ABSTRACT
Four CreER lines that are commonly used in the auditory field to label cochlear supporting cells (SCs) are expressed in multiple SC subtypes, with some lines also showing reporter expression in hair cells (HCs). We hypothesized that altering the tamoxifen dose would modify CreER expression and target subsets of SCs. We also used two different reporter lines, ROSA26 tdTomato and CAG-eGFP, to achieve the same goal. Our results confirm previous reports that Sox2 CreERT2 and Fgfr3-iCreER T2 are not only expressed in neonatal SCs but also in HCs. Decreasing the tamoxifen dose did not reduce HC expression for Sox2 CreERT2, but changing to the CAG-eGFP reporter decreased reporter-positive HCs sevenfold. However, there was also a significant decrease in the number of reporter-positive SCs. In contrast, there was a large reduction in reporter-positive HCs in Fgfr3-iCreER T2 mice with the lowest tamoxifen dose tested yet only limited reduction in SC labeling. The targeting of reporter expression to inner phalangeal and border cells was increased when Plp-CreER T2 was paired with the CAG-eGFP reporter; however, the total number of labeled cells decreased. Changes to the tamoxifen dose or reporter line with Prox1 CreERT2 caused minimal changes. Our data demonstrate that modifications to the tamoxifen dose or the use of different reporter lines may be successful in narrowing the numbers and/or types of cells labeled, but each CreER line responded differently. When the ROSA26 tdTomato reporter was combined with any of the four CreER lines, there was no difference in the number of tdTomato-positive cells after one or two injections of tamoxifen given at birth. Thus, tamoxifen-mediated toxicity could be reduced by only giving one injection. While the CAG-eGFP reporter consistently labeled fewer cells, both reporter lines are valuable depending on the goal of the study.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
The mammalian cochlea is a highly organized structure containing sensory hair cells (HCs) surrounded by supporting cells (SCs), which are divided into subtypes based on location (Fig. 1A). Cells of the greater epithelial ridge (GER), inner phalangeal cells (IPhCs), and border cells (BCs) are located medial to inner HCs (IHCs). Inner pillar cells (IPCs) and outer pillar cells (OPCs) separate IHCs and outer HCs (OHCs). Deiters’ cells (DCs) surround OHCs with Hensen cells (HeCs) and Claudius cells (CCs) located lateral to the last row of OHCs (Raphael and Altschuler 2003). While HCs and SCs are distinct cell types, they are derived from the same pool of progenitor cells (Yang et al. 2010; Cai et al. 2013; Driver et al. 2013).
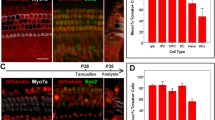
F gfr3-iCreER T2 is expressed in the majority of PCs/DCs. A Cross-sectional view of the mouse organ of Corti with SC subtypes highlighted in different colors. The tunnel of Corti is partially open to represent the immature state of the neonatal cochlea. GER, greater epithelial ridge; SGN, spiral ganglion neuron. B Quantification of total reporter-positive SCs in Fgfr3-iCreER T2/+ ::ROSA26 tdTomato/+ mice or Fgfr3-iCreER T2/+ ::CAG-eGFP +/loxP mice given different doses of tamoxifen (TAM). Cells were quantified in two randomly chosen 200 μm regions per cochlear turn, averaged, and expressed as a percentage of labeled cells compared to total cells. (**P < 0.01, ***P < 0.001 as determined by a two-way ANOVA F(3, 24) = 274.5 with a Tukey’s post hoc test) (N = 3–4). Representative confocal images of tdTomato (red) expression in Fgfr3-iCreER T2/+ ::ROSA26 tdTomato/+ controls without tamoxifen (C–E) and those given various doses of tamoxifen (F–H″′). Representative confocal images of eGFP (green) expression in Fgfr3-iCreER T2/+ ::CAG-eGFP +/loxP controls without tamoxifen (I–K) and those given various doses of tamoxifen (L–N″′). Optical cross sections showing that the location of tdTomato (H′–H″′) and eGFP (N′–N″′) expression is primarily in SCs. Scale bar in C is 25 μm and in H′ and N′ is 6.25 μm.
Much research has focused on HCs including studies of planar cell polarity (Denman-Johnson and Forge 1999; Lewis and Davies 2002), mechanotransduction (Hudspeth and Corey 1977; LeMasurier and Gillespie 2005), otoprotection (Huang et al. 2000; Lefebvre et al. 2002), and regeneration (Corwin and Warchol 1991; Rubel et al. 1995). While less is known about SCs, several functions have been described including protection of HCs from excitotoxicity (Spicer and Schulte 1996; Kikuchi et al. 2000; Boettger et al. 2002; Furness et al. 2002), phagocytosis of dying HCs (Abrashkin et al. 2006; Taylor et al. 2008; Anttonen et al. 2014), and sealing the epithelial surface after HC death (McDowell et al. 1989; Raphael and Altschuler 1991a, b). Studies have also shown that SCs release factors which promote the formation of synapses and survival of HCs and auditory nerves (Pirvola et al. 1992; Flores-Otero et al. 2007; Sugawara et al. 2007; Tritsch et al. 2007; Gomez-Casati et al. 2010; Tritsch and Bergles 2010; Zuccotti et al. 2012). In the neonatal mouse cochlea, SCs are the source of regenerated HCs (Bramhall et al. 2014; Cox et al. 2014). These functions have been attributed to SCs as a group, with little known about the function of individual SC subtypes, primarily because tools are limited.
Genetically modified mouse models that target cochlear cells have provided critical tools to increase our understanding of inner ear physiology and response to damage. The CreER/loxP system, which allows cell type-specific and temporal control of gene expression, has been used to delete or overexpress genes, to label cell populations for fate-mapping, and to ablate specific cell types. Here, we used four CreER alleles known to target broad populations of SCs. The CreER enzyme driven by both the Fgfr3-iCreER T2 and Sox2 CreERT2 lines is also expressed in neonatal HCs (Cox et al. 2012; Bramhall et al. 2014; Walters et al. 2015). We hypothesized that modifying the tamoxifen induction paradigm and/or changing the paired reporter line would label subpopulations of SCs, as well as decrease HC labeling in Fgfr3-iCreER T2 and Sox2 CreERT2 lines. This hypothesis is based, in part, on previous studies showing that the ROSA26 tdTomato reporter produces a brighter fluorescent protein that is more stable than the fluorophores produced by other reporter lines such as CAG-eGFP or ROSA26 LacZ (Madisen et al. 2010). With the Fgfr3-iCreER T2 line, a 10-fold reduction in tamoxifen or changing to the CAG-eGFP reporter produced a large reduction of labeled OHCs. However, the number of reporter-positive SCs also decreased. Reducing the tamoxifen dose in Sox2 CreERT2+/− ::ROSA26 tdTomato/+ mice produced a minimal effect. However, changing to the CAG-eGFP reporter reduced reporter-positive HCs and SCs. Plp-CreER T2 is largely expressed in IPhCs/BCs of the neonatal cochlea with some PCs/DCs also labeled. Reduction in tamoxifen dose had a minimal effect when the ROSA26 tdTomato reporter was used. However, using the CAG-eGFP reporter with a reduced tamoxifen dose, fewer PCs/DCs were labeled, but this also decreased the number of reporter-positive IPhCs/BCs. As previously described, Prox1 CreERT2 is limited to PCs/DCs in the neonatal cochlea, with relatively fewer IPCs labeled. Alterations in tamoxifen dose caused few changes in labeled cells.
MATERIALS AND METHODS
Mice
Sox2 CreERT2 (stock no. 17593; Arnold et al. 2011), Plp-CreER T2 (stock no. 5975; Doerflinger et al. 2003), and ROSA26 CAG-loxP-stop-loxP-tdTomato (ROSA26 tdTomato ) mice, also referred to as Ai14, (stock no. 7914; Madisen et al. 2010) were obtained from The Jackson Laboratory (Bar Harbor, ME). Fgfr3-iCreER T2 mice (Rivers et al. 2008; Young et al. 2010) were provided by Dr. William Richardson (University College London, UK), Prox1 CreERT2 mice (Srinivasan et al. 2007) were provided by Dr. Guillermo Oliver (St. Jude Children’s Research Hospital, Memphis, TN) and CAG-loxP-stop-loxP-eGFP (CAG-eGFP) mice (Nakamura et al. 2006) were provided by Dr. Jeffery Robbins (Cincinnati Children’s Hospital, Cincinnati, OH). Genotyping for all mouse lines was performed by Transnetyx, Inc. (Cordova, TN). Mice of both genders were used and all animal work was performed in accordance with approved animal protocols from the Institutional Animal Care and Use Committee at Southern Illinois University School of Medicine.
