Abstract
Most of the known 4-coumarate:coenzyme A ligase (4CL) isoforms lack CoA-ligation activity for sinapic acid. Therefore, there is some doubt as to whether sinapic acid contributes to sinapyl alcohol biosynthesis. In this study, we characterized the enzyme activity of a protein mixture extracted from the developing xylem of Robinia pseudoacacia. The crude protein mixture contained at least two 4CLs with sinapic acid 4-CoA ligation activity. The crude enzyme preparation displayed negligible sinapaldehyde dehydrogenase activity, but showed ferulic acid 5-hydroxylation activity and 5-hydroxyferulic acid O-methyltransferase activity; these activities were retained in the presence of competitive substrates (coniferaldehyde and 5-hydroxyconiferaldehyde, respectively). 5-Hydroxyferulic acid and sinapic acid accumulated in the developing xylem of R. pseudoacacia, suggesting, in part at least, sinapic acid is a sinapyl alcohol precursor in this species.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
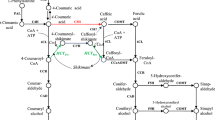
Lignin is a complex aromatic heteropolymer that is derived principally from three primary monolignols (p-coumaryl, coniferyl, and sinapyl alcohols) in angiosperms. Studies on the mechanism of monolignol biosynthesis have revealed that the biosynthetic pathway is more of a “metabolic grid” (Higuchi 2003; Shi et al. 2010). To date, 11 enzyme families including Arabidopsis caffeoyl shikimate esterase (CSE), a recently discovered enzyme that converts caffeoyl shikimic acid to caffeic acid (Barros et al. 2015; Kumar et al. 2016; Vanholme et al. 2013), have been shown to be involved in the metabolic grid of monolignol biosynthesis (Fig. 1). One of these is the 4-coumarate:coenzyme A ligase (4CLs) family, whose members convert p-coumaric acid and other substituted cinnamic acids into corresponding coenzyme A (CoA) thiolesters, and exhibit distinct substrate specificity (Chen et al. 2013; Costa et al. 2005; Lindermayr et al. 2002). The 4CL family is generally encoded by a small number of genes. For example, there are four isoforms in Arabidopsis (Hamberger and Hahlbrock 2004) and five in Populus trichocarpa (Souza Cde et al. 2008). The 4CLs play roles in the biosynthesis of monolignols and other phenylpropanoids (Allina et al. 1998; Hamberger and Hahlbrock 2004; Hu et al. 1998; Lindermayr et al. 2002; Lozoya et al. 1988) and their distinct substrate specificity directs metabolic flux through different pathways to synthesize specific compounds (Ehlting et al. 1999).
Proposed pathway of lignin biosynthesis. Gray arrows show principal pathway of sinapyl alcohol biosynthesis in Arabidopsis and poplar. Pathway through sinapic acid is shown in white arrows. Circled P and R indicate monolignol intermediates detected in developing xylem of Populus alba and Robinia pseudoacacia, respectively. PAL phenylalanine ammonia-lyase, SMT sinapoylglucose:malate sinapoyltransferase, C4H cinnamate 4-hydroxylase, SGT sinapate glucosyltransferase, C3H p-coumarate 3-hydroxylase, COMT caffeic acid O-methyltransferase, F5H ferulate 5-hydroxylase, 4CL 4-coumarate:CoA ligase, CSE caffeoyl shikimate esterase, HCT p-hydroxycinnamoyl-CoA:quinate/shikimate p-hydroxycinnamoyltransferase, CCoAOMT caffeoyl-CoA O-methyltransferase, ALDH aldehyde dehydrogenase, CCR cinnamoyl-CoA reductase, CAD cinnamyl alcohol dehydrogenase
