Abstract
Peanut, like most legumes, develops a symbiotic relationship with rhizobia to overcome nitrogen limitation. Rhizobial infection of peanut roots occurs through a primitive and poorly characterized intercellular mechanism. Knowledge of the molecular determinants of this symbiotic interaction is scarce, and little is known about the molecules implicated in the recognition of the symbionts. Here, we identify the LysM extracellular domain sequences of two putative peanut Nod factor receptors, named AhNFR1 and AhNFP. Phylogenetic analyses indicated that they correspond to LjNFR1 and LjNFR5 homologs, respectively. Transcriptional analysis revealed that, unlike LjNFR5, AhNFP expression was not induced at 8 h post bradyrhizobial inoculation. Further examination of AhNFP showed that the predicted protein sequence is identical to GmNFR5 in two positions that are crucial for Nod factor perception in other legumes. Analysis of the AhNFP LysM2 tridimensional model revealed that these two amino acids are very close, delimiting a zone of the molecule essential for Nod factor recognition. These data, together with the analysis of the molecular structure of Nod factors of native peanut symbionts previously reported, suggest that peanut and soybean could share some of the determinants involved in the signalling cascade that allows symbiosis establishment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In order to avoid nitrogen limitation, legume plants are able to establish a mutualistic symbiotic association with soil bacteria collectively known as rhizobia. Symbiosis development depends on a molecular dialogue between plants and bacterial symbionts, and specific molecular signals are necessary for mutual recognition between the partners. Rhizobial Nod factors (NFs) are among the best characterized of these molecules. NFs consist of a backbone of 3–5 units of N-acetyl-glucosamine acylated on the non-reducing glucosamine with a fatty acid of variable length and degree of unsaturation. In addition, different species specific NFs substitutions (glycosylations, sulfations, acetylations, methylations) determine host specificity (Perret et al. 2000).
In most known legumes, perception of specific NFs structure triggers a signalling cascade that allows the establishment of nitrogen fixing symbiosis. Genetic analyses allowed the identification of NFs receptors in model legumes. In Lotus japonicus, NFs are perceived by two Receptor-Like Kinases (RLKs), LjNFR1 and LjNFR5, which are predicted to form a heterocomplex (Madsen et al. 2011). Direct binding of NFs to receptors LjNFR1 and LjNFR5 has been experimentally demonstrated (Broghammer et al. 2012). LjNFR1 and LjNFR5 belong to the lysin motif class of RLKs (LysM-RLKs). They are localized in the plasma membrane and possess a conserved structure consisting of an intracellular region exhibiting homology to a serine/threonine kinase, a single pass transmembrane portion and an extracellular region containing three LysM domains separated by characteristic CXC (Cysteine-any amino acid-Cysteine) motifs (Madsen et al. 2003; Radutoiu et al. 2003). LjNFR1 and its orthologs LYK3 of Medicago truncatula and SYM37 of Pisum sativum contain an active protein kinase domain. Moreover, LjNFR1 is capable of autophosphorylating in 24 residues and also phosphorylates LjNFR5 in vitro (Madsen et al. 2011). In contrast, LjNFR5 and its orthologs MtNFP and PsSYM10 contain an inactive kinase domain (Arrighi et al. 2006; Madsen et al. 2003; Radutoiu et al. 2003).
LysM domains are protein motifs of about 44–65 amino acid originally described in bacterial autolysins (Birkeland 1994; Joris et al. 1992) and later found in many eukaryotic proteins, associated with diverse domains (Buist et al. 2008). However, association between LysM and protein kinase domains seems to takes place exclusively in plants (Ponting et al. 1999). LysM domains are implicated in the binding of peptidoglycan and chitin, most likely recognizing the N-acetylglucosamine moiety. In legumes, LysM domains of RLKs mediate rhizobial NFs perception. In addition, it was recently shown that NFP gene also participates in M. truncatula resistance to pathogens (Gough and Jacquet 2013; Rey et al. 2013). Interestingly, LysM domain sequences within RLKs diverge considerably. Bensmihen et al. (2011) reported that each LysM domain of MtNFP contributes with different relevance to the function of the receptor. The authors also demonstrated the importance of the NFP LysM2 domain for rhizobial infection and identified a single leucine residue within LysM2 (Leu154) that is essential for nodulation. Similarly, a single leucine residue of LjNFR5 (Leu118 within LysM2) is required for specific recognition of Rhizobium leguminosarum DZL NFs (Radutoiu et al. 2007).