Tamoxifen Injections
CreER recombination was induced by intraperitoneal (IP) injections of tamoxifen (Sigma-Aldrich – St. Louis, MO) dissolved in 100 % corn oil. Mice received one injection per day on either postnatal day (P) 0, P1, or on both P0 and P1 with the dose per injection ranging from 0.3 mg/40 g to 5 mg/40 g. Stock solutions were diluted so that the total volume of each injection remained the same. To measure CreER leakiness, samples that were CreER-positive and reporter-positive but did not receive tamoxifen were analyzed as controls. Samples were evaluated between P5-P7.
Immunostaining
Samples were post-fixed in 4 % paraformaldehyde overnight (Polysciences, Inc. – Warrington, PA) and subsequently stored in 10 mM Phosphate Buffered Saline (Sigma-Aldrich – St. Louis, MO) at 4 °C. Cochleae were dissected into apical, middle, and basal sections using a whole-mount or surface preparation method. Routine immunostaining was performed on free-floating cochlear turns as previously described (Montgomery and Cox 2016) with the following primary antibodies: anti-myosin VIIa (1:200, cat no. 25-6790, Proteus BioSciences, Ramona, CA), anti-Sox2 (1:500, cat no. sc-17320, Santa Cruz, Dallas, TX), anti-GFP (1:1000, cat no. ab13970, Abcam, Cambridge, MA), and anti-GFP conjugated to Alexa 488 (1:50 cat no. A21311, Invitrogen, Grand Island, NY). All secondary antibodies were Alexa-conjugated from Invitrogen (Waltham, MA) and were used at a 1:1000 dilution. Images were taken using a Leica SP5 confocal microscope, and image analysis was performed using Leica LAS AF LITE software.
Cell Counts
HCs and SCs were identified by immunostaining for myosin VIIa and Sox2, respectively. Myosin VIIa is expressed in the cytoplasm of HCs and Sox2 is expressed in SC nuclei, while both tdTomato and eGFP are expressed throughout the cell in both cytoplasm and nuclei. SC subtypes that expressed either the tdTomato or eGFP reporter were quantified in two randomly chosen 200 μm regions per cochlear turn, averaged, and expressed as a percentage of labeled cells compared to total cells. For the Fgfr3-iCreER T2 and Sox2 CreERT2 lines, all labeled SC subtypes were pooled into one value. For the Plp-CreER T2 and Prox1 CreERT2 lines, SC subtypes were counted individually for IPCs, OPCs, DCs, and IPhC/BCs.
Reporter labeling in HCs was detected in Fgfr3-iCreER T2 and Sox2 CreERT2 lines. For the ROSA26 tdTomato reporter, the entire organ of Corti was imaged, measured, and divided into six equal sections for quantification of tdTomato-positive HCs to determine if a gradient of tdTomato expression was present. For the CAG-eGFP reporter, eGFP-positive HCs were quantified in two sections: a 250 μm region at the most apical tip (where the majority of eGFP-positive cells were found) and the rest of the organ of Corti. Data for both reporter lines are expressed as a percentage of the reporter-positive HCs compared to the total HCs within each region.
Statistical Analysis
All data are presented as mean ± SEM. One-way or two-way ANOVA followed by Tukey’s post hoc tests, Student’s t test, and Pearson’s correlations was performed using GraphPad Prism 6.0 2 (GraphPad Software Inc., La Jolla, CA).
RESULTS
The CreER/loxP system allows cell type-specific gene expression through the use of cell type-specific promoters that drive expression of the Cre enzyme. Temporal control is achieved by fusing a modified estrogen receptor (ER) to Cre, which restricts the CreER protein to the cytoplasm. Only in the presence of tamoxifen will CreER translocate to the nucleus for excision of loxP sites (Feil et al. 1996; Hayashi and McMahon 2002). Using four independent CreER mouse lines (Fgfr3-iCreER T2 , Sox2 CreERT2 , Plp-CreER T2, and Prox1 CreERT2) that have previously been shown to label broad populations of SCs in the neonatal cochlea, we sought to label subpopulations of SCs by altering the tamoxifen induction paradigm and/or reporter line.
The ROSA26 tdTomato reporter line was developed to improve the fluorescent labeling of cells in a Cre-dependent manner over standard reporter lines such as CAG-eGFP. It uses two ubiquitously expressed promoters (ROSA26 and CAG) to drive expression of tdTomato, as well as the woodchuck hepatitis virus post-transcriptional regulatory element (WPRE) to enhance mRNA stability. Endogenous fluorescence of tdTomato is ∼2.8 times brighter than eGFP and is readily detected, whereas detection of eGFP often requires amplification with immunostaining prior to imaging (Madisen et al. 2010, 2015). Therefore, different patterns of reporter labeling are seen when ROSA26 tdTomato and CAG-eGFP reporter lines are used with the same CreER allele and tamoxifen induction paradigm. We also tested for Cre leakage, which can occur when some of the CreER enzyme enters the nucleus in the absence of tamoxifen. To determine the leakiness of the four CreER lines, we performed control experiments using CreER-positive::reporter-positive mice that did not receive tamoxifen.
Fgfr3-iCreER T2
Fibroblast growth factor receptor 3 (Fgfr3) is necessary for development of PCs in the organ of Corti (Colvin et al. 1996). At P0, Fgfr3 expression is thought to be confined to PCs and DCs in the lateral compartment of the cochlea (Peters et al. 1993; Mueller et al. 2002; Pirvola et al. 2002; Hayashi et al. 2007). However, recent single-cell RNA-seq analyses have detected a low level of Fgfr3 expression in some HCs at P1–2 (Burns et al. 2015; Waldhaus et al. 2015). The Fgfr3-iCreER T2 mouse line is a transgenic allele, where CreER is driven by the Fgfr3 promoter (Young et al. 2010). Previous characterization of the Fgfr3-iCreER T2 allele showed that after tamoxifen injection at neonatal ages, reporter expression was detected in the vast majority of PCs and DCs within the organ of Corti (Cox et al. 2012). Specifically when Fgfr3-iCreER T2/+ ::ROSA26 tdTomato/+ mice were injected with tamoxifen (3 mg/40 g, IP) at both P0 and P1, 100 % of PCs and DCs expressed tdTomato. However, there were also 25–75 % of OHCs labeled (Cox et al. 2012). To verify this expression pattern and to establish a basis for comparison, we repeated this induction protocol in Fgfr3-iCreER T2/+ ::ROSA26 tdTomato/+ mice and found that 94.4 ± 5.5 % of PCs and DCs were labeled throughout the organ of Corti (Fig. 1B and Table 1). Similar to previously published results, this dosing paradigm also showed robust labeling of OHCs (43.8 ± 2.9 %; Fig. 2B and Table 1). To investigate whether there was a gradient of HC labeling across cochlear turns, the organ of Corti was divided into six sections of equal length (Fig. 2A) and the percentage of labeled OHCs was counted within each section. The percentage of tdTomato-positive OHCs declined in an apical to basal gradient (98.5 ± 0.7 % in the first segment of apex and 13.2 ± 3.1 % in the last segment of the base; Pearson correlation coefficient r 2 = 0.81, P = 0.049; Fig. 2B and Table 1). Three tdTomato-positive IHCs were detected in the apical tip of one sample (data not shown). In addition, no tdTomato-positive SCs or HCs were detected in controls that were not treated with tamoxifen (Figs. 1B–E and 2B–E).
Fgfr3-iCreER T2/+ ::ROSA26 tdTomato/+ is robustly expressed in OHCs in an apical to basal gradient. A For quantification with the ROSA26 tdTomato/+ reporter line, the entire cochlea was imaged, measured, and divided into six equal sections. A, apex and B, base. B Quantification of reporter-positive OHCs in Fgfr3-iCreER T2/+ ::ROSA26 tdTomato/+ mice given different doses of tamoxifen (TAM). All cells were quantified in each of the six segments, averaged, and expressed as a percentage of labeled cells compared to total cells. (N = 3–4). Representative confocal images of tdTomato (red) expression in Fgfr3-iCreER T2/+ ::ROSA26 tdTomato/+ controls without tamoxifen (C–E) and those given various doses of tamoxifen (F–K). L–M Higher magnification images from regions marked in H and K, respectively. Scale bars = 25 μm.
To attenuate HC labeling and target only PCs and DCs, Cre-mediated recombination was induced with a 10-fold lower dose of tamoxifen (0.3 mg/40 g, IP) at P1 only. The percentage of tdTomato-positive HCs was reduced to 3.9 ± 2.2 % of total OHCs across the entire cochlea (Fig. 2B, F–M and Table 1). Yet the apical to basal gradient remained with 14.6 ± 10.6 % OHCs labeled in the first apical section and 0.2 ± 0.2 % in the most basal section (Pearson correlation coefficient r 2 = 0.81, P = 0.049; Fig. 2B, F–M and Table 1). However, this dosing paradigm also reduced total PC/DC labeling to 77.7 ± 4.9 % (Fig. 1B, F–H″′ and Table 1).