Functional characterizations using recombinant proteins have revealed most of the known 4CLs related to monolignol biosynthesis, such as Pt4CL1 and 2 in Populus tremuloides (Hu et al. 1998), Ptr4CL3, and 17 in P. trichocarpa (Chen et al. 2013), 4CL9 in hybrid poplar (Allina et al. 1998; Cukovic et al. 2001), Gm4CL2, 3, and 4, in soybean, and At4CL1 and 2 in Arabidopsis. These 4CLs cannot convert sinapic acid to sinapoyl CoA. In addition to the poor substrate selectivity of 4CLs towards sinapic acid, coniferaldehyde, which is the preferred substrate for ferulate 5-hydroxylase (F5H), prevents hydroxylation of ferulic acid (Humphreys et al. 1999; Li et al. 2000; Osakabe et al. 1999). Similarly, 5-hydroxyconiferaldehyde, which is the preferred substrate for caffeic acid O-methyltransferase (COMT), inhibits O-methylation of caffeic acid and 5-hydroxyferulic acid (Li et al. 2000). Therefore, sinapyl alcohol is thought to be synthesized mainly through coniferaldehyde (Fig. 1), and it has been questioned whether sinapic acid can serve as a precursor of sinapyl alcohol. On the other hand, a very limited number of 4CLs in some species can convert sinapic acid to sinapoyl-CoA (Hamada et al. 2004; Hamberger and Hahlbrock 2004; Li et al. 2015; Lindermayr et al. 2002). Furthermore, a tracer experiment showed that heptadeuterosinapic acid fed to shoots of Robinia pseudoacacia and Nerium indicum was incorporated into syringyl lignin (Hamada et al. 2004, 2003; Yamauchi et al. 2002).
In this study, we focused on enzymatic activities that are essential for sinapyl alcohol biosynthesis through sinapic acid. The activities of 4CL, cinnamyl alcohol dehydrogenase (CAD), aldehyde dehydrogenase (ALDH), F5H, and COMT in a protein mixture extracted from the developing xylem of R. pseudoacacia were characterized in vitro. We also detected monolignol intermediates in the developing xylem of R. pseudoacacia to provide in vivo evidence that sinapic acid is a precursor of sinapyl alcohol in this species.
Materials and methods
4CL assay
Stems and branches of R. pseudoacacia were harvested from Kyushu University Forest from May to June. Developing xylem tissue (approx. 10 g) was ground in liquid nitrogen using a mortar and pestle. To prepare the crude enzyme mixture including 4CL, extraction buffer (5 ml g−1 xylem) containing 200 mM Tris–HCl (pH 7.8), 5% (v/v) β-mercaptoethanol, 20 mM ascorbic acid, 30% glycerol, 3% (w/v) polyvinylpolypyrrolidone and 2% AG 1-X2 Resin (Bio-Rad, Hercules, CA, USA) was added and the mixture was further homogenized. The homogenates were centrifuged at 10,000g for 20 min, and the supernatants were recovered and filtered through a 0.45-μM syringe filter. The reactions were performed at 30 °C in a 1-ml volume containing 100 mM Tris–HCl (pH 7.5), 5 mM MgCl2, 5 mM ATP, 330 μM CoA, and 50 μM substrate (4-coumaric acid, caffeic acid, ferulic acid, 5-hydroxyferulic acid, or sinapic acid). The change in absorbance was monitored with a UV–visible spectrophotometer (V-530; Jasco, Tokyo, Japan). The CoA esters were quantified using the following extinction coefficients: cinnamoyl-CoA (ε333 nm = 21 mM−1 cm−1) (Stöckigt and Zenk 1975), caffeoyl-CoA (ε346 nm = 18 mM−1 cm−1) (Stöckigt and Zenk 1975), feruloyl-CoA (ε345 nm = 19 mM−1 cm−1) (Gross and Zenk 1966), 5-hydroxyferuloyl-CoA (ε356 nm = 20 mM−1 cm−1) (Knobloch and Hahlbrock 1975; Lüderitz et al. 1982), and sinapoyl-CoA (ε352 nm = 20 mM−1 cm−1) (Stöckigt and Zenk 1975).