Different root invasion processes and nodule morphogenetic programmes have been described in legume–rhizobia symbiosis (Oldroyd and Downie 2008). The most studied invasion mechanism involves infection thread formation, and occurs in most legumes (Lotus japonicus, Medicago truncatula, Vicia, Trifolium, Pisum, Phaseolus, Glycine, Vigna, Macroptilium among others). An alternative and less investigated mode of infection involves intercellular penetration of roots. This mechanism is considered more primitive, and remains poorly characterized. Interestingly, in the usually root-hair infected L. japonicus, unusual intercellular crack entry infection process and formation of trans-cellular infection threads within the root nodules have been reported in absence of functional NFR1 and NFR5 receptors (Madsen et al. 2010). Similarly, a NF independent crack entry invasion of Glycine max roots has also been described (Okazaki et al. 2013). This process depends on the presence of an intact type III secretion system (T3SS) in the microsymbiont, which is used to deliver into the host molecules that activate the symbiotic signalling cascade. These findings support the hypothesis that alternative invasion modes have been maintained during evolution and that they are not mutually exclusive (Madsen et al. 2010). Arachis hypogaea L. (peanut) exhibits a particular mode of intercellular invasion and nodule morphogenetic programme. In this legume, infection threads never form, not for epidermal or cortical invasion or for nodular dissemination (Boogerd and van Rossum 1997). Peanut rhizobia penetrate the root at the junction of a hair cell with epidermal and cortical cells through structurally altered cell walls (Uheda et al. 2001). Within cortical cells, rhizobia multiply and the infected cells divide repeatedly, giving raise to the typical aeschynomenoid nodules whose core infected zone is not interspersed with uninfected cells (Boogerd and van Rossum 1997). In previous work, we demonstrated that development of such particular nodule morphogenetic programme depends on the perception of rhizobial NFs for the reinitiation of meristematic activity of cortical cells to form the nodule primordia (Ibañez and Fabra 2011). However, peanut NFs receptors and the components of the symbiotic signalling pathway initiated by their recognition have not been studied in detail, despite some exceptions such as CCaMK (Sinharoy and DasGupta 2009). To a certain extent, this missing information is due to the lack of genomic and molecular-genetic resources associated with peanut.
With the aim of gaining insight into the determinants involved in early recognition events in peanut-bradyrhizobium symbiosis, in this work we identify putative peanut Nod factor receptors that are named AhNFR1 and AhNFP, as well as NFR1 paralog copies. We also examined the three dimensional structure of the deduced amino acid sequence of AhNFP and analyzed its expression profile after bradyrhizobial inoculation. Advances in the knowledge of the molecular basis of intercellular infection will contribute to the understanding of the evolution of symbiosis, and may allow the identification of target molecules for further improving of the association through biotechnological applications.
Materials and methods
Identification of Arachis hypogaea L. Nod factor receptors in sequence databases
In order to identify LjNRF1 and LjNFR5 homologs in available peanut sequences, BLASTN searches (Altschul et al. 1997) were performed against a consensus transcriptome assembly developed for tetraploid peanut (Pandey et al. 2012, available at http://nespal.org/oziasakinslab) and PeanutDB 1.0 (Duan et al. 2012).
Plant growth conditions and DNA extraction
A. hypogaea L. (var. Granoleico) seeds were surface sterilized by soaking in ethanol for 30 s followed by 6 % H2O2 for 15 min, and then washed six times with sterile distilled water. Seeds were germinated at 28 °C in sterilized Petri dishes with one layer of Whatman No. 1 filter paper and moist cotton, until the radicle reached approximately 2 cm. Seedlings were sown in previously sterilized plastic cups filled with sand. Plants were grown under controlled environment (light intensity of 200 µE m−2 s−1, 16-h day/8-h night cycle, at a constant temperature of 28 °C and a relative humidity of 50 %), and watered regularly with Hoagland’s solution (Hoagland and Arnon 1950). Plants were harvested 3 weeks after emergence and DNA was extracted from young leaves with NucleoSpin Plant II Genomic DNA extraction kit (Macherey–Nagel, Düren, Germany) according to the manufacturer’s instructions.
For inoculation experiments, plants were grown and spot-inoculated as described previously (Ibañez and Fabra 2011). Briefly, plantlets grown between the wall of a beaker and a rolled filter paper were inoculated with a Bradyrhizobium sp. SEMIA6144 bacterial culture (MIRCEN, Brazil) in the junction of a first- and second-order lateral roots. Inoculated roots were sampled at 8 h PI (post inoculation). Uninoculated roots were used as controls.
