells and fused egg cells (Fig. 5e–h, S6). It is therefore possible that the increasing Ca2+ levels in rice zygotes could contribute to the increase in velocity of actin filaments that was observed in zygotes at 5–85 min after fusion (Fig. 7b). A second increase in velocity was detected at 85–95 min after fusion (Fig. 7c), and this increase continued until at least 4 h after fusion, but the mechanism of this second increase remains unknown.
Our present study suggests that an active convergence of the actin meshwork can be detected in the zygote, in which the migration of the sperm nucleus is completed and gamete nuclei are united. Furthermore, egg cells converge the actin meshwork before fusion with the sperm cell. These results indicate the possibility that, in addition to nuclear migration in fused egg cells, the convergence of the actin meshwork has a biological function in egg cells and early zygotes, which show characteristic cell architectures. In these cells, the cytoplasm, the mitochondria and possibly the starch granules are densely distributed around the egg nucleus and vacuoles exist at the peripheries of the cells (Uchiumi et al. 2006, 2007). These characteristic architectures have been shown to disappear around 4 h after fusion (Uchiumi et al. 2007; Ueda and Laux 2012). This along with the results of our study, implicates that the actin meshwork may have a major function in maintaining these cell architectures. The loss of function of the actin meshwork may coincide with the remodeling of cell polarity and asymmetric cell division of the zygotes, resulting in the production of daughter cells with a different cell fate (Sato et al. 2010; Ueda and Laux 2012).
References
Abiko M, Maeda H, Tamura K, Hara-Nishimura I, Okamoto T (2013) Gene expression profiles in rice gametes and zygotes: Identification of gamete-enriched genes and up- or down-regulated genes in zygotes after fertilization. J Exp Bot 64:1927–1940
Antoine AF, Faure JE, Cordeiro S, Dumas C, Rougier M, Feijo JA (2000) A calcium influx is triggered and propagates in the zygote as a wavefront during in vitro fertilization of flowering plants. Proc Natl Acad Sci USA 97:10643–10648
Chambers EL (1939) The movement of the egg nucleus in relation to the sperm aster in the echinoderm egg. J Exp Bot 16:409–424
Denninger P, Bleckmann A, Lausser A, Vogler F, Ott T, Ehrhardt DW, Frommer WB, Sprunck S, Dresselhaus T, Grossmann G (2014) Male–female communication triggers calcium signatures during fertilization in Arabidopsis. Nat Commun 5:4645
Digonnet C, Aldon D, Leduc N, Dumas C, Rougier M (1997) First evidence of a calcium transient in flowering plants at fertilization. Development 124:2867–2874
Endow SA, Kang SJ, Satterwhite LL, Rose MD, Skeen VP, Salmon ED (1994) Yeast Kar3 is a minus-end microtubule motor protein that destabilizes microtubules preferentially at the minus ends. EMBO J 13:2708–2713
Gibeaux R, Politi AZ, Nedelec F, Antony C, Knop M (2013) Spindle pole body-anchored Kar3 drives the nucleus along microtubules from another nucleus in preparation for nuclear fusion during yeast karyogamy. Genes Dev 27:335–349
Granumann K, Runions J, Evans DE (2010) Characterization of SUN-domain proteins at the higher plant nuclear envelope. Plant J 61:134–144
Guignard ML (1899) Sur les antherozoides et la double copulation sexuelle chez les vegetaux angiosperms. Rev Gén de Botanique 11:129–135
Gundersen GG, Worman HJ (2013) Nuclear positioning. Cell 152:1376–1389
Hamamura Y, Nishimaki M, Takeuchi H, Geitmann A, Kurihara D, Higashiyama T (2014) Live imaging of calcium spikes during double fertilization in Arabidopsis. Nat Commun 5:4722
Higa T, Suetsugu N, Wada M (2014) Plant nuclear photorelocation movement. J Exp Bot 65:2873–2881
Ingouff M, Rademacher S, Holec S, Soljić L, Xin N, Readshaw A, Foo SH, Lahouze B, Sprunck S, Berger F (2010) Zygotic resetting of the HISTONE 3 variant repertoire participates in epigenetic reprogramming in Arabidopsis. Curr Biol 20:2137–2143
Kawashima T, Maruyama D, Shagirov M, Li J, Hamamura Y, Yelagandula R, Toyama Y, Berger F (2014) Dynamic F-actin movement is essential for fertilization in Arabidopsis thaliana. eLife 3:e04501
Kurihara LJ, Beh CT, Latterich M, Schekman R, Rose MD (1994) Nuclear congression and membrane fusion: two distinct events in the yeast karyogamy pathway. J Cell Biol 126:911–923