CAD and ALDH assays
To prepare the crude enzyme solution including ALDH and CAD, the ground xylem tissue was mixed with buffer (5 ml g−1 xylem) containing 100 mM NaH2PO4-NaOH (pH 7.5), 250 mM sucrose, 1 mM EDTA, 5% (v/v) β-mercaptoethanol, 20 mM ascorbic acid, 1% (w/v) polyvinylpolypyrrolidone, and 2 μM leupeptin, and the mixture was further homogenized. The homogenates were centrifuged at 10,000g for 20 min, and the supernatants were recovered and filtered through a syringe filter. The CAD and ALDH activity assays were conducted using 15–150 μg protein in 200 μl buffer containing 50 mM HEPES–KOH (pH 8.0), 5 mM dithiothreitol, 10% glycerol, 5% ethanol, and 0.2 mM substrate (coniferaldehyde, sinapaldehyde). After preheating the mixture at 30 °C for 3 min, NADPH (for the CAD activity assay) or NAD+ (for the ALDH activity assay) was added to a final concentration of 0.5 mM to start the reaction. The reactions were mixed every 5 min for 25 min, and the reaction was stopped by adding 15 μl 36% HCl. The sample was extracted three times with ethyl acetate and then the solvent mixture was dried. Trimethylsilylation derivatization was carried out by adding 15 μl N,O-bis (trimethylsilyl) trifluoroacetamide (BSTFA) and 10 μl pyridine to the residue and heating the mixture at 75 °C for 30 min. Then, the trimethylsilylated samples were subjected to GC–MS analysis. The GC–MS analysis was performed using a Shimadzu GC-17A gas chromatograph coupled with a Shimadzu QP5050 mass spectrometer (Shimadzu Co., Ltd., Kyoto, Japan) equipped with a DB-Wax column (0.25 mm × 60 cm; J&W Scientific, Folsom, CA, USA). The column temperature was programmed to increase at 5 °C min−1 from 200 to 320 °C. The injector and detector temperatures were 280 °C. The amounts of coniferyl alcohol, sinapyl alcohol, ferulic acid, and sinapic acid were determined using calibration curves derived from trimethylsilylated pure authentic samples.
F5H and COMT assays
The protein mixture containing F5H and COMT was prepared in the same way as the mixture including ALDH and CAD up to the 0.45-μM filtration step. After centrifugation at 100,000g for 90 min at 4 °C, the supernatant was recovered as a crude enzyme solution containing COMT, and the pellet containing F5H was suspended in buffer containing 50 mM NaH2PO4-NaOH (pH 7.5) and 0.1 mM EDTA. Each enzyme solution was flushed with nitrogen gas and stored at 4 °C until use. The F5H activity assay was conducted using 350 μg protein in 200 μl oxygen-saturated buffer containing 50 mM NaH2PO4-NaOH (pH 7.5), 1 mM β-mercaptoethanol, 5% ethanol, and 0.2 mM substrate (ferulic acid, coniferaldehyde, or coniferyl alcohol). After preheating the mixture at 30 °C for 3 min, 0.5 mM NADPH was added to a final concentration of 1 mM. The COMT activity assay was conducted using 15 µg protein in 200 µl buffer containing 50 mM Tris–HCl (pH 7.5), 5 mM β-mercaptoethanol, 5% ethanol, 2 mM MgCl2, and substrate (5-hydroxyferulic acid, 5-hydroxyconiferaldehyde, or 5-hydroxyconiferyl alcohol). After preheating the mixture at 30 °C for 3 min, S-adenosyl-l-methionine was added to a final concentration of 0.3 mM. The reactions were mixed every 5 min for 25 min, and then stopped by adding 15 μl 36% HCl. Then, GC–MS analysis was performed as described above for the CAD and ALDH assays. For the COMT kinetics assay, substrates were supplied at various concentrations (5, 10, 15, 30, 50, 100, 100 µM of 5-hydroxyferulic acid; 1.5, 2.5, 5, 10, 15, 20, 30 µM of 5-hydroxyconiferaldehyde; or 1.5, 2.5, 5, 10, 15, 30, 50 µM coniferyl alcohol).