Amplification of Arachis hypogaea L. NFP gene
PCR primers were designed based on conserved sequences of Nod factor perception proteins from the legumes G. max (ABQ59609), M. truncatula (CAO02950 and AET03984), Melilotus dentatus (AEN71536), and L. japonicus (CAE02597). These primers, named AhNFP-F and AhNFP-R (Table S1), amplify a fragment of approximately 509 bp of NFP gene (AhNFP). PCR was performed in a 20 μl reaction mixture containing 1X PCR buffer, 1.5 mM MgCl2, 200 μM of each nucleotide (Promega), 1 μM of each primer, 1 U of Taq DNA polymerase (Promega) and 1 μl of template DNA solution. Temperature profile was as follows: initial denaturation at 95 °C for 3 min; 35 cycles of denaturation at 94 °C for 1 min, annealing at 57 °C for 1 min, extension at 72 °C for 1 min, and a final extension step at 72 °C for 10 min.
Amplification products in 5 μl sub-samples were separated according to molecular size by horizontal electrophoresis on 1.2 % (w/v) agarose gels stained with SYBR green (molecular probes).
Sequence analysis
A PCR product of the expected size (509 bp) was cloned into pGEM-T vector (Promega). Afterwards, the cloned fragment was sequenced with M13 primers by Macrogen (Korea).
Homologous sequences were identified using BLASTN and BLASTX algorithms (Altschul et al. 1997). For sequence analysis, LysM-RLKs sequences were downloaded from the GenBank database and aligned using ClustalW (Larkin et al. 2007).
Phylogenetic analysis was conducted using PhyML (Guindon and Gascuel 2003). Best fit substitution models were selected according to MEGA 5 software (Tamura et al. 2011).
AhNFP LysM2 model building
Modeling of the three dimensional structure of the LysM2 domain of AhNFP protein was performed by Swissmodel homology modeling programs (Arnold et al. 2006). Tertiary structure was predicted using homology modeling by taking as template the NMR structure of a LysM domain from E. coli MLTD, a membrane-bound lytic murein transglycosylase D (Bateman and Bycroft 2000).
RNA isolation and preparation of peanut cDNA
RNA was prepared from three replicates samples of 4 plants each that were pooled to reduce noise arising from biological variations. Susceptible root zones from spot inoculated and non-inoculated plants were ground with sterile mortars and pestles under liquid nitrogen. Total RNA was isolated using the NucleoSpin RNA Clean-Up Kit (Macherey–Nagel) according to the manufacturer’s protocol. First strand cDNA synthesis was carried out using MMLV Reverse Transcriptase 1st Strand cDNA Synthesis Kit (epicentre) according to the manufacturer’s specifications with oligodT primers. The reaction conditions for cDNA synthesis consisted of 2 min at 65 °C, followed by 60 min at 37 °C and finally 5 min at 85 °C.
Quantitative real-time PCR (q-PCR) of AhNFP
Real Time-PCR was performed in 20 μl reaction mixtures containing 1X Brilliant SYBR green QPCR master mix (Stratagene), 1:10 dilution of synthesized cDNA and 200 nM of AhNFP139F and AhNFP139R primers (Table S1). Results obtained from the different treatments were standardized to the Actin mRNA level, which was amplified with 200 nM of the primers PA1-Right and PA-1Left (Table S1). Real-Time PCR determinations were carried out with cDNA synthesized from RNA that was extracted from three independent biological samples, and the threshold cycle (Ct) was determined in triplicates. Relative levels of transcription were calculated using the 2−ΔΔCt method (Livak and Schmittgen 2001). Melting curves were recorded after cycle 40 by heating from 55 to 95 °C.
Statistical analysis
Data were analysed using SigmaStat Software v3.5. A p ≤ 0.05 significance level was used throughout.
Accession numbers
Nucleotide sequence of AhNFP gene was deposited in GenBank databank under accession number JX860404.
Results
LjNFR1 orthologs are present in peanut genome
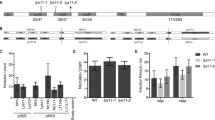
Fragments of mRNA sequences encoding LysM extracellular domains from LjNFR1 and LjNFR5 (Genbank AJ575248 and AJ575254, respectively) were used to search within A. hypogaea sequence databases. A total of four EST contigs showing significant similarity to LjNFR1 were identified (c5561, c6035, c18043 and c85956). Contig sequences were translated in silico and BLASTP analysis reported significant alignments with extracellular domains of RLKs from several legumes. Further analyses indicated that identity between proteins sequences predicted from contigs c85956 and c5561 was 99 %. Therefore, only c6035, c85956 and c18043 were used for additional studies. Three extracellular LysM domains were detected in the predicted protein sequences, with typical CXC motifs in the interspacers domains lying between LysM1-LysM2 and LysM2-LysM3 (Fig. 1a). Phylogenetic analysis (Fig. 1b) revealed that the protein encoded by contig c85956 belongs to a group of LjNFR1 orthologs. Hence, it was named AhNFR1. This predicted protein sequence shows high identity and similarity percentages with extracellular domains of RLKs from several legumes (Table 1). On the other hand, protein sequences predicted from c6035 to c18043 contigs belong to a group containing LjNFR1 paralog copies.