Maruyama D, Endo T, Nishikawa S (2010) BiP-mediated polar nuclei fusion is essential for the regulation of endosperm nuclei proliferation in Arabidopsis thaliana. Proc Natl Acad Sci USA 107:1684–1689
Nawaschin S (1898) Revision der Befruchtungsvorgange bei Lilium martagon und Fritillaria tenella. Bull Acad Imp Sci St Pétersbourg 9:377–382
Ohnishi Y, Okamoto T (2015) Karyogamy in rice zygotes: Actin filament-dependent migration of sperm nucleus, chromatin dynamics, and de novo gene expression. Plant Signal Behav 10:e989021
Ohnishi Y, Hoshino R, Okamoto T (2014) Dynamics of male and female chromatin during karyogamy in rice zygotes. Plant Physiol 165:1533–1543
Ohtsuki I, Maruyama K, Ebashi S (1986) Regulatory and cytoskeletal proteins of vertebrate skeletal muscle. Adv Protein Chem 38:1–67
Payne C, Rawe V, Ramalho-Santos J, Simerly C, Schatten G (2003) Preferentially localized dynein and perinuclear dynactin associate with nuclear pore complex proteins to mediate genomic union during mammalian fertilization. J Cell Sci 116:4727–4738
Reinsch S, Gönczy P (1998) Mechanisms of nuclear positioning. J Cell Sci 111:2283–2295
Rose MD (1996) Nuclear fusion in the yeast Saccharomyces cerevisiae. Annu Rev Cell Dev Biol 12:663–695
Russell SD (1992) Double fertilization. Int Rev Cytol 140:357–390
Sato A, Toyooka K, Okamoto T (2010) Asymmetric cell division of rice zygotes located in embryo sac and produced by in vitro fertilization. Sex Plant Reprod 23:211–217
Schatten G (1994) The centrosome and its mode of inheritance: the reduction of the centrosome during gametogenesis and its restoration during fertilization. Dev Biol 165:299–335
Schatten G, Schatten H (1981) Effects of motility inhibitors during sea urchin fertilization: microfilament inhibitors prevent sperm incorporation and restructuring of fertilized egg cortex, whereas microtubule inhibitors prevent pronuclear migrations. Exp Cell Res 135:311–330
Scholten S, Lörz H, Kranz E (2002) Paternal mRNA and protein synthesis coincides with male chromatin decondensation in maize zygotes. Plant J 32:221–231
Stricker SA (1999) Comparative biology of calcium signaling during fertilization and egg activation in animals. Dev Biol 211:157–176
Uchiumi T, Komatsu S, Koshiba T, Okamoto T (2006) Isolation of gametes and central cells from Oryza sativa L. Sex Plant Reprod 19:37–45
Uchiumi T, Uemura I, Okamoto T (2007) Establishment of an in vitro fertilization system in rice (Oryza sativa L.). Planta 226:581–589
Ueda M, Laux T (2012) The origin of the plant body axis. Curr Opin Plant Biol 15:578–584
van der Honing HS, van Bezouwen LS, Emons AM, Ketelaar T (2011) High expression of Lifeact in Arabidopsis thaliana reduces dynamic reorganization of actin filaments but does not affect plant development. Cytoskeleton (Hoboken) 68:578–587
Wilson EB (1928) The cell in development and heredity, 3rd edn. The Macmillan Co, New York, pp 377
Wühr M, Dumont S, Groen AC, Needleman DJ, Mitchison TJ (2009) How does a millimeter-sized cell find its center? Cell Cycle 8:1115–1121
Wühr M, Tan ES, Parker SK, Detrich HW 3rd, Mitchison TJ (2010) A model for cleavage plane determination in early amphibian and fish embryos. Curr Biol 20:2040–2045
Acknowledgements
We thank Ms. H. Maeda for help in preparing transformed plants, and Ms. N. Koiso, Ms. A. Matsuura and Ms. M. Shinada (Tokyo Metropolitan University) for help with data entry for quantitative analysis. Thank you to Dr. T. Igawa (Chiba University) for the DD45 promoter::H2B-TagRFP in pENTRTM3C and the DD45 promoter::Lifeact-TagRFP in pGWB1, Dr. K. Tamura (Kyoto University) for the GFP–SUN2 in pGWB405, and Dr. T. Nakagawa (Shimane University) for the pGWB1 and pGWB405 vectors. We also thank Dr. HS. Weiner and Mr. MH. Rahman for the critical reading of this manuscript. This work was supported by a research fellowship from the Japan Society for the Promotion of Science (15J05885 to Y.O.). This work was also partly supported by the Ministry of Education Culture Sports Science and Technology of Japan (26113715 to T.O.) and the Japan Society for the Promotion of Science (25650083 to T.O.).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ohnishi, Y., Okamoto, T. Nuclear migration during karyogamy in rice zygotes is mediated by continuous convergence of actin meshwork toward the egg nucleus. J Plant Res 130, 339–348 (2017). https://doi.org/10.1007/s10265-016-0892-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10265-016-0892-2