Analysis of ethyl acetate-soluble metabolites
Branches of P. alba and R. pseudoacacia were harvested at Kyushu University nursery in Fukuoka city (Fukuoka, Japan) in May. Developing xylem tissues from P. alba and R. pseudoacacia (approx. 3 g) were ground in liquid nitrogen using a mortar and pestle. Ethyl acetate-soluble compounds were extracted three times in 33 ml ethyl acetate at 4 °C, and then the mixtures were pooled and dried. The residue was dissolved in 100 µl pyridine, and then a 10-µl aliquot was subjected to trimethylsilylation derivatization by adding 15 µl BSTFA and incubating the mixture at 75 °C for 30 min. Then, GC–MS analyses were performed as described above for the CAD and ALDH assays.
Results
Enzyme activities to modify phenylpropanoid side chains
A previous study showed that the developing xylem of R. pseudoacacia contains at least three 4CLs (Rp4CL1, Rp4CL2, and Rp4CL3), and that Rp4CL2 and Rp4CL3 are able to convert sinapic acid to sinapoyl-CoA (Hamada et al. 2004). In this study, the crude enzyme mixture prepared from the developing xylem of R. pseudoacacia was able to convert sinapic acid to sinapoyl-CoA. The specific activity for sinapic acid (4.6 nmol/min/mg protein) was about one-third that for p-coumaric acid (12.3 nmol/min/mg protein), the highest specific activity among the five substrates used in these analyses (Table 1). To evaluate whether there is a functional pathway to biosynthesize sinapyl alcohol from sinapic acid, the ALDH and CAD activities of the crude enzyme were measured using coniferaldehyde and sinapaldehyde e as the substrates. A previous study showed that CAD catalyzes the reduction of both coniferaldehyde and sinapaldehyde to coniferyl alcohol and sinapyl alcohol, respectively, while ALDH catalyzes the oxidation of both coniferaldehyde and sinapaldehyde to ferulic acid and sinapic acid, respectively (Nair et al. 2004). The CAD activities for coniferaldehyde and sinapaldehyde were 7.7 and 8.5 nmol min–1 mg protein–1, respectively (Table 2), whereas the ALDH activities for coniferaldehyde and sinapaldehyde were 9.6 and 36.2 pmol min–1 mg protein–1, respectively (Table 2). The ALDH activities were a few-hundredths to a few one-thousandths of the 4CL and CAD activities. Therefore, we concluded that ALDH activities have almost no impact on monolignol biosynthesis in R. pseudoacacia.
Enzyme activities to catalyze aromatic hydroxylation and methylation
Next, to determine whether a path via sinapic acid contributes to sinapyl alcohol biosynthesis, we investigated the activities of F5H and COMT, which catalyze aromatic hydroxylation of ferulic acid, coniferaldehyde, and coniferyl alcohol, and the following methylation reaction. The hydroxylation activities of xylem microsomes for ferulic acid, coniferaldehyde, and coniferyl alcohol were measured by monitoring the formation of 5-hydroxyferulic acid, 5-hydroxyconiferaldehyde, and 5-hydroxyconiferyl alcohol, respectively. The xylem microsomes showed hydroxylation activity for all three substrates with a mole ratio of 1/3.1/0.9 for ferulic acid/coniferaldehyde/coniferyl alcohol (Table 3). We also measured the hydroxylation activities for ferulic acid in the presence of coniferaldehyde or coniferyl alcohol, because coniferaldehyde has been shown to completely inhibit ferulic acid 5-hydroxylation, blocking the production of sinapic acid from ferulic acid. This has been demonstrated in various angiosperm species using in vitro experiments (Li et al. 2000; Osakabe et al. 1999). In our experiments using xylem microsomes from R. pseudoacacia, the presence of mixed substrates resulted in the reduction of a rate that converts ferulic acid to 5-hydroxyferulic acid, but did not abolish the conversion, unlike recombinant F5H and xylem microsomes derived from many other species, including aspen, as previously reported (Li et al. 2000). On the other hand, in presence of ferulic acid, 71 and 87% of 5-hydroxylation activities for coniferaldehyde and sinapyl alcohol, respectively, were lost when mixed substrates were supplied, compared with the reaction containing individual substrates.