Because HC labeling was still present with the reduced tamoxifen dose, Fgfr3-iCreER T2 mice were next paired with the CAG-eGFP reporter. Using the previously published tamoxifen induction paradigm (3 mg/40 g, IP at P0/P1) in Fgfr3-iCreER T2/+ ::CAG-eGFP +/loxP mice, we observed 37.8 ± 3.4 % eGFP-positive PCs and DCs throughout the cochlea (Fig. 1B, L–N″′ and Table 1). However, similar to the 0.3 mg/40 g tamoxifen paradigm in Fgfr3-iCreER T2/+ ::ROSA26 tdTomato/+ mice, 5.8 ± 2.6 % eGFP-positive OHCs were still detected (Fig. 3B, F–M and Table 1). Because the apical tip of the cochlea contained a large number of eGFP-positive HCs which quickly declined, the cochlea was divided into two sections for quantification: the first 250 μm of the apex and the remainder of the cochlea (Fig. 3A). Upon quantification, there was a significant difference between the two regions with the largest amount of OHC labeling in the most apical section (40.5 ± 6.1 % compared to 4.4 ± 2.5 %; paired Student’s t test t(2) = 9.822, P = 0.0102; Fig. 3B and Table 1). No eGFP-positive SCs or HCs were observed in control animals that were not treated with tamoxifen (Figs. 1B, I–K and 3B–E).
Fgfr3-iCreER T2/+ ::CAG-eGFP +/loxP labels fewer HCs except for the apical tip of the cochlea. A For quantification with the CAG-eGFP reporter line, the cochlea was divided into two sections: the apical most 250 μm and the rest of the cochlea. A, apex and B, base. B Quantification of reporter-positive OHCs in Fgfr3-iCreER T2/+ ::CAG-eGFP +/loxP mice given different doses of tamoxifen (TAM). All cells were quantified in the two segments, averaged, and expressed as a percentage of labeled cells compared to total cells. (*P = 0.0101, as determined by a paired Student’s t test t(2) = 9.867) (N = 3–4). Representative confocal images of eGFP (green) expression in Fgfr3-iCreER T2/+ ::CAG-eGFP +/loxP controls without tamoxifen (C–E) and those given various doses of tamoxifen (F–K). L–M Higher magnification images from regions marked in H and K, respectively. Scale bars = 25 μm.
Sox2 CreERT2
Sox2 is a transcription factor expressed in all SCs throughout the organ of Corti as well as in immature HCs (Hume et al. 2007; Dabdoub et al. 2008). Low levels of Sox2 expression have been detected in some HCs at P1–2 using single-cell RNA-seq analyses (Burns et al. 2015; Waldhaus et al. 2015). The Sox2 CreERT2 mouse line is a knock-in allele, where CreER was inserted into the endogenous Sox2 locus (Arnold et al. 2011). Sox2 CreERT2 expression has previously been reported in both HCs and SCs in the neonatal organ of Corti (Bramhall et al. 2014; Walters et al. 2015). When paired with the ROSA26 tdTomato reporter line and tamoxifen (3 mg/40 g, IP) given at P0/P1, more than 85 % of SCs and more than 50 % of HCs throughout the cochlea were labeled (Walters et al. 2015). To determine whether HC labeling could be reduced, we performed a tamoxifen dose–response analysis. For comparison, we repeated the dosing paradigm from Walters et al. (2015). In Sox2 CreERT2+/− ::ROSA26 tdTomato/+ mice with tamoxifen (3 mg/40 g, IP) injected at P0/P1, 100 ± 0.0 % of SCs from the GER to HeCs throughout the cochlea expressed tdTomato (Fig. 4A and Table 1). tdTomato-positive HCs (35.3 ± 4.8 %) were also observed which appeared to decrease in a gradient from the apical to basal turn of the cochlea (Fig. 5A and Table 1). To quantify this gradient, we divided the organ of Corti into six segments of equal length as before (Fig. 2A). No difference was observed between the percentage of IHCs and OHCs labeled in any of the six segments (Fig. 5A). However, the percentage of tdTomato-positive HCs was 97.2 ± 1.7 % in the most apical segment which declined to 2.7 ± 1.2 % in the most basal segment (Fig. 5A and Table 1; Pearson correlation coefficient r 2 = 0.996, P = 0.00002). In addition, Sox2 CreERT2+/− ::ROSA26 tdTomato/+ mice showed some Cre leakiness with approximately 20 tdTomato-positive SCs detected in the entire cochlea in controls that did not receive tamoxifen (Figs. 4A–D and 5A–D).
Sox2 CreERT2 is expressed in all SCs within the organ of Corti. A Quantification of total reporter-positive SCs in Sox2 CreERT2+/− ::ROSA26 tdTomato /+ mice or Sox2 CreERT2+/− ::CAG-eGFP +/loxP mice given different doses of tamoxifen (TAM). Cells were quantified in two randomly chosen 200 μm regions per cochlear turn, averaged, and expressed as a percentage of labeled cells compared to total cells. (***P < 0.0001 as determined by a two-way ANOVA F(4, 33) = 462.3 with a Tukey’s post hoc test) (N = 3–4). Representative confocal images of tdTomato (red) expression in Sox2 CreERT2+/− ::ROSA26 tdTomato /+ controls without tamoxifen (B–D) and those given various doses of tamoxifen (E–G″′). Representative confocal images of eGFP (green) expression in Sox2 CreERT2+/− ::CAG-eGFP +/loxP controls without tamoxifen (H–J) and those given various doses of tamoxifen (K–M″′). Optical cross sections showing location of tdTomato (G′–G″′) and eGFP (M′–M″′) expression is in SCs and some HCs. Scale bar in B is 25 μm and in G′ and M′ is 6.25 μm.
Sox2 CreERT2+/− ::ROSA26 tdTomato /+ is expressed in a large number of HCs in a descending apical to basal gradient. A Quantification of reporter-positive HCs in Sox2 CreERT2+/− ::ROSA26 tdTomato /+ mice given different doses of tamoxifen (TAM). All cells were quantified in each of the six segments labeled in Fig. 2A, averaged, and expressed as a percentage of labeled cells compared to total cells. (For comparison of OHCs ***P = 0.00005 as determined by a Pearson correlation coefficient r 2 = 0.994, for comparison of IHCs ***P = 0.0003 as determined by a Pearson correlation coefficient r 2 = 0.985) (N = 3–4). Representative confocal images of tdTomato (red) expression in Sox2 CreERT2+/− ::ROSA26 tdTomato /+ controls without tamoxifen (B–D) and those given various doses of tamoxifen (E–J). K–L Higher magnification images from regions marked in G and J, respectively. Scale bars = 25 μm.
To determine if a reduced tamoxifen dose leads to a reduced number of labeled HCs, Sox2 CreERT2+/− ::ROSA26 tdTomato/+ mice were injected with tamoxifen (3 mg/40 g, IP) at P0 only. Again, 99.9 ± 0 % of SCs throughout the cochlea were labeled (Fig. 4A, E–G″′ and Table 1). There was minimal change in the percentage of tdTomato-labeled HCs (34.5 ± 4.7 %) with no significant reduction seen compared to the dual injection of tamoxifen (Fig. 5A, E–L and Table 1).
We next used CAG-eGFP mice to investigate whether a different reporter would minimize HC labeling. Sox2 CreERT2+/− ::CAG-eGFP +/loxP mice were injected with tamoxifen (3 mg/40 g, IP) at P1 only. Unlike the ROSA26 tdTomato reporter, no eGFP-positive cells were observed in controls that did not receive tamoxifen (Figs. 4A, H–J and 6A–D). As predicted, there was a large reduction in the total number of eGFP-positive HCs with the majority of the labeled cells located in the apex (Fig. 6A, E–L). Therefore, we quantified eGFP-positive HCs using the same parameters used for Fgfr3-iCreER T2/+::CAG-eGFP +/loxP samples (Fig. 3A). Throughout the entire organ of Corti, only 3.2 ± 0.4 % HCs were labeled with eGFP (Fig. 6A and Table 1), which is far less than the number of tdTomato-positive HCs observed with tamoxifen induction at either P0 only (34.5 ± 4.7 %) or P0/P1 (35.3 ± 4.8 %; Fig. 5A and Table 1). Within the first 250 μm of the apical tip in Sox2 CreERT2+/− ::CAG-eGFP +/loxP mice, 44.8 ± 10.8 % of HCs were eGFP-positive; however, only 1.6 ± 0.5 % eGFP-positive HCs were observed throughout the remainder of the cochlea (paired Student’s t test t(3.967) = 2, P = 0.0581; Fig. 6A and Table 1). Parallel to the reduction in eGFP-positive HCs, fewer SCs were labeled with eGFP throughout the cochlea (66.0 ± 5.9 % compared to 100 ± 0.0 % tdTomato-positive SCs with tamoxifen given at P0/P1 and 99.9 ± 0.0 % tdTomato-positive SCs with tamoxifen given at P0 only; Fig. 4A, K–M″′ and Table 1).