Similarity and phylogenetic relationship of NFR1-like Nod factor perception proteins among different legumes. a Alignment of NFR1-like Nod factor perception proteins from different legumes. Grey boxes indicate LysM domains. Empty boxes indicate CXC motifs in the inter-domain spacer regions. Genbank accession numbers of the provided sequences are as follows: GmNFR1A (ABB30246), MtLYK3 (XP_003616972), PsSYM37A (ACE81772), LjNFR1a (CAE02589), LjNFR1b (CAE02590). b Phylogenetic tree of different LYK proteins inferred using the maximum-likelihood method and JTT matrix-based model (Jones et al. 1992). A discrete Gamma distribution was used to model evolutionary rate differences among sites (+G = 1.003). References: Gm, Glycine max; Lj, Lotus japonicus; Lp, Lotus pedunculatus; Mt, Medicago truncatula, Ps, Pisum sativum. Denomination of phylogenetic groups was performed according to Zhang et al. (2007)
In contrast, no sequences showing significant alignments to the portion encoding for the extracellular LysM domains of LjNFR5 were obtained in any of the two databases analyzed. The same result was obtained using GmNFR5 as query (Indrasumunar et al. 2010).
Identification and sequencing of AhNFR5 (LjNFR5 homolog) in peanut
A PCR-based strategy was used for identifying LjNFR5 peanut ortholog. In a first approach, the primer pair RosidNFP0-F/R was used for PCR amplification. Although a band of the expected size was obtained using G. max genomic DNA as template (positive control), no PCR-products were obtained for A. hypogaea. Further sequencing of the amplicon from soybean confirmed its similarity with GmNFR5A (data not shown).
In a second approach, a primer pair (AhNFP-F/R) based on conserved sequences of NF perception proteins from different legumes was designed. By using these primers, a single amplicon of the expected size (509 bp) was obtained. The PCR product was cloned and sequenced. Analysis of the obtained sequence showed high identity percentages with genes encoding LjNFR5-like RLKs of several legumes. Afterwards, the DNA sequence was translated in silico and the predicted protein sequence obtained revealed high similarity and identity percentages with NF perception proteins from several legumes (Table 2). Subsequently, it was determined that the sequence obtained corresponds to a portion of the gene that encodes for 169 amino acids of the extracellular region of AhNFP, and includes the typical three extracellular LysM domains of NF receptors and CXC motifs in the two spacers lying between LysM1-LysM2 and LysM2-LysM3 domains (Fig. 2a).
Similarity of NFR5-like Nod factor perception proteins among different legumes and a model of LysM2 of AhNFP. a Alignment of NFR5-like Nod factor perception proteins from different legumes. Grey boxes indicate LysM domains. Asterisk indicates position 118 of Lotus japonicus NFR5 sequence (CAE02597). Dragger indicates position 154 of Medicago truncatula NFP sequence (AET75784). Empty boxes indicate CXC motifs in the inter-domain spacer regions. b Model of LysM2 of AhNFP. Residue in grey represents Q (glutamine) in position 63 of AhNFP. Residue in black represents I (Isoleucine) in position 98 of AhNFP. Genbank accession numbers of the provided sequences are as follows: MtNFP (AET75784), LpNFR5 (CAZ66917), LjNFR5 (CAE02597), GsNFR5 (BAG85157), GmNFR5A (BAG85175)
The general topology of an unrooted phylogenetic tree constructed with AhNFP and different RLKs of M. truncatula, G. max, L. japonicus and Arabidopsis thaliana indicated that they grouped in three clades (Fig. 3). One of them (LjNFR5 paralogs I) contains GmLYK8, AtLYK5 and GmLYK4 sequences. The second one (LjNFR5 paralogs II) includes AtLYK2 and GmLYK10, while LjNFR5 and its orthologs MtNFP and GmNFR5A and B are included in the third clade. Interestingly, AhNFP falls within this group, closely related to LjNFR5.