The methylation activities of the xylem extract for 5-hydroxyferulic acid, 5-hydroxyconiferaldehyde, and 5-hydroxyconiferyl alcohol were measured by monitoring the formation of sinapic acid, sinapaldehyde, and sinapyl alcohol, respectively. Although the specific activity was higher for 5-hydroxyferulic acid than for 5-hydroxyconiferaldehyde (Table 4), the highest affinity was for 5-hydroxyconiferaldehyde, with a V max /K m value that was 10 and 3.1 times higher than those for 5-hydroxyferulic acid and 5-hydroxyconiferyl alcohol, respectively. This result indicated that 5-hydroxyconiferaldehyde is the best COMT substrate (Fig. 2). An inhibitory effect of mixed substrates has also been reported for 5-hydroxyferulic acid methylation (Li et al. 2000). When 5-hydroxyconiferaldehyde (K m = 3.81 µM) or 5-hydroxyconiferyl (K m = 7.99 µM) alcohol were present with 5-hydroxyferulic acid (K m = 62.8 µM), methylation activities for 5-hydroxyferulic acid were retained, but were lower than those recorded when each substrate was provided individually (Table 4). Although the in vitro reaction with the mixed substrates may not necessarily represent the conditions in vivo, the existence of F5H and COMT activities in the developing xylem of R. pseudoacacia does not exclude the possibility of a pathway producing sinapic acid through ferulic acid and 5-hydroxyferulic acid.
Accumulation of monolignol intermediates in developing xylem of P. alba and R. pseudoacacia
To obtain additional information about the pathway producing sinapyl alcohol, we analyzed the accumulation of monolignol intermediates in the developing xylem by GC–MS. As well as detecting enzymatic activities, detecting certain monolignol intermediates that serve as enzyme substrates can predict a practical pathway for monolignol biosynthesis. Table 5 shows the contents of monolignol intermediates in the developing xylem of P. alba and R. pseudoacacia. It was suggested that sinapic acid was not a precursor of sinapyl alcohol in P. alba by tracer experiments using deutero-labeled sinapic acid (Hamada et al. 2003). While the contents of sinapyl alcohol in P. alba and R. pseudoacacia were approximately the same, the contents of 5-hydroxyferulic acid, sinapic acid, and coniferaldehyde differed markedly between the two plant species. We detected 5-hydroxyferulic acid and sinapic acid only in R. pseudoacacia, and the concentration of coniferaldehyde in R. pseudoacacia (18.2 nmol g dry weight–1 of xylem) was much higher than that in P. alba (0.17 nmol g dry weight–1 of xylem).
Discussion
In this study, we focused on the enzymatic activities and monolignol intermediates in the developing xylem to demonstrate that sinapyl alcohol biosynthesis in R. pseudoacacia occurs via a pathway in which sinapic acid is a precursor. The substrate selectivity of the enzymes related to monolignol biosynthesis provides information about the pathway(s) that produces monolignols. A well-studied enzyme with highly selective substrate specificity is 4CL. Cinnamic acids related to monolignol biosynthesis, such as p-coumaric acid, caffeic acid, ferulic acid, 5-hydroxyferulic acid, and sinapic acid are candidates for Co-A-ligation catalyzed by 4CL. To our knowledge, A. thaliana At4CL4 (Hamberger and Hahlbrock 2004; Li et al. 2000), G. max Gm4CL1 (Lindermayr et al. 2002), Oryza sativa Os4CL5 (Gui et al. 2011; Sun et al. 2013) and R. pseudoacacia Rp4CL2 and Rp4CL3 (Hamada et al. 2004) can effectively convert sinapic acid to sinapoyl-CoA, unlike most of the 4CLs related to monolignol biosynthesis. Recent gene expression and T-DNA insertion mutant analyses revealed that At4CL4 makes a modest contribution to lignin biosynthesis (Li et al. 2015). Because the localization and substrate selectivity of enzymes related to monolignol biosynthesis differ among different species and isoforms, the pathway responsible for monolignol production may differ depending on the plant species, organ, and even cell type.