Sox2 CreERT2+/−::CAG-eGFP +/loxP labels fewer HCs except for the most apical tip of the cochlea. A Quantification of reporter-positive HCs in Sox2 CreERT2+/−::CAG-eGFP +/loxP mice given different doses of tamoxifen (TAM). All cells were quantified in the two segments labeled in Fig. 3A, averaged, and expressed as a percentage of labeled cells compared to total cells. (N = 3–4). Representative confocal images of eGFP (green) expression in Sox2 CreERT2+/−::CAG-eGFP +/loxP controls without tamoxifen (B–D) and those given various doses of tamoxifen (E–J). K–L Higher magnification images from regions marked in G and J, respectively. Scale bars = 25 μm.
Plp-CreER T2
Proteolipid protein (Plp) is expressed in oligodendrocytes and Schwann cells of the central and peripheral nervous systems (Fuss et al. 2000; Mallon et al. 2002). In the mammalian cochlea, Plp is expressed in PCs and DCs in the late embryonic stage (Morris et al. 2006). Plp-CreER T2 is a transgene that has previously been reported to label IPhCs and BCs, as well as some PCs and DCs in the neonatal organ of Corti (Doerflinger et al. 2003; Gomez-Casati et al. 2010; Cox et al. 2012; Mellado Lagarde et al. 2014). Specifically, Plp-CreER T2/+− ::ROSA26 eYFP/+ mice injected with tamoxifen (3 mg/40 g, IP) at P0/P1 had 47 % eYFP-positive IPhs/BCs, 3.4 % eYFP-positive PCs, and 5.2 % eYFP-positive DCs with no statistical differences detected among cochlear turns (Liu et al. 2014). When paired with the ROSA26 LacZ reporter and tamoxifen (33 mg/kg, IP) given from P0 to P7, Plp-CreER T2 was expressed only in SCs with the majority of labeled cells being IPhCs/BCs, but no quantification was given (Gomez-Casati et al. 2010). In Plp-CreER T2/+ ::ROSA26 tdTomato/+ mice given tamoxifen (3 mg/40 g, IP) at P0/P1, tdTomato expression was observed in 50 % of IPhCs/BCs in the apex, 80 % of IPhCs/BCs in the middle and base, and 5–10 % of PCs and DCs throughout the whole cochlea (Cox et al. 2012). Similar to Cox et al. (2012), Mellado Lagarde et al. (2014) found fewer tdTomato-positive IPhCs/BCs in the apex (25.9 %) compared to middle and basal turns (71.9 and 86.1 %, respectively) with the same tamoxifen induction paradigm. In order to set a standard with which to compare alternate tamoxifen dosing paradigms, we repeated the tamoxifen dose published in Cox et al. (2012) and Mellado Lagarde et al. (2014). Similar to the previous reports, there was an apical to basal gradient of increasing numbers of tdTomato-positive SCs (Fig. 7A and Table 1). Additionally, fewer IPhCs/BCs expressed tdTomato in the apex (48.2 ± 4.7 %) compared to the middle (79.9 ± 2.5 %) and base (89.2 ± 3.7 %; one-way ANOVA F(1.313, 3.940) = 71.03, P = 0.0010 with Tukey’s multiple comparisons test: Fig. 8A and Table 1). Interestingly, this paradigm also showed SC labeling lateral to IHCs in PCs and DCs. Specifically, 13.5 ± 4.6 % of IPCs and 10.6 ± 2.0 % OPCs were labeled throughout the organ of Corti (Fig. 8A–D and Table 1). There also appeared to be a gradient for DC labeling with fewer tdTomato-positive DCs in the apex (5.8 ± 1.9 %) and middle (9.8 ± 2.8 %) than in the base (29.3 ± 4.7 %; one-way ANOVA F(1.193, 3.579) = 39.29, P = 0.0044 with Tukey’s multiple comparisons test; Fig. 8D and Table 1).
Targeting of Plp-CreER T2 to IPhCs/BCs increased when the CAG-eGFP reporter was used. A Quantification of total reporter-positive SCs in the three cochlear turns of Plp-CreER T2/+ ::ROSA26 tdTomato /+ or Plp-CreER T2/+ ::CAG-eGFP +/loxP mice given different doses of tamoxifen (TAM). Cells were quantified in two randomly chosen 200 μm regions per cochlear turn, averaged, and expressed as a percentage of labeled cells compared to total cells. Representative confocal images of tdTomato (red) expression in Plp-CreER T2/+ ::ROSA26 tdTomato /+ controls without tamoxifen (B–D) and those given various doses of tamoxifen (E–J). Representative confocal images of eGFP (green) expression in Plp-CreER T2/+ ::CAG-eGFP +/loxP controls without tamoxifen (K–M) and those given various doses of tamoxifen (N–S). Scale bar = 25 μm. (For comparison among cochlear turns within the same tamoxifen dosing paradigm: 3 mg/40 g tamoxifen dose at P0 only *P < 0.05 as determined by a one-way ANOVA F(1.016, 2.031) = 7.435 with a Tukey’s post hoc test, for the 3-mg/40-g tamoxifen dose at P0/P1 **P < 0.01 as determined by a one-way ANOVA F(1.346, 4.038) = 129.5 with a Tukey’s post hoc test. For comparison across tamoxifen dosing paradigms: *P < 0.05 and ***P < 0.001 as determined by a two-way ANOVA F(6, 44) = 40.9 with a Tukey’s post hoc test.) (N = 3–4).
Quantification of reporter expression among SC subtypes in Plp-CreER T2 mice. SC subtypes that expressed the tdTomato or eGFP reporter were quantified as described in 7A. A IPhCs/BCs (For comparison among cochlear turns within the same tamoxifen dosing paradigm: ROSA26 tdTomato reporter with tamoxifen at P0/P1 ***P < 0.001 as determined by a one-way ANOVA F(1.313, 3.940) = 71.03 with a Tukey’s post hoc test and CAG-eGFP +/loxP reporter with tamoxifen at P0 *P < 0.05, ***P < 0.001 as determined by a one-way ANOVA F(1.012, 2.025) = 268.8 with a Tukey’s post hoc test. For comparison across tamoxifen dosing paradigms: **P < 0.01 and ***P < 0.001 as determined by a two-way ANOVA F(6, 45) = 97.06 with a Tukey’s post hoc test). B IPCs (For comparison across tamoxifen dosing paradigms: *P < 0.05 as determined by a two-way ANOVA F(6, 45) = 6.017 with a Tukey’s post hoc test). C OPCs (For comparison across tamoxifen dosing paradigms: *P < 0.05 as determined by a two-way ANOVA F(6, 60) = 8.524 with a Tukey’s post hoc test). D DCs (For comparison among cochlear turns within the same tamoxifen dosing paradigm: ROSA26 tdTomato reporter with tamoxifen at P0/P1 *P < 0.05 as determined by a one-way ANOVA F(1.193, 3.579) = 39.29 with a Tukey’s post hoc test and ROSA26 tdTomato reporter with tamoxifen at P1 *P < 0.05 as determined by a one-way ANOVA F(2, 6) = 5.189 with a Tukey’s post hoc test. For comparison across tamoxifen dosing paradigms: *P < 0.05 and **P < 0.01 as determined by a two-way ANOVA F(6, 45) = 6.313 with a Tukey’s post hoc test) (N = 3–4).
We attempted to label only IPhCs/BCs by reducing the tamoxifen dose to one injection given at either P0 only or P1 only, while maintaining the same concentration (3 mg/40 g). Expression of tdTomato in IPhCs/BCs of Plp-CreER T2/+ ::ROSA26 tdTomato/+ mice that received tamoxifen at P0 only did not differ from tamoxifen injections given at P1 only or at both P0 and P1 (Figs. 7E–J and 8A and Table 1). There was also no difference in the number of tdTomato-positive IPCs, OPCs, and DCs with a P0 only or P1 only injection compared to the P0/P1 injection (Figs. 7E–J and 8A–D and Table 1).
Because reduction in tamoxifen dose did not reduce labeling of PCs/DCs, we next generated Plp-CreER T2/+− ::CAG-eGFP +/loxP mice. When Cre recombination was induced with tamoxifen (3 mg/40 g, IP) at P0/P1, fewer IPhC/BC were labeled by eGFP than tdTomato in all turns of the cochlea (apex = 21.8 ± 10.9 % eGFP-positive IPhCs/BCs compared to 48.2 ± 4.7 % tdTomato-positive IPhCs/BCs; middle = 28.2 ± 2.1 % eGFP-positive IPhCs/BCs compared to 79.9 ± 2.5 % tdTomato-positive IPhCs/BCs; base = 38.1 ± 3.9 % eGFP-positive IPhCs/BCs compared to 89.2 ± 3.7 % tdTomato-positive IPhCs/BCs; two-way ANOVA F(2, 45) = 30.39, P < 0.0001 with Tukey’s multiple comparisons test; Figs. 7Q–S and 8A and Table 1). There was no difference in IPC, OPC, or DC labeling between eGFP and tdTomato reporter lines with tamoxifen induction (3 mg/40 g, IP) at P0/P1 (Figs. 7Q–S and 8B–D and Table 1).