Unrooted phylogenetic tree of different LYK proteins. The tree was inferred using the maximum-likelihood method and Whelan and Goldman model (Whelan and Goldman 2001). A discrete Gamma distribution was used to model evolutionary rate differences among sites (+G = 2.717). References: At, Arabidopsis thaliana, Gm, Glycine max; Lj, Lotus japonicus; Mt, Medicago truncatula. Denomination of phylogenetic groups was performed according to Zhang et al. (2007)
Tridimensional modeling of AhNFP LysM2
A tridimensional homology model was obtained for AhNFP LysM2 (Fig. 2b), a domain crucial for NFs perception in L. japonicus and M. truncatula (Bensmihen et al. 2011; Radutoiu et al. 2007). Examination of the model reveals close proximity between two positions that proved to be essential for the recognition of specific NFs in these legumes (Leu 118 in L. japonicus and Leu 154 in M. truncatula, representing positions 63 and 98 of AhNFP). Proximity predicted between these positions in the model suggests that they define a well-delimited area of the molecule directly related to NFs perception. In these positions, AhNFP possesses two residues that are identical to those found in GmNFR5 (glutamine and isoleucine, respectively).
Transcriptional analysis of AhNFP
In order to analyse the AhNFP expression profile at early times post-bradyrhizobial inoculation, qRT-PCR experiments were performed. Results demonstrated that at 8, 16 and 24 h post-inoculation (PI), no significant differences in AhNFP transcript levels were observed when compared to uninoculated roots (Fig. 4).
Discussion
Peanut is a very economically important legume that belongs to the family Fabaceae, subfamily Papilionoideae and to the aeschynomenoid/dalbergioid clade (Doyle and Luckow 2003). This particular legume clade includes species such as Aeschynomene indica and A. sensitiva that can be nodulated without NFs and others such as A. afraspera which depend on NFs for nodulation (Bonaldi et al. 2011; Giraud et al. 2007). Interestingly, peanut roots are invaded through a NF dependent intercellular mechanism (Ibañez and Fabra 2011), and it constitutes a very interesting model for studying evolution of nitrogen fixing symbiosis. In contrast to other crops, the peanut genome is not fully sequenced yet. Moreover, it lacks taxonomic proximity to any major model legume and there are few studies dealing with the molecular determinants involved in peanut-rhizobia symbiotic association. In this work, we obtained and analyzed the sequence, tridimensional modelling and expression profile of putative peanut NF receptors.
It is known that LysM-RLKs belong to an important family that has undergone duplication and functional diversification in legumes, leading to the emergence of different members of the family (Arrighi et al. 2006). In order to identify peanut LysM-RLKs, homology searches were performed against a transcriptome assembly developed for tetraploid peanut plants. This database includes 211,244 contigs and was generated by combining publicly-available sequences with data from more than 350 Mb of transcript sequences generated by the University of Georgia (UGA), USA (Pandey et al. 2012). In addition, searches were also performed against PeanutDB 1.0 (Duan et al. 2012), which contains a total of 198,156 sequences including peanut core nucleotide sequences, ESTs and Genome Survey Sequences. Three different EST contigs showing significant similarity to LjNFR1 and typical features of this receptor (three extracellular LysM domains with CXC motifs in the interspacers domains between LysM1-LysM2 and LysM2-LysM3) were identified. Phylogenetic analysis indicated that one of them (named AhNFR1) represents LjNFR1 peanut ortholog and the others correspond to paralog copies. In contrast, no sequences showing significant alignments to the extracellular LysM domains of LjNFR5 or GmNFR5 were obtained. Absence of these sequences within EST databases could be related to a low level of expression of the gene. Therefore, a PCR based approach using primer pair RosidNFP0-F/R (Op den Camp et al. 2011) was used to identify the LjNFR5 ortholog in peanut. However, no amplification product was obtained using peanut DNA as template. Lack of amplification in peanut was unexpected, since this primer pair was shown to be useful for LysM-RLK amplification not only in a legume (soybean) but also in the phylogenetically distant non-legume