Arabidopsis accumulates a hydroxycinnamoyl ester primarily found in members of the Brassicaceae that serves as a UV protectant (Chapple et al. 1992; Landry et al. 1995). The biosynthesis of sinapoylmalate is thought to require 4CL and ALDH activities. Therefore, sinapic acid, a precursor of sinapoyl malate, can be produced through sinapaldehyde in Arabidopsis. However, there have been no reports to date that R. pseudoacacia accumulates sinapoyl malate. The ALDH activity was much lower in R. pseudoacacia (9.6 and 36.2 pmol min–1 mg protein–1 for coniferaldehyde and sinapaldehyde e, respectively, Table 2) than in Arabidopsis (2118 and 2886 pmol min–1 mg protein–1 coniferaldehyde and sinapaldehyde, respectively) (Nair et al. 2004). This result indicated that ALDH with cinnamic aldehyde substrates contributes little to the production of sinapic acid in R. pseudoacacia, and therefore has little effect on monolignol production. In addition, F5H retained its activity for ferulic acid and COMT retained its activity for 5-hydroxyferulic acid in the presence of competitive substrates. These observations did not contradict the hypothesis that the sinapyl alcohol biosynthetic pathway through ferulic acid, 5-hydroxyferulic acid, and sinapic acid operates in R. pseudoacacia.
As shown in Fig. 1, the main route to produce sinapyl alcohol in P. trichocarpa, whose relative 4CL activity for sinapic acid is extremely low, passes through coniferaldehyde (Chen et al. 2013). The monolignol intermediates detected by the GC–MS analysis of the developing xylem of P. alba (which is closely related to P. trichocarpa), were substantially coincident with the predicted metabolites in the proposed principal monolignol biosynthetic pathway in P. trichocarpa (Fig. 2). Therefore, the detected monolignol intermediates reflect the metabolic flux to a certain degree.
5-Hydroxyferulic acid and sinapic acid were detected in the developing xylem of R. pseudoacacia but not in P. alba. This result suggested that the pathway to produce sinapyl alcohol through sinapic acids plays a certain role in R. pseudoacacia but not poplar. Interestingly, a large amount of coniferaldehyde accumulated in R. pseudoacacia. Because of the low ALDH activity in R. pseudoacacia, coniferaldehyde may not recycle coniferaldehyde into ferulic acid. Also, in R. pseudoacacia, sinapic acid can serve as a sinapyl alcohol precursor, but this is not the main route of sinapyl alcohol production. The main route may pass through coniferaldehyde, like in Arabidopsis and P. trichocarpa.
In summary, these provide evidence that in R. pseudoacacia, sinapyl alcohol can be synthesized in a pathway that uses sinapic acid as substrates. Yamauchi et al. (2003) reported that heptadeuterosinapic acid was incorporated into sinapyl alcohol in R. pseudoacacia and N. indicum but not in Magnolia kobus and Arabidopsis. The fact that two of these four plants can incorporate sinapic acid into sinapyl alcohol indicates that there is some diversity or flexibility in sinapyl alcohol biosynthesis in angiosperms, and that some plant species can utilize sinapic acid as a sinapyl alcohol precursor. It is likely that R. pseudoacacia can use these pathways separately depending on the developmental stage, organ, or cell type.