Since minimal changes in the Plp-CreER T2 expression pattern occurred with the CAG-eGFP reporter, we also investigated a lower dose of tamoxifen to reduce PC/DC labeling. Reducing the amount of tamoxifen given to Plp-CreER T2/+ ::CAG-eGFP +/loxP mice to a single injection (3 mg/40 g, IP) at P0 only reduced the number of eGFP-positive cells only in specific turns of the cochlea. The eGFP-positive IPhCs/BCs in the middle turn were reduced (24.3 ± 5.9 %) compared to Plp-CreER T2/+ ::ROSA26 tdTomato/+ mice (79.9 ± 2.5 %; two-way ANOVA F(6, 45) = 97.06, P < 0.0001 with Tukey’s multiple comparisons test; Fig. 8A and Table 1). OPCs were also reduced in the middle turn (0.0 ± 0.0 % eGFP-positive OPCs compared to 15.7 ± 3.4 % tdTomato-positive OPCs; two-way ANOVA F(6, 60) = 8.524, P < 0.0001 with Tukey’s multiple comparisons test) and DCs were reduced in the base (5.9 ± 4.8 % eGFP-positive DCs compared to 29.3 ± 4.7 % tdTomato-positive DCs; two-way ANOVA F(6, 45) = 6.313, P < 0.0001 with Tukey’s multiple comparisons test; Figs. 7N–P and 8D and Table 1). Unlike Fgfr3-iCreER T2 and Sox2 CreERT2, neither Plp-CreER T2/+ ::ROSA26 tdTomato/+ nor Plp-CreER T2+/− ::CAG-eGFP +/loxP mice showed reporter expression in HCs throughout the cochlea. Finally, no tdTomato-positive or eGFP-positive cells were detected in Plp-CreER T2 control mice which did not receive tamoxifen (Figs. 7A–D, K–M and 8A–D).
Prox1 CreERT2
Prox1 is a transcription factor that is expressed in IPCs, OPCs, and DCs throughout the neonatal organ of Corti but is downregulated by ∼P14 (Bermingham-McDonogh et al. 2006). The Prox1 CreERT2 mouse line is a knock-in allele, where CreER was inserted into the endogenous Prox1 locus (Srinivasan et al. 2007). Previously, Yu et al. (2010) characterized Prox1 CreERT2 mice using ROSA26 LacZ and ROSA26 eYFP reporters and induced Cre recombination with tamoxifen injections (3 mg/40 g, IP) at P0 and P1. LacZ and eYFP expressions were found to only label PCs and DCs; however, a small number of cells were labeled (∼13 % in the apex, ∼10 % in the middle, and ∼7 % in the base, Yu et al. 2010). When the same tamoxifen induction paradigm was used with the ROSA26 tdTomato reporter, tdTomato also labeled only PCs and DCs, but many more cells were labeled (Mellado Lagarde et al. 2013). As a basis for comparison, we replicated this paradigm and quantified tdTomato expression at P6. As previously reported, Prox1 CreERT2 expression was limited to PCs and DCs and no HC labeling was observed. The tdTomato labeling of PCs and DCs combined was higher in the apex (71.1 ± 1.2 %) and middle (57.2 ± 1.5 %) compared to basal turns (44.3 ± 3.3 %; one-way ANOVA F(1.018, 2.036) = 37.08, P = 0.0248 with Tukey’s multiple comparisons test; Fig. 9A). Similar to the previous report, there was differential labeling among SC subtypes. Specifically, tdTomato was expressed in fewer IPCs (25.1 ± 2.9 %) compared to OPCs (70.1 ± 2.6 %) and DCs (76.1 ± 0.1 %; one-way ANOVA F(1.122, 2.245) = 141.1, P = 0.0045 with Tukey’s multiple comparisons test; Fig. 9N and Table 1). In addition, the number of tdTomato-positive IPCs was higher in the apex (45.9 ± 1.6 %) compared to the base (7.4 ± 1.2 %; one-way ANOVA F(1.021, 2.042) = 35.11, P = 0.0260 with Tukey’s multiple comparisons test; Fig. 9N and Table 1). There was no difference in tdTomato-positive OPCs across turns; however, significantly fewer DCs expressed tdTomato in the base (61.7 ± 4.0 %) compared to the apex (89.6 ± 3.0 %) and middle (80.7 ± 0.6 %; one-way ANOVA F(1.302, 2.603) = 18.53, P = 0.0299 with Tukey’s multiple comparisons test; Fig. 9P and Table 1). Our results are quite similar to Mellado Lagarde et al. (2013). No tdTomato-positive cells were detected in Prox1 CreERT2+/− ::ROSA26 tdTomato/+ control samples that did not receive tamoxifen (Fig. 9A–D, N–P).
Targeting of Prox1 CreERT2 to PCs and DCs was not altered with changes in tamoxifen induction paradigm. A Quantification of total reporter-positive SCs in the three cochlear turns of Prox1 CreERT2+/− ::ROSA26 tdTomato /+ or Prox1 CreERT2+/− ::CAG-eGFP +/loxP mice given different doses of tamoxifen (TAM). Cells were quantified in two randomly chosen 200 μm regions per cochlear turn, averaged, and expressed as a percentage of labeled cells compared to total cells. (For comparison among cochlear turns within the same tamoxifen dosing paradigm: 3 mg/40 g tamoxifen dose at P0/P1 *P < 0.05 as determined by a one-way ANOVA F(1.018, 2.036) = 37.08 with a Tukey’s post hoc test and 5 mg/40 g tamoxifen dose *P < 0.05 and **P < 0.01 as determined by a one-way ANOVA F(1, 2) = 144.1 with a Tukey’s post hoc test. For comparison across tamoxifen dosing paradigms: **P < 0.01 and ***P < 0.001 as determined by a two-way ANOVA F(4, 30) = 151.8 with a Tukey’s post hoc test) (N = 3–4). Representative confocal images of tdTomato (red) expression in Prox1 CreERT2+/− ::ROSA26 tdTomato /+ controls without tamoxifen (B–D) and those given various doses of tamoxifen (E–M). N–P SC subtypes that expressed the tdTomato reporter were quantified as described in A. N IPCs (For comparison among cochlear turns within the same tamoxifen dosing paradigm: 3 mg/40 g tamoxifen dose at P0/P1 **P < 0.01 as determined by a one-way ANOVA F(1.021, 2.042) = 35.11 with a Tukey’s post hoc test and 5 mg/40 g tamoxifen dose *P < 0.05 as determined by a one-way ANOVA F(1.445, 2.890) = 47.77 with a Tukey’s post hoc test. For comparison across tamoxifen dosing paradigms: *P < 0.05, **P < 0.01, and ***P < 0.001 as determined by a two-way ANOVA F(4, 30) = 24.63 with a Tukey’s post hoc test). O OPCs (For comparison among cochlear turns within the same tamoxifen dosing paradigm: 5 mg/40 g tamoxifen dose *P < 0.05 as determined by a one-way ANOVA F(1.427, 2.854) = 37.35 with a Tukey’s post hoc test. For comparison across tamoxifen dosing paradigms: **P < 0.01 and ***P < 0.001 as determined by a two-way ANOVA F(4, 30) = 139.5 with a Tukey’s post hoc test). P DCs (For comparison among cochlear turns within the same tamoxifen dosing paradigm: 5 mg/40 g tamoxifen dose *P < 0.05, **P < 0.01 as determined by a one-way ANOVA F(1.156, 2.911) = 94.69 with a Tukey’s post hoc test. For comparison across tamoxifen dosing paradigms: ***P < 0.001 as determined by a two-way ANOVA F(4, 30) = 231.2 with a Tukey’s post hoc test). (N = 3–4). O–Q Representative confocal images of a tdTomato-positive HC (arrow) in Prox1 CreERT2+/− ::ROSA26 tdTomato /+ mice given the highest dose of tamoxifen (5 mg/40 g, P0/P1). Scale bars = 25 μm.
Because so few IPCs were labeled with this induction paradigm, we attempted to increase the number of tdTomato-positive IPCs by using a higher dose of tamoxifen. In Prox1 CreERT2+/− ::ROSA26 tdTomato/+ mice injected (IP) with 5 mg/40 g tamoxifen at P0/P1, we again observed fewer tdTomato-positive IPCs (24.5 ± 0.9 %) than OPCs (71.4 ± 1.3 %) or DCs (79.5 ± 0.4 %; one-way ANOVA F(1.038, 2.077) = 927.6, P = 0.0009 with Tukey’s multiple comparisons test; Fig. 9K–P and Table 1) across the whole cochlea. We did not observe an increase in tdTomato labeling in IPCs at this dose (24.5 ± 0.9 %) compared to the 3 mg/40 g dose (25.1 ± 2.9 %) nor did increasing the dose increase tdTomato expression in OPCs or DCs (Fig. 9K–M, O–P and Table 1). Interestingly, however, 17 OHCs and 1 IHC in the apical tip of one sample expressed tdTomato with the 5 mg/40 g dose (Fig. 9Q–S).