P. andersonii. Therefore, degenerated primers were constructed from conserved regions of LysM-RLKs of different legumes. Due to the degenerated sequence of these primers and the fact that they were designed from conserved regions of LysM-RLKs from several legumes, they could be helpful for identification and sequencing of genes encoding for NF perception proteins in other species that, as peanut, lack of many sequences deposited in databases. By using this primer pair, we obtained a sequence corresponding to a putative peanut NF receptor denominated AhNFP. Phylogenetic analysis confirmed that it corresponds to a peanut ortholog of LjNFR5, and is closely related to this receptor. Relatedness between A. hypogaea and L. japonicus sequences is not unexpected, since previous data indicated similarity between genomic sequences and the codon usage pattern of these legumes (Jayashree et al. 2005). Further analysis of the AhNFP sequence revealed that it shows typical features of LysM-RLKs, including three LysM extracellular domains and CXC motifs in the spacer region between them. Cysteine residues of AhNFP CXC motifs aligned with those of MtNFP found in positions 102, 104, 164 and 166. In MtNFP, Cys166 is essential for NFP activity, and either Cys102 or Cys104 should be present for proper NFP activity, being Cys164 required if Cys104 is mutated (Lefebvre et al. 2012). Considering that LysM2 is crucial for specific NF perception in M. truncatula and L. japonicus (Bensmihen et al. 2011; Radutoiu et al. 2007), a tridimensional model for this domain was obtained. Analysis of the model indicated that the region for NFs perception has similar molecular features to those of GmNFR5, and led us to speculate that AhNFP and GmNFR5 could recognize structurally related lipochitooligosaccharides. Therefore, we compared the previously described structure of NFs from the native peanut symbionts Bradyrhizobium sp. NLH25, Bradyrhizobium sp. NDEHE and Bradyrhizobium sp. NOD31 (Taurian et al. 2008) with those produced by known rhizobia. In accordance with our interpretation, the structure of NFs produced by native peanut symbionts is similar to that synthesized by soybean symbiont Bradyrhizobium elkanii USDA61 (Carlson et al. 1993; Stokkermans et al. 1996). Moreover, these similarities (fatty acid moieties C18:1 and C16:0 and methyl and carbamoyl substituents) are mostly found in the non-reducing end of the NFs backbone, which is predicted to be recognized by NFs perception proteins in M. truncatula and L. japonicus (Bensmihen et al. 2011; Radutoiu et al. 2007).
The expression profile of AhNFP gene revealed that it was not induced at 8, 16 or 24 h PI. Interestingly, for Lotus-Mesorhizobium interaction, Lohmann et al. (2010) reported a slight but statistically significant increase in NFR5 expression at 8 h PI. Considering that both Lotus and peanut develop determinate nodules after interaction with their specific microsymbionts, variations in the receptor expression pattern between these legumes could not be attributed to the nodule morphogenetic programme, but could instead reflect differences between two dissimilar mechanisms of rhizobial invasion (infection thread vs. intercellular invasion). However, further work will be performed in order to test this hypothesis.
Taken together, results from this work allowed identifying sequences coding for LysM-RLKs representing putative NF receptors in an intercellularly infected legume lacking of many genomic and molecular resources. Further sequence and modelling analysis indicated that, despite considerable overall sequence similarity between L. japonicus and A. hypogaea genomes, peanut could share with soybean some of the determinants involved in the symbiotic signalling cascade implicated in nitrogen fixing symbiosis development. Moreover, transcriptional analysis of AhNFP suggests differences in the NF receptor expression pattern between A. hypogaea intercellular and L. japonicus infection thread mechanisms. Considering the particular characteristics of the nodule morphogenetic programme that occurs in peanut, it will be interesting to determine the cortical or epidermal localization of NFs receptors and their functional implication in symbiosis development. These results could shed light on the evolution of symbiosis, and the transition from the more primitive intercellular mode to the more elaborated infection thread via of rhizobial invasion.
