Abstract
The impact of immune checkpoint molecule inhibitors on cancer treatment is significant. At the same time, further improvement in their efficacy has become essential. For this reason, there has been increasing interest in investigating the state of the cancer microenvironment in which efficacy can be demonstrated. The gut microbiota plays an important role in the cancer microenvironment. Recent developments in the study of gut microbiota have been explosive, benefiting from technological innovations in next-generation sequencing. Gut microbiota are specific enough to identify an individual and change gently with age. Disruptions in the gut microbiota have been extensively studied in relation to a variety of diseases. In addition to monotherapy with anti-PD-1/PD-L1 antibodies, combination therapy with chemotherapy and molecular target therapy, as well as combination therapy with anti-PD-1 and anti-CTLA-4 antibodies, is now widely used in cancer treatment with immune checkpoint inhibitors. Therefore, the development of biomarkers that can predict anti-tumor and adverse events is urgently required due to the complexity of the treatment, and research on gut microbiota is expected in this respect.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
It is said that about 1000 kinds of 100 trillion bacteria coexist in the human intestine, forming an intestinal microflora (also called intestinal flora), weighing approximately 1.5 to 2 kg. However, the number of gut microbiota in the body is now being disputed due to advances in technology, and the number of bacteria may be revised in the future. In fact, it is not known how these gut microbiota coexist with humans. However, it is an extremely important partner, and it has become known in recent years that disruption of the relationship with this partner causes inflammatory bowel disease (IBD), rheumatic diseases, obesity, diabetes, atopy, and allergies. This disordered state of the intestinal microflora is called dysbiosis, which means a breakdown of the composition of gut microbiota [1].
Rapid progress in the analysis of intestinal microflora began with the advent of next-generation sequencing, which was born as a result of the instantaneous availability of a large amount of genetic analysis. Before the full-scale appearance of next-generation sequencers, it was known that the bacterial genome, which consists of several million base pairs, contains a polymorphic region of approximately 1600 base pairs called the “16S ribosomal RNA region.” The 16S ribosomal RNA region contains nine hypervariable regions consisting of tens to hundreds of base pairs, which have characteristic sequences depending on the type of bacteria. The sequences of these hypervariable regions are conserved among the same bacterial species, and it is thought that it is possible to identify bacterial species by reading and analyzing the entire length or part of the 16S region without sequencing the entire bacterial genome. Technological innovations using next-generation sequencing have dramatically advanced research on the classification and analysis of bacteria living in the intestine. It is now possible to decode the human genome, which consists of approximately 3 billion base pairs, in less than a week using only one machine, whereas it took more than 10 years to decode it.
Next-generation sequencers read a large amount of gene sequence information from gene fragments of several hundred base pairs, although there are differences depending on the equipment used and the detection method. The hypervariable region in the 16S region can be covered by one or two identical reads in this unit of reading, which is several hundred base pairs. Therefore, the information of each read fragment was identified as a type of bacteria, and the 16S metagenomic analysis method, which simultaneously analyzes the genomes of bacteria extracted from feces, can be implemented. In principle, it is now possible to reveal up to 100 million or more types of intestinal microflora in a single examination.
Approximately half to one-third of feces is considered to be bacterial in origin, and approximately 10 billion bacteria per gram, or 2 to 3 trillion bacteria per day, are discharged in humans. In the analysis of intestinal microflora by the 16S metagenomic method, DNA, the main body of genes of gut microbiota in feces, was extracted and purified as a template, and the gene was amplified using primers for gene amplification that were set to amplify a part of the 16S region. Gene amplification does not need to be performed more than tens of millions of times as in the normal PCR method, and one of the purposes is to attach sequences necessary for subsequent analysis. DNA extracted and purified from feces includes not only bacterial DNA but also DNA from the host, such as human DNA and food residues, which do not have the bacterial 16S region and, therefore, do not participate in the amplification. The gene amplification products of several hundred base pairs were purified and quantified to create a library. The gene sequences contained in the library were read using a next-generation sequencer, and the types of bacteria were identified by comparing them with a database. Using the genetic testing method, it is possible to detect not only the DNA of dead bacteria, which could not be detected by the culture method, but also DNA fragments derived from living bacteria that exist in feces, and it is thought to provide information closer to the gut microbiome. It takes about three days to complete the following processes: gene extraction and purification from feces, gene amplification, purification and quantification of amplified products, library preparation, sequencing, and identification and calculation of the percentage of bacteria using analysis software. In the future, it is expected that the obtained information will be incorporated into a database one after another. In this way, information on gut microbiota is accumulating due to recent technological innovations, and information on gut microbiota and diseases, especially cancer, is progressing, [3].