We next attempted to eliminate IPC labeling to make Prox1 CreERT2 label just OPCs and DCs using two different adjustments to the tamoxifen regimen. In Prox1 CreERT2+/ ::ROSA26 tdTomato/+ mice injected with tamoxifen (3 mg/40 g, IP) at P0 only, there were no differences in the number of tdTomato-positive IPCs, OPCs, or DCs compared to the P0/P1 injection with the same dose (Fig. 9H–J, N–P and Table 1). Similarly, when the tamoxifen dose was reduced to 0.75 mg/40 g (IP) at P0 only, there was no significant difference in the number of tdTomato-positive IPCs (16.8 ± 4.0 %) compared to the 3 mg/40 g dose given at P0/P1 (25.1 ± 2.9 %); however, this number was significantly lower compared to the 3 mg/40 g dose given at P0 only (33.4 ± 2.4 %; one-way ANOVA F(4, 10) = 25.88, P < 0.0001 with Tukey’s multiple comparisons test, Fig. 9E–G, N and Table 1). Additionally, tdTomato expression was reduced in OPCs (37.9 ± 2.5 %; one-way ANOVA F(4, 10) = 201.3, P < 0.0001 with Tukey’s multiple comparisons test) and DCs (41.5 ± 2.4 %; one-way ANOVA F(4, 10) = 519.5, P < 0.0001 with Tukey’s multiple comparisons test; Fig. 9E–G, O–P and Table 1) compared to 3 mg/40 g given at either P0 (77.7 ± 3.4 % OPCs and 77.9 ± 2.4 % DCs) or P0/P1 (70.1 ± 2.6 % OPCs and 76.1 ± 0.1 % DCs).
DISCUSSION
Advances in the science of genetic modification have produced an arsenal of tools that can be used to study the expression or deletion of specific genes in specific cell types. This has allowed the development of more targeted therapeutics, as well as broadened our understanding of living organisms on a cellular and genetic level. To better employ these tools, we should understand their capabilities and limitations. To that end, we have endeavored to provide a guide for the use of four CreER mouse lines that are commonly used in the investigation of the neonatal organ of Corti. We were able to reduce HC labeling and increase labeling of SC subpopulations by reducing the tamoxifen dose when Fgfr3-iCreER T2 mice were paired with the tdTomato reporter. In addition, changing to the CAG-eGFP reporter achieved this goal for both Fgfr3-iCreER T2 and Sox2 CreERT2 mice; however, this also reduced the total amount of SC labeling. The remaining two CreER lines only labeled SCs with the previously used tamoxifen doses and no changes in tamoxifen dose altered the expression pattern in Prox1 CreERT2 mice. However, a handful of HCs in the apical tip of the cochlea were labeled when we increased the tamoxifen dose. Altering tamoxifen dose had little impact on Plp-CreER T2 activity. Although when paired with the CAG-eGFP reporter, the number of labeled PCs/DCs decreased and fewer IPhCs/BCs were labeled as well. Importantly, we learned that for all CreER lines, a single dose of tamoxifen at P0 is equivalent to doses given at both P0 and P1 using the tdTomato reporter. This is an important finding because reduced tamoxifen exposure may reduce toxicity and mortality.
The four CreER lines we chose to study use promoters expressed at high levels in postnatal SCs to drive expression of CreER, and therefore their expression patterns were previously expected to label only those cell types. However, low levels of Fgfr3 and Sox2 expression have been detected in some HCs at P1–2 using single-cell RNA-seq analyses (Burns et al. 2015; Waldhaus et al. 2015), which correspond with our results using Sox2 CreERT2 and Fgfr3-iCreER T2 . Interestingly, Fgfr3-iCreER T2 HC expression occurred primarily in OHCs, with only a handful of labeled IHC in the apical tip. Fgfr3 expression begins at approximately embryonic day (E) 15.5 in the region where PCs, DCs, and OHCs will form (Peters et al. 1993; Pirvola et al. 1995; Mueller et al. 2002; Hayashi et al. 2007). Therefore it is possible that Fgfr3 is not expressed in the progenitor cells that give rise to IHCs. Alternatively, since Fgfr3-iCreER T2 is a transgenic allele, expression could be affected by the location of the transgene insertion site in the genome. Importantly, reducing the tamoxifen dose 10-fold (from 3 mg/40 g given at both P0/P1 to 0.3 mg/40 g given at P1 only) caused a significant reduction in the amount of labeled HCs (from ∼44 to ∼4 %), while there was a minimal reduction in PC and DC labeling. In contrast, Sox2 CreERT2 activity was detected in both IHCs and OHCs. This was expected since Sox2 is expressed throughout the developing cochlea (Kiernan et al. 2005; Hume et al. 2007; Dabdoub et al. 2008). In addition, immature HCs are known to express Sox2 in the first postnatal week (Kiernan et al. 2005; Hume et al. 2007; Walters et al. 2015) which overlaps with the ages when tamoxifen was injected. When the ROSA26 tdTomato reporter was paired with Sox2 CreERT2, the number of tdTomato-positive HCs did not change with varying doses of tamoxifen. Switching to the CAG-eGFP reporter was able to reduce HC expression (from ∼35 to ∼3 %); however, it also reduced SC labeling (∼100 to ∼66 %).
Two of the four CreER lines investigated (Sox2 CreERT2 and Fgfr3-iCreER T2) expressed an apical to basal decreasing gradient of reporter expression in HCs with both ROSA26 tdTomato and CAG-eGFP reporter lines. Prox1 CreERT2 also showed an apical to basal decreasing gradient of reporter expression in SC subtypes. Cochlear HCs and SCs are derived from the same pool of progenitor cells (Yang et al. 2010; Cai et al. 2013; Driver et al. 2013), and the organ of Corti is still maturing during the first postnatal week. Therefore, genes necessary for progenitor cells are being downregulated, while those required for the transition from a progenitor cell fate to a HC or SC fate are being upregulated during the first few days after birth. Atoh1 is one of the first genes to be expressed prior to HC commitment and is upregulated at ∼E13.5 in the basal turn of the cochlea (Woods et al. 2004). Between ∼E13.5 and E15.5, Atoh1 expression extends throughout the rest of the cochlea allowing IHCs to differentiate first, followed by OHCs and then SCs (Kelley 2007). Therefore, cells in the basal turn of the cochlea are approximately 2 days more mature than cells in the apical turn. Genes that are expressed in early cochlear differentiation like Sox2, Prox1, and Fgfr3 would have higher expression levels in the less mature cells of the apical turn at P0, matching the gradient of CreER activity we observed. However, the lack of gradient seen for SCs in Sox2 CreERT2 mice likely occurred because expression of Sox2 persists in SCs throughout the life of the animal (Oesterle et al. 2008). Interestingly, Plp-CreER T2 showed the opposite gradient with increased numbers of labeled IPhCs/BCs and DCs in middle and basal turns compared to the apex. Maturation of the organ of Corti also occurs in a wave from the base to the apex of the cochlea and therefore genes required for differentiation are first expressed in the base. Little is known about how Plp expression patterns change in the developing cochlea. However, our data may suggest that Plp is expressed later than Sox2, Prox1, and Fgfr3 and only in cells that have committed to a SC fate as part of the maturation process.
Of note, Prox1 CreERT2 labeled more OPCs and DCs compared to IPCs. This was somewhat surprising since PROX1 is detected in all IPCs using immunostaining (Bermingham-McDonogh et al. 2006; Cox et al. 2014); however, there are known differences between IPCs located in the medial compartment of the cochlea and OPCs/DCs located in the lateral compartment. For example, Hes5 is detected in OPCs, DCs, and IPhCs/BCs, but not IPCs (Cox et al. 2014) and CD44 is expressed only in OPCs (Hertzano et al. 2010). Similarly, Lgr5 is expressed in IPCs, IPhCs/BCs, and the third row of DCs, but not in the rest of the DC pool or in OPCs (Chai et al. 2011; Shi et al. 2012). Taken together, even though IPCs and OPCs are both classified as PCs and have similar morphologies, they are very distinct cell types. Another interesting result obtained with Prox1 CreERT2 was the expression of the tdTomato reporter in a handful of HCs in the apical tip when the highest tamoxifen dose (5 mg/40 g, P0/P1) was given. In the embryonic cochlea, Prox1 is expressed in progenitor cells beginning at ∼E14.5 and is downregulated in cells as they take on a HC fate. By ∼E18.5, Prox1 is not detectable in HCs using immunostaining (Bermingham-McDonogh et al. 2006); however, the sensitivity of antibodies is known to be limited and low levels of Prox1 may persist in neonatal HCs of the apical turn that is revealed when a high dose of tamoxifen is combined with the ROSA26 tdTomato reporter.