References
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201
Arrighi JF, Barre A, Ben Amor B, Bersoult A, Soriano LC, Mirabella R, de Carvalho-Niebel F, Journet EP, Ghérardi M, Huguet T, Geurts R, Dénarié J, Rougé P, Gough C (2006) The Medicago truncatula lysin motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol 142:265–279
Bateman A, Bycroft M (2000) The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D MltD. J Mol Biol 299:1113–1119
Bensmihen S, de Billy F, Gough C (2011) Contribution of NFP LysM domains to the recognition of Nod factors during the Medicago truncatula/Sinorhizobium meliloti symbiosis. PLoS One 6:e26114. doi:10.1371/journal.pone.0026114
Birkeland NK (1994) Cloning, molecular characterization, and expression of the genes encoding the lytic functions of lactococcal bacteriophage phi LC3: a dual lysis system of modular design. Can J Microbiol 40:658–665
Bonaldi K, Gargani D, Prin Y, Fardoux J, Gully D, Nouwen N, Goormachtig S, Giraud E (2011) Nodulation of Aeschynomene afraspera and A. indica by photosynthetic Bradyrhizobium sp. strain ORS285: the nod-dependent versus the nod-independent symbiotic interaction. Mol Plant Microbe Interact 24:1359–1371
Boogerd F, van Rossum D (1997) Nodulation of groundnut by Bradyrhizobium: a simple infection process by crack entry. FEMS Microbiol Rev 21:5–27
Broghammer A, Krusell L, Blaise M, Sauer J, Sullivan JT, Maolanon N, Vinther M, Lorentzen A, Madsen EB, Jensen KJ, Roepstorff P, Thirup S, Ronson CW, Thygesen MB, Stougaard J (2012) Legume receptors perceive the rhizobial lipochitin oligosaccharide signal molecules by direct binding. Proc Natl Acad Sci USA 109:13859–13864
Buist G, Steen A, Kok J, Kuipers OP (2008) LysM, a widely distributed protein motif for binding to peptidoglycans. Mol Microbiol 68:838–847
Carlson RW, Sanjuan J, Bhat UR, Glushka J, Spaink HP, Wijfjes AH, van Brussel AA, Stokkermans TJ, Peters NK, Stacey G (1993) The structures and biological activities of the lipo-oligosaccharide nodulation signals produced by type I and II strains of B. japonicum. J Biol Chem 268:18372–18381
Doyle JJ, Luckow MA (2003) The rest of the iceberg. Legume diversity and evolution in a phylogenetic context. Plant Physiol 131:900–910
Duan X, Schmidt E, Li P, Lenox D, Liu L, Shu C, Zhang J, Liang C (2012) PeanutDB: an integrated bioinformatics web portal for Arachis hypogaea transcriptomics. BMC Plant Biol 19(12):94
Giraud E, Moulin L, Vallenet D, Barbe V, Cytryn E, Avarre JC, Jaubert M, Simon D, Cartieaux F, Prin Y, Bena G, Hannibal L, Fardoux J, Kojadinovic M, Vuillet L, Lajus A, Cruveiller S, Rouy Z, Mangenot S, Segurens B, Dossat C, Franck WL, Chang WS, Saunders E, Bruce D, Richardson P, Normand P, Dreyfus B, Pignol D, Stacey G, Emerich D, Verméglio A, Médigue C, Sadowsky M (2007) Legumes symbioses: absence of Nod genes in photosynthetic bradyrhizobia. Science 316:1307–1312
Gough C, Jacquet C (2013) Nod factor perception protein carries weight in biotic interactions. Trends Plant Sci 18:566–574
Guindon S, Gascuel O (2003) PhyML-A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Hoagland D, Arnon D (1950) Water culture method for growing plants without soil. Calif Agric Exp Stn 347:1–32
Ibañez F, Fabra A (2011) Rhizobial Nod factors are required for cortical cell division in the nodule morphogenetic programme of the Aeschynomeneae legume Arachis. Plant Biol 13:794–800
Indrasumunar A, Kereszt A, Searle I, Miyagi M, Li D, Nguyen CD, Men A, Carroll BJ, Gresshoff PM (2010) Inactivation of duplicated nod factor receptor 5 (NFR5) genes in recessive loss-of-function non-nodulation mutants of allotetraploid soybean (Glycine max L. Merr.). Plant Cell Physiol 51:201–214
Jayashree B, Ferguson M, Ilut D, Doyle J, Crouch JH (2005) Analysis of genomic sequences from peanut (Arachis hypogaea). Electron J Biotech 8:226–237
Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282
Joris B, Englebert S, Chu CP, Kariyama R, Daneo-Moore L, Shockman GD, Ghuysen JM (1992) Modular design of the Enterococcus hirae muramidase-2 and Streptococcus faecalis autolysin. FEMS Microbiol Lett 70:257–264
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Lefebvre B, Klaus-Heisen D, Pietraszewska-Bogiel A, Hervé C, Camut S, Auriac MC, Gasciolli V, Nurisso A, Gadella TW, Cullimore J (2012) Role of N-glycosylation sites and CXC motifs in trafficking of Medicago truncatula Nod factor perception protein to the plasma membrane. J Biol Chem 287:10812–10823
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–408