Composition of intestinal microflora
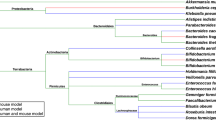
The formation of the human intestinal microbiota begins immediately after birth. The intestinal flora formed in the neonatal period is not invariant throughout life, and the composition of the constituent bacteria changes with age [2]. It has been reported that the formation process of intestinal flora is not invariant, but is affected by various environmental factors such as duration of fetal life, mode of delivery, and mode of lactation [3]. It has been reported that the process of disbiosis is not invariant and is influenced by various environmental factors such as duration of fertility, mode of delivery, and mode of lactation. According to the research on dysbiosis described above, it is important that the so-called good bacteria and bad bacteria in the gut microbiota exist in good balance. The oblate anaerobic bacteria (fermentative bacteria) are called good bacteria, and the commensal anaerobic bacteria are called bad bacteria. However, with the recent progress of research on disbiosis, this rule does not necessarily apply, or the terms “good bacteria” and “bad bacteria” themselves are becoming less commonly used. [4,5,6,7,8,9,10]. The etymology of the word flora is not certain and there are many theories. However, the diagram describing the phylogeny of intestinal bacteria appears, coincidentally, as if it were a flora [11] (Fig. 1).
Diagram of data showing the phylogenetic composition of the intestinal microbiota, which is just like a flora (flower garden). The figure demonstrated that the gut microbiota of 65 hepatobiliary cancer patients was associated with clinical response to anti-PD-1 antibodies. Seventy-four taxa were significantly enriched in the clinical benefit response (CBR) group, and 40 taxa were significantly enriched in the non-clinical benefit (NCB) group [11].
Mechanism of action of anti-PD-1/PD-L1 and anti-CTLA-4 antibodies and gut microbiota
Anti-PD-1/PD-L1 antibodies primarily suppress the negative regulatory mechanisms between tumors and T cells. This is referred to as the effector phase. In contrast, anti-CTLA-4 antibodies maintain T cell activation by blocking inhibitory signals from dendritic cells in lymph nodes [12]. This phase is called the priming phase. The main mechanism by which CTLA-4 suppresses T cell activation is by trapping the ligand CD86 from T cells and sending a negative signal to T cells through transendocytosis. This transendocytosis can be avoided by anti-CTLA-4 antibody, which maintains the expression of the ligand [13]. Transendocytosis of CTLA-4 on regulatory T cells (Treg) results in the removal of the ligand from antigen-presenting cells (APCs) and prevents normal T-cell activation, resulting in the absence of anti-tumor effects (Fig. 2) [14].
CTLA-4, which is abundantly expressed on Tregs, removes the ligand CD86 from antigen-presenting cells by transendocytosis. This result indicates that Teff cannot obtain the CD86 signal from APCs and cannot be activated [14].
The influence of gut microbiota on ICI, groups in the USA and France [4, 5, 9] have reported that certain gut microbiota may modulate the clinical effects of anti-PD-1 antibodies. However, the gut microbiota reported by each group is different and has not yet been identified. In addition, the pattern of gut microbiota differs among countries and diets.
Attenuation of ICI efficacy via the effects of antibiotics on gut microbiota
In the field of cancer, data are emerging suggesting that there may be a high correlation between cancer immunotherapy and therapeutic effects [15,16,17]. It is also thought that gut microbiota may be involved in cancer of the esophagus, stomach, and many other organs [18].
Interestingly, a growing body of data suggests that antibiotic administration has a strong negative impact on the therapeutic effect of ICI on the gut microbiota [19].
Here, we compared the overall survival rates of patients treated with immune checkpoint inhibitors for renal cell carcinoma and non-small cell lung cancer, as well as those of patients treated with antibiotics and without antibiotics in cases including esophageal cancer, gastric cancer, non-small cell lung cancer, and urothelial carcinoma, which are also carcinomas that use anti-PD-1 antibodies antibiotics within 3 weeks before and after the start of anti-PD-1 antibody administration (Fig. 3). This shocking fact that inhibits the efficacy of immune checkpoint inhibitors has been revealed [20].
Kaplan–Meier curves comparing the groups that used antibiotics at least once within 21 days before and after the start of ICI treatment with those that did not. The left panel shows OS and the right panel shows PFS. Antibiotics reduced the therapeutic efficacy of anti-PD-1 antibodies [20].
Effects of gut microbiota on cancer
In addition, various studies have been conducted on how these intestinal bacteria act on the immune system. Among them, the involvement of single-chain fatty acids is a major mechanism of action. It is thought that the action of single-chain fatty acids may change due to differences in their receptors, and the details of this will be clarified in future research [21].
It is important to note that several researchers, including our own, are currently conducting research aimed at estimating the immune state in which irAEs are likely to occur and to be effective by analyzing intestinal bacteria through various treatments, including modification of the intestinal microflora. In addition, research is underway to induce a state in which irAEs are less likely to occur. If these conditions can be inferred as biomarkers, treatment strategies, especially side effect management, will be facilitated.