Interestingly, Sox2 CreERT2 showed differential Cre leakiness when paired with the two different reporter lines. Cre leakiness refers to the phenomenon where some CreER molecules are able to enter the nucleus and excise loxP sites in the absence of tamoxifen. With the ROSA26 tdTomato reporter, there were several tdTomato-positive SCs in the organ of Corti, but no eGFP-positive SCs were detected when the CAG-eGFP +/loxP reporter was used. Because ROSA26 tdTomato produces a more stable and brighter fluorescent protein than CAG-eGFP (Madisen et al. 2010), it is possible that smaller quantities of CreER enzyme that translocated into the nucleus in the absence of tamoxifen were sufficient to induce tdTomato expression. However, no tdTomato-positive cells were detected in the controls for Fgfr3-iCreER T2, Plp-CreER T2, or Prox1 CreERT2.
For this study, we used two reporter lines to assess the activity of four CreER lines in the neonatal organ of Corti. However, each floxed allele is different because the location of the loxP sites within the gene and the distance between loxP sites affects the efficiency of the Cre enzyme to bind and excise DNA located between loxP sites (Sauer and Henderson 1989; Kuhn et al. 1995; Feltri et al. 1999; Kellendonk et al. 1999). While our study is primarily useful for fate-mapping purposes, it is difficult to translate the reporter expression patterns we measured to other floxed alleles. Some alleles may be similar to the ROSA26 tdTomato reporter, while others may closely resemble the CAG-eGFP reporter line. This illustrates the importance of using more than one reporter line to understand the expression pattern of any CreER line used to delete or overexpress a gene. In addition, it would be wise to use immunostaining to confirm that the gene deletion/overexpression pattern matches the reporter data. Overall, the best method for CreER induction when using the ROSA26 tdTomato reporter appears to be one injection of tamoxifen at 3 mg/40 g given at P0 because it induces similar CreER activity as two injections without the increased risk of toxicity. Depending on the goal of the study, either the ROSA26 tdTomato reporter or CAG-eGFP reporter is valuable; however, the CAG-eGFP labeled fewer cells with all CreER lines investigated.
References
Abrashkin KA, Izumikawa M, Miyazawa T, Wang CH, Crumling MA, Swiderski DL, Beyer LA, Gong TW, Raphael Y (2006) The fate of outer hair cells after acoustic or ototoxic insults. Hear Res 218:20–29
Anttonen T, Belevich I, Kirjavainen A, Laos M, Brakebusch C, Jokitalo E, Pirvola U (2014) How to bury the dead: elimination of apoptotic hair cells from the hearing organ of the mouse. J Assoc Res Otolaryngol 15:975–992
Arnold K, Sarkar A, Yram MA, Polo JM, Bronson R, Sengupta S, Seandel M, Geijsen N, Hochedlinger K (2011) Sox2(+) adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell 9:317–329
Bermingham-McDonogh O, Oesterle EC, Stone JS, Hume CR, Huynh HM, Hayashi T (2006) Expression of Prox1 during mouse cochlear development. J Comp Neurol 496:172–186
Boettger T, Hubner CA, Maier H, Rust MB, Beck FX, Jentsch TJ (2002) Deafness and renal tubular acidosis in mice lacking the K-Cl co-transporter Kcc4. Nature 416:874–878
Bramhall NF, Shi F, Arnold K, Hochedlinger K, Edge AS (2014) Lgr5-positive supporting cells generate new hair cells in the postnatal cochlea. Stem Cell Rep 2:311–322
Burns JC, Kelly MC, Hoa M, Morell RJ, Kelley MW (2015) Single-cell RNA-Seq resolves cellular complexity in sensory organs from the neonatal inner ear. Nat Commun 6:8557
Cai T, Seymour ML, Zhang H, Pereira FA, Groves AK (2013) Conditional deletion of Atoh1 reveals distinct critical periods for survival and function of hair cells in the organ of Corti. J Neurosci 33:10110–10122
Chai R, Xia A, Wang T, Jan TA, Hayashi T, Bermingham-McDonogh O, Cheng AG (2011) Dynamic expression of Lgr5, a Wnt target gene, in the developing and mature mouse cochlea. J Assoc Res Otolaryngol 12:455–469
Colvin JS, Bohne BA, Harding GW, McEwen DG, Ornitz DM (1996) Skeletal overgrowth and deafness in mice lacking fibroblast growth factor receptor 3. Nat Genet 12:390–397
Corwin JT, Warchol ME (1991) Auditory hair cells: structure, function, development, and regeneration. Annu Rev Neurosci 14:301–333
Cox BC, Liu Z, Lagarde MM, Zuo J (2012) Conditional gene expression in the mouse inner ear using Cre-loxP. J Assoc Res Otolaryngol 13:295–322
Cox BC, Chai R, Lenoir A, Liu Z, Zhang L, Nguyen D, Chalasani K, Steigelman KA, Fang J, Rubel EW, Cheng AG, Zuo J (2014) Spontaneous hair cell regeneration in the neonatal mouse cochlea in vivo. Development 141:816–829
Dabdoub A, Puligilla C, Jones JM, Fritzsch B, Cheah KS, Pevny LH, Kelley MW (2008) Sox2 signaling in prosensory domain specification and subsequent hair cell differentiation in the developing cochlea. Proc Natl Acad Sci U S A 105:18396–18401
Denman-Johnson K, Forge A (1999) Establishment of hair bundle polarity and orientation in the developing vestibular system of the mouse. J Neurocytol 28:821–835
Doerflinger NH, Macklin WB, Popko B (2003) Inducible site-specific recombination in myelinating cells. Genesis 35:63–72
Driver EC, Sillers L, Coate TM, Rose MF, Kelley MW (2013) The Atoh1-lineage gives rise to hair cells and supporting cells within the mammalian cochlea. Dev Biol 376:86–98
Feil R, Brocard J, Mascrez B, LeMeur M, Metzger D, Chambon P (1996) Ligand-activated site-specific recombination in mice. Proc Natl Acad Sci U S A 93:10887–10890
Feltri ML, D’Antonio M, Previtali S, Fasolini M, Messing A, Wrabetz L (1999) P0-Cre transgenic mice for inactivation of adhesion molecules in Schwann cells. Ann N Y Acad Sci 883:116–123
Flores-Otero J, Xue HZ, Davis RL (2007) Reciprocal regulation of presynaptic and postsynaptic proteins in bipolar spiral ganglion neurons by neurotrophins. J Neurosci 27:14023–14034
Furness DN, Hulme JA, Lawton DM, Hackney CM (2002) Distribution of the glutamate/aspartate transporter GLAST in relation to the afferent synapses of outer hair cells in the guinea pig cochlea. J Assoc Res Otolaryngol 3:234–247
Fuss B, Mallon B, Phan T, Ohlemeyer C, Kirchhoff F, Nishiyama A, Macklin WB (2000) Purification and analysis of in vivo-differentiated oligodendrocytes expressing the green fluorescent protein. Dev Biol 218:259–274
Gomez-Casati ME, Murtie J, Taylor B, Corfas G (2010) Cell-specific inducible gene recombination in postnatal inner ear supporting cells and glia. J Assoc Res Otolaryngol 11:19–26
Hayashi S, McMahon AP (2002) Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol 244:305–318
Hayashi T, Cunningham D, Bermingham-McDonogh O (2007) Loss of Fgfr3 leads to excess hair cell development in the mouse organ of Corti. Dev Dyn 236:525–533
Hertzano R, Puligilla C, Chan SL, Timothy C, Depireuz DA, Ahmed Z, Wolf J, Eisenman DJ, Friedman TB, Riazuddin S, Kelley MW, Strome SE (2010) CD44 is a marker for the outer pillar cells in the early postnatal mouse inner ear. J Assoc Res Otolaryngol 11(3):407–418
Huang T, Cheng AG, Stupak H, Liu W, Kim A, Staecker H, Lefebvre PP, Malgrange B, Kopke R, Moonen G, Van De Water TR (2000) Oxidative stress-induced apoptosis of cochlear sensory cells: otoprotective strategies. Int J Dev Neurosci 18:259–270
Hudspeth AJ, Corey DP (1977) Sensitivity, polarity, and conductance change in the response of vertebrate hair cells to controlled mechanical stimuli. Proc Natl Acad Sci U S A 74:2407–2411
Hume CR, Bratt DL, Oesterle EC (2007) Expression of LHX3 and SOX2 during mouse inner ear development. Gene Expr Patterns 7:798–807
Kellendonk C, Tronche F, Casanova E, Anlag K, Opherk C, Schutz G (1999) Inducible site-specific recombination in the brain. J Mol Biol 285:175–182
Kelley MW (2007) Cellular commitment and differentiation in the organ of Corti. Int J Dev Biol 51:571–583
Kiernan AE, Pelling AL, Leung KK, Tang AS, Bell DM, Tease C, Lovell-Badge R, Steel KP, Cheah KS (2005) Sox2 is required for sensory organ development in the mammalian inner ear. Nature 434:1031–1035