Lohmann GV, Shimoda Y, Nielsen MW, Jørgensen FG, Grossmann C, Sandal N, Sørensen K, Thirup S, Madsen LH, Tabata S, Sato S, Stougaard J, Radutoiu S (2010) Evolution and regulation of the Lotus japonicus LysM receptor gene family. Mol Plant Microbe Interact 23:510–521
Madsen EB, Madsen LH, Radutoiu S, Olbryt M, Rakwalska M, Szczyglowski K, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J (2003) A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425:637–640
Madsen LH, Tirichine L, Jurkiewicz A, Sullivan JT, Heckmann AB, Bek AS, Ronson CW, James EK, Stougaard J (2010) The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat Commun 1:10
Madsen EB, Antolín-Llovera M, Grossmann C, Ye J, Vieweg S, Broghammer A, Krusell L, Radutoiu S, Jensen ON, Stougaard J, Parniske M (2011) Autophosphorylation is essential for the in vivo function of the Lotus japonicus Nod factor receptor 1 and receptor-mediated signalling in cooperation with Nod factor receptor 5. Plant J 65:404–417
Okazaki S, Kaneko T, Sato S, Saeki K (2013) Hijacking of leguminous nodulation signaling by the rhizobial type III secretion system. Proc Natl Acad Sci USA 110:17131–17136
Oldroyd G, Downie J (2008) Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol 59:519–546
Op den Camp R, Streng A, De Mita S, Cao Q, Polone E, Liu W, Ammiraju JS, Kudrna D, Wing R, Untergasser A, Bisseling T, Geurts R (2011) LysM-type mycorrhizal receptor recruited for rhizobium symbiosis in nonlegume Parasponia. Science 331:909–912
Pandey MK, Monyo E, Ozias-Akins P, Liang X, Guimarães P, Nigam SN, Upadhyaya HD, Janila P, Zhang X, Guo B, Cook DR, Bertioli DJ, Michelmore R, Varshney RK (2012) Advances in Arachis genomics for peanut improvement. Biotechnol Adv 30:639–651
Perret X, Staehelin C, Broughton W (2000) Molecular basis of symbiotic promiscuity. Microbiol Mol Biol Rev 64:180–201
Ponting CP, Aravind L, Schultz J, Bork P, Koonin EV (1999) Eukaryotic signalling domain homologues in archaea and bacteria. Ancient ancestry and horizontal gene transfer. J Mol Biol 289:729–745
Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Grønlund M, Sato S, Nakamura Y, Tabata S, Sandal N, Stougaard J (2003) Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425:585–592
Radutoiu S, Madsen LH, Madsen EB, Jurkiewicz A, Fukai E, Quistgaard EM, Albrektsen AS, James EK, Thirup S, Stougaard J (2007) LysM domains mediate lipochitin-oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO J 26:3923–3935
Rey T, Nars A, Bonhomme M, Bottin A, Huguet S, Balzergue S, Jardinaud MF, Bono JJ, Cullimore J, Dumas B, Gough C, Jacquet C (2013) NFP, a LysM protein controlling Nod factor perception, also intervenes in Medicago truncatula resistance to pathogens. New Phytol 198:875–886
Sinharoy S, DasGupta M (2009) RNA interference highlights the role of CCaMK in dissemination of endosymbionts in the Aeschynomeneae legume Arachis. Mol Plant Microbe Interact 22:1466–1475
Stokkermans TJW, Orlando R, Kolli VSK, Carlson RW, Peters NK (1996) Biological activities and structures of Bradyrhizobium elkanii low abundance lipo chitin-oligosaccharides. Mol Plant Microbe Interact 9:298–304
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol 28:2731–2739
Taurian T, Morón B, Soria-Díaz ME, Angelini JG, Tejero-Mateo P, Gil-Serrano A, Megías M, Fabra A (2008) Signal molecules in the peanut-bradyrhizobia interaction. Arch Microbiol 189:345–356
Uheda E, Daimon H, Yoshizako F (2001) Colonization and invasion of peanut Arachis hypogea L. roots by gusA-marked Bradyrhizobium sp. Can J Bot 79:733–738
Whelan S, Goldman N (2001) A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol 18:691–699
Zhang XC, Wu X, Findley S, Wan J, Libault M, Nguyen HT, Cannon SB, Stacey G (2007) Molecular evolution of lysin motif-type receptor-like kinases in plants. Plant Physiol 144:623–636
Acknowledgments
The authors would like to thank Dr. Peggy Ozias-Akins and Dr. Yuofang Guo (UGA, Athens) for performing BLAST searches and providing peanut contig sequences. This study was financially supported by the SECyT-UNRC, CONICET, Ministerio de Ciencia y Tecnología de Córdoba and ANPCyT grants. V. Muñoz and M. Figueredo are recipients of scholarships from CONICET. F. Ibáñez, J. Angelini, M. L. Tonelli and A. Fabra are members of the Research Career from CONICET.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ibáñez, F., Angelini, J., Figueredo, M.S. et al. Sequence and expression analysis of putative Arachis hypogaea (peanut) Nod factor perception proteins. J Plant Res 128, 709–718 (2015). https://doi.org/10.1007/s10265-015-0719-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10265-015-0719-6