References
Allina SM, Pri-Hadash A, Theilmann DA, Ellis BE, Douglas CJ (1998) 4-Coumarate:coenzyme A ligase in hybrid poplar. Properties of native enzymes, cDNA cloning, and analysis of recombinant enzymes. Plant Physiol 116:743–754
Barros J, Serk H, Granlund I, Pesquet E (2015) The cell biology of lignification in higher plants. Ann Bot 115:1053–1074
Chapple CC, Vogt T, Ellis BE, Somerville CR (1992) An Arabidopsis mutant defective in the general phenylpropanoid pathway. Plant Cell 4:1413–1424
Chen HC et al (2013) Monolignol pathway 4-coumaric acid:coenzyme A ligases in Populus trichocarpa: novel specificity, metabolic regulation, and simulation of coenzyme A ligation fluxes. Plant Physiol 161:1501–1516
Costa MA et al (2005) Characterization in vitro and in vivo of the putative multigene 4-coumarate:CoA ligase network in Arabidopsis: syringyl lignin and sinapate/sinapyl alcohol derivative formation. Phytochemistry 66:2072–2091
Cukovic D, Ehlting J, VanZiffle JA, Douglas CJ (2001) Structure and evolution of 4-coumarate:coenzyme A ligase (4CL) gene families. Biol Chem 382:645–654
Ehlting J, Buttner D, Wang Q, Douglas CJ, Somssich IE, Kombrink E (1999) Three 4-coumarate:coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. Plant J 19:9–20
Gross GG, Zenk MH (1966) Darstellung und Eigenschaften von Coenzym A-Thioestern substituierter Zimtsäuren. Z Naturforsch B 21:683–690
Gui J, Shen J, Li L (2011) Functional characterization of evolutionarily divergent 4-coumarate:coenzyme a ligases in rice. Plant Physiol 157:574–586
Hamada K, Tsutsumi Y, Nishida T (2003) Treatment of poplar callus with ferulic and sinapic acids II: effects on related monolignol biosynthetic enzyme activities. J Wood Sci 49:366–370
Hamada K, Nishida T, Yamauchi K, Fukushima K, Kondo R, Tsutsumi Y (2004) 4-Coumarate:coenzyme A ligase in black locust (Robinia pseudoacacia) catalyses the conversion of sinapate to sinapoyl-CoA. J Plant Res 117:303–310
Hamberger B, Hahlbrock K (2004) The 4-coumarate:CoA ligase gene family in Arabidopsis thaliana comprises one rare, sinapate-activating and three commonly occurring isoenzymes. Proc Natl Acad Sci 101:2209–2214
Higuchi T (2003) Pathways for monolignol biosynthesis via metabolic grids: coniferyl aldehyde 5-hydroxylase, a possible key enzyme in angiosperm syringyl lignin biosynthesis. Proc Jpn Acad B Phys 79:227–236
Hu WJ et al (1998) Compartmentalized expression of two structurally and functionally distinct 4-coumarate:CoA ligase genes in aspen (Populus tremuloides). Proc Natl Acad Sci 95:5407–5412
Humphreys JM, Hemm MR, Chapple C (1999) New routes for lignin biosynthesis defined by biochemical characterization of recombinant ferulate 5-hydroxylase, a multifunctional cytochrome P450-dependent monooxygenase. Proc Natl Acad Sci 96:10045–10050
Knobloch KH, Hahlbrock K (1975) Isoenzymes of p-coumarate: CoA ligase from cell suspension cultures of Glycine max. Eur J Biochem 52:311–320
Kumar M, Campbell L, Turner S (2016) Secondary cell walls: biosynthesis and manipulation. J Exp Bot 67:515–531
Landry LG, Chapple CC, Last RL (1995) Arabidopsis mutants lacking phenolic sunscreens exhibit enhanced ultraviolet-B injury and oxidative damage. Plant Physiol 109:1159–1166
Li L, Popko JL, Umezawa T, Chiang VL (2000) 5-Hydroxyconiferyl aldehyde modulates enzymatic methylation for syringyl monolignol formation, a new view of monolignol biosynthesis in angiosperms. J Biol Chem 275:6537–6545