Kikuchi T, Kimura RS, Paul DL, Takasaka T, Adams JC (2000) Gap junction systems in the mammalian cochlea. Brain Res Brain Res Rev 32:163–166
Kuhn R, Schwenk F, Aguet M, Rajewsky K (1995) Inducible gene targeting in mice. Science 269:1427–1429
Lefebvre PP, Malgrange B, Lallemend F, Staecker H, Moonen G, Van De Water TR (2002) Mechanisms of cell death in the injured auditory system: otoprotective strategies. Audiol Neurootol 7:165–170
LeMasurier M, Gillespie PG (2005) Hair-cell mechanotransduction and cochlear amplification. Neuron 48:403–415
Lewis J, Davies A (2002) Planar cell polarity in the inner ear: how do hair cells acquire their oriented structure? J Neurobiol 53:190–201
Liu Z, Fang J, Dearman J, Zhang L, Zuo J (2014) In vivo generation of immature inner hair cells in neonatal mouse cochleae by ectopic atoh1 expression. PLoS One 9:e89377
Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H (2010) A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nature Neurosci 13:133–140
Madisen L et al (2015) Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron 85:942–958
Mallon BS, Shick HE, Kidd GJ, Macklin WB (2002) Proteolipid promoter activity distinguishes two populations of NG2-positive cells throughout neonatal cortical development. J Neurosci 22:876–885
McDowell B, Davies S, Forge A (1989) The effect of gentamicin-induced hair cell loss on the tight junctions of the reticular lamina. Hear Res 40:221–232
Mellado Lagarde MM, Cox BC, Fang J, Taylor R, Forge A, Zuo J (2013) Selective ablation of pillar and deiters’ cells severely affects cochlear postnatal development and hearing in mice. J Neurosci 33:1564–1576
Mellado Lagarde MM, Wan G, Zhang L, Gigliello AR, McInnis JJ, Zhang Y, Bergles D, Zuo J, Corfas G (2014) Spontaneous regeneration of cochlear supporting cells after neonatal ablation ensures hearing in the adult mouse. Proc Natl Acad Sci U S A 111:16919–16924
Montgomery SC, Cox BC (2016) Whole mount dissection and immunofluorescence of the adult mouse cochlea. J Vis Exp 107:e53561
Morris JK, Maklad A, Hansen LA, Feng F, Sorensen C, Lee KF, Macklin WB, Fritzsch B (2006) A disorganized innervation of the inner ear persists in the absence of ErbB2. Brain Res 1091:186–199
Mueller KL, Jacques BE, Kelley MW (2002) Fibroblast growth factor signaling regulates pillar cell development in the organ of corti. J Neurosci 22:9368–9377
Nakamura T, Colbert MC, Robbins J (2006) Neural crest cells retain multipotential characteristics in the developing valves and label the cardiac conduction system. Circ Res 98:1547–1554
Oesterle EC, Campbell S, Taylor RR, Forge A, Hume CR (2008) Sox2 and JAGGED1 expression in normal and drug-damaged adult mouse inner ear. J Assoc Res Otolaryngol 9:65–89
Peters K, Ornitz D, Werner S, Williams L (1993) Unique expression pattern of the FGF receptor 3 gene during mouse organogenesis. Dev Biol 155:423–430
Pirvola U, Ylikoski J, Palgi J, Lehtonen E, Arumae U, Saarma M (1992) Brain-derived neurotrophic factor and neurotrophin 3 mRNAs in the peripheral target fields of developing inner ear ganglia. Proc Natl Acad Sci U S A 89:9915–9919
Pirvola U, Cao Y, Oellig C, Suoqiang Z, Pettersson RF, Ylikoski J (1995) The site of action of neuronal acidic fibroblast growth factor is the organ of corti of the rat cochlea. Proc Natl Acad Sci U S A 92:9269–9273
Pirvola U, Ylikoski J, Trokovic R, Hebert JM, McConnell SK, Partanen J (2002) FGFR1 is required for the development of the auditory sensory epithelium. Neuron 35:671–680
Raphael Y, Altschuler RA (1991a) Scar formation after drug-induced cochlear insult. Hear Res 51:173–183
Raphael Y, Altschuler RA (1991b) Reorganization of cytoskeletal and junctional proteins during cochlear hair cell degeneration. Cell Motil Cytoskeleton 18:215–227
Raphael Y, Altschuler RA (2003) Structure and innervation of the cochlea. Brain Res Bull 60:397–422
Rivers LE, Young KM, Rizzi M, Jamen F, Psachoulia K, Wade A, Kessaris N, Richardson WD (2008) PDGFRA/NG2 glia generate myelinating oligodendrocytes and piriform projection neurons in adult mice. Nature Neurosci 11:1392–1401
Rubel EW, Dew LA, Roberson DW (1995) Mammalian vestibular hair cell regeneration. Science 267:701–707
Sauer B, Henderson N (1989) Cre-stimulated recombination at loxP-containing DNA sequences placed into the mammalian genome. Nucleic Acids Res 17:147–161
Shi F, Kempfle JS, Edge AS (2012) Wnt-responsive lgr5-expressing stem cells are hair cell progenitors in the cochlea. J Neurosci 32:9639–9648
Spicer SS, Schulte BA (1996) The fine structure of spiral ligament cells relates to ion return to the stria and varies with place-frequency. Hear Res 100:80–100
Srinivasan RS, Dillard ME, Lagutin OV, Lin FJ, Tsai S, Tsai MJ, Samokhvalov IM, Oliver G (2007) Lineage tracing demonstrates the venous origin of the mammalian lymphatic vasculature. Genes Dev 21:2422–2432
Sugawara M, Murtie JC, Stankovic KM, Liberman MC, Corfas G (2007) Dynamic patterns of neurotrophin 3 expression in the postnatal mouse inner ear. J Comp Neurol 501:30–37
Taylor RR, Nevill G, Forge A (2008) Rapid hair cell loss: a mouse model for cochlear lesions. J Assoc Res Otolaryngol 9:44–64
Tritsch NX, Bergles DE (2010) Developmental regulation of spontaneous activity in the mammalian cochlea. J Neurosci 30:1539–1550
Tritsch NX, Yi E, Gale JE, Glowatzki E, Bergles DE (2007) The origin of spontaneous activity in the developing auditory system. Nature 450:50–55
Waldhaus J, Durruthy-Durruthy R, Heller S (2015) Quantitative high-resolution cellular map of the organ of Corti. Cell Rep 11:1385–1399
Walters BJ, Yamashita T, Zuo J (2015) Sox2-CreER mice are useful for fate mapping of mature, but not neonatal, cochlear supporting cells in hair cell regeneration studies. Sci Rep 5:11621
Woods C, Montcouquiol M, Kelley MW (2004) Math1 regulates development of the sensory epithelium in the mammalian cochlea. Nature Neurosci 7:1310–1318
Yang H, Xie X, Deng M, Chen X, Gan L (2010) Generation and characterization of Atoh1-Cre knock-in mouse line. Genesis 48:407–413
Young KM, Mitsumori T, Pringle N, Grist M, Kessaris N, Richardson WD (2010) An Fgfr3-iCreER(T2) transgenic mouse line for studies of neural stem cells and astrocytes. Glia 58:943–953
Yu Y, Weber T, Yamashita T, Liu Z, Valentine MB, Cox BC, Zuo J (2010) In vivo proliferation of postmitotic cochlear supporting cells by acute ablation of the retinoblastoma protein in neonatal mice. J Neurosci 30:5927–5936
Zuccotti A, Kuhn S, Johnson SL, Franz C, Singer W, Hecker D, Geisler HS, Kopschall I, Rohbock K, Gutsche K, Dlugaiczyk J, Schick B, Marcotti W, Ruttiger L, Schimmang T, Knipper M (2012) Lack of brain-derived neurotrophic factor hampers inner hair cell synapse physiology, but protects against noise-induced hearing loss. J Neurosci 32:8545–8553
Acknowledgments
We thank other members of the Cox lab for discussion. We thank Dr. William Richardson (University College London) for providing Fgfr3-iCreER T2 mice; Dr. Guillermo Oliver (St. Jude Children’s Research Hospital) for providing Prox1 CreERT2 mice; and Dr. Jeffery Robbins (Cincinnati Children’s Hospital) for providing CAG-eGFP mice. This work was supported by a grant from the Office of Naval Research (N000141310569). Southern Illinois University School of Medicine Research Imaging facility equipment was supported by Award Number S10RR027716 from the National Center for Research Resources-Health.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
All authors declare no conflict of interest on the present manuscript.
Rights and permissions
About this article
Cite this article
McGovern, M.M., Brancheck, J., Grant, A.C. et al. Quantitative Analysis of Supporting Cell Subtype Labeling Among CreER Lines in the Neonatal Mouse Cochlea. JARO 18, 227–245 (2017). https://doi.org/10.1007/s10162-016-0598-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10162-016-0598-0