Li Y, Kim JI, Pysh L, Chapple C (2015) Four isoforms of Arabidopsis 4-Coumarate:CoA ligase have overlapping yet distinct roles in phenylpropanoid metabolism. Plant Physiol 169:2409–2421
Lindermayr C et al (2002) Divergent members of a soybean (Glycine max L.) 4-coumarate:coenzyme A ligase gene family. Eur J Biochem 269:1304–1315
Lozoya E, Hoffmann H, Douglas C, Schulz W, Scheel D, Hahlbrock K (1988) Primary structures and catalytic properties of isoenzymes encoded by the two 4-coumarate: CoA ligase genes in parsley. Eur J Biochem 176:661–667
Lüderitz T, Schatz G, Grisebach H (1982) Enzymic synthesis of lignin precursors. Purification and properties of 4-coumarate:CoA ligase from cambial sap of spruce (Picea abies L.). Eur J Biochem 123:583–586
Nair RB, Bastress KL, Ruegger MO, Denault JW, Chapple C (2004) The Arabidopsis thaliana REDUCED EPIDERMAL FLUORESCENCE1 gene encodes an aldehyde dehydrogenase involved in ferulic acid and sinapic acid biosynthesis. Plant Cell 16:544–554
Osakabe K et al (1999) Coniferyl aldehyde 5-hydroxylation and methylation direct syringyl lignin biosynthesis in angiosperms. Proc Natl Acad Sci 96:8955–8960
Shi R, Sun YH, Li Q, Heber S, Sederoff R, Chiang VL (2010) Towards a systems approach for lignin biosynthesis in Populus trichocarpa: transcript abundance and specificity of the monolignol biosynthetic genes. Plant Cell Physiol 51:144–163
Souza Cde A, Barbazuk B, Ralph SG, Bohlmann J, Hamberger B, Douglas CJ (2008) Genome-wide analysis of a land plant-specific acyl:coenzyme A synthetase (ACS) gene family in Arabidopsis, poplar, rice and Physcomitrella. New Phytol 179:987–1003
Stöckigt J, Zenk MH (1975) Chemical syntheses and properties of hydroxycinnamoyl-coenzyme A derivatives. Z Naturforsch C 30:352–358
Sun H, Li Y, Feng S, Zou W, Guo K, Fan C, Si S, Peng L (2013) Analysis of five rice 4-coumarate:coenzyme A ligase enzyme activity and stress response for potential roles in lignin and flavonoid biosynthesis in rice. Biochem Biophys Res Commun 430:1151–1156
Vanholme R et al (2013) Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science 341:1103–1106
Yamauchi K, Yasuda S, Fukushima K (2002) Evidence for the biosynthetic pathway from sinapic acid to syringyl lignin using labeled sinapic acid with stable isotope at both methoxy groups in Robinia pseudoacacia and Nerium indicum. J Agric Food Chem 50:3222–3227
Yamauchi K, Yasuda S, Hamada K, Tsutsumi Y, Fukushima K (2003) Multiform biosynthetic pathway of syringyl lignin in angiosperms. Planta 216:496–501
Acknowledgements
We thank Prof. Umezawa T. and Dr. Nakatsubo T. (Research Institute for Sustainable Humanosphere, Kyoto Uniyersity) for providing the substrates (5-hydroxyfelulic acid, 5-hydoroxyconiferyl aldehyde and 5-hydroxyconiferyl alcohol). This work was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI Scientific Research (B) Grant Number 26292097 (Y.T.), JSPS KAKENHI Exploratory Research Grant Number 15K14774 (Y.T.) and JSPS KAKENHI Young Scientists (B) Grant Number 15K18724 (J.S.).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shigeto, J., Ueda, Y., Sasaki, S. et al. Enzymatic activities for lignin monomer intermediates highlight the biosynthetic pathway of syringyl monomers in Robinia pseudoacacia . J Plant Res 130, 203–210 (2017). https://doi.org/10.1007/s10265-016-0882-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10265-016-0882-4