Metabolites of gut microbiota affect immunity
The aforementioned beneficial bacteria produce short-chain fatty acids (SCFAs) by fermentation using dietary fiber as a nutrient source. These short-chain fatty acids, such as propionate, acetate, and butyrate, are representative of SCFAs and are thought to be involved in the activity and regulation of immunity, mainly through their receptors. SCFAs play important roles in human immunity and homeostasis, such as induction of regulatory T cells, induction of type 1 helper T cells, and maintenance of intestinal epithelial cell proliferation [21]. However, the relationship between dietary fiber and anti-tumor effects remains to be elucidated. Interestingly, SCFAs, which ferment dietary fiber as a nutritional source, are certainly one of the keys, and research on the importance of fiber in the diet and the effects of each SCFA on immunity is becoming increasingly important. Recently, in addition to SCFAs, metabolites produced by the gut microbiota have been actively studied. Commensal anaerobes are known to have low expression of digestive enzymes that digest dietary fiber and utilize nutrient sources such as monosaccharides, disaccharides, fats, proteins, and alcohols, which are abundant in the westernized diet, rather than dietary fiber [22].
These short-chain fatty acids are generally recognized to increase anti-tumor activity, but there are also data that they may inhibit some conditions and types of cancer. A representative example is a mouse study in which sodium butyrate inhibited anti-CTLA-4-induced DC maturation and T-cell priming [23], suggesting that further studies are needed to elucidate the mechanisms underlying the effects of individual SCFAs on cancer immunity.
Immune-related adverse events and gut microbiota
irAEs are a serious problem in the treatment of ICI that can impair treatment continuity. On the other hand, there are reports that patients who did not continue treatment due to irAE had a longer OS than those who did [23]. On the other hand, it is still difficult to predict irAE in ICI treatment, and microbiota is expected to be a biomarker for irAE or to elucidate its mechanism, but the evidence is not clear. Interestingly, functional similarities were reported between the inflammatory regions of irAE colitis and UC: The non-inflamed mucosa of patients with irAE colitis was characterized by the mobilization of immune cells, unlike that of UC patients. Similarities in microbiota profiles were also observed: 16S rDNA sequencing revealed a decrease in Bacteroides spp. in the inflammatory zone of irAE colitis and UC [24].
Stool transplantation increases efficacy of anti-PD-1 antibodies
Transplantation of stool samples from patients with advanced, treatment refractory melanoma who had responded to anti-PD-1 therapy produced high clinical benefit, according to a study. In this clinical trial, investigators sought to determine whether modifying the gut microbiota could overcome anti-PD-1 resistance. Responder-derived stool transplantation (FMT) was combined with anti-PD-1 antibody therapy to evaluate its safety and efficacy [25, 26]. Davar et al. reported that it was well tolerated and provided clinical benefits to 6 of 15 patients [25] (ORR 20% 3 patients, SD > 12M 3 patients). Responders experienced increased CD8 + T cell activation, increased activation of CD8 + T cells, and decreased frequency of myeloid cells expressing interleukin-8. The gut microbiota modulates proteomic and metabolomic changes, and FMT and anti-PD-1 altered the gut microbiota and reprogrammed the tumor microenvironment to overcome resistance to anti-PD-1 in advanced melanoma [27]. On the other hand, anti-PD-1 antibody therapy combined with stool transplantation is due to one of the three circumstances: (i) The patient is immunocompromised or lacks tumor immunogenicity and is unable to respond to the tumor regardless of the composition of the flora; (ii) the patient lacks the flora necessary for the efficacy of anti-PD-1 therapy with FMT; or (iii) FMT cannot be successfully transplanted into the recipient. Although it is unclear whether stool transplantation will be established as a treatment in the future, we believe that this study demonstrates that the gut microbiota has a strong influence on anti-tumor immunity [27] (Fig. 4).
Spider plot showing that 6 of 16 patients with anti-PD-1 antibody resistance achieved a clinical response when anti-PD-1 antibodies were combined with FMT derived from patients who had previously benefited from anti-PD-1 antibody treatment[27].
Summary
In this article, we describe the research on intestinal microflora and ICI, focusing on the combined therapy of anti-CTLA-4 and anti-PD-1 antibodies in the treatment of malignant melanoma, its mechanism, concerns, and the current status of research on intestinal microflora. It is thought that the skeleton of the intestinal microflora, which forms the main component of the flora, is formed during the period from weaning to early elementary school age. Bacteria are also thought to be supplied by the oral microflora, which is not as diverse and numerous as the intestinal microflora, as well as by attachment to food and drinks. Unlike the intestine, oxygen is present in the oral cavity, and aerobic bacteria form the majority of the bacterial flora, which is completely different from that in the intestine, which is mainly anaerobic. However, even in the oral cavity, there are bacteria known as the red complex, which includes the mutans bacteria that form plaque and cause tooth decay, and gingivalis bacteria that cause periodontal disease.
We are currently studying the relationship between the efficacy of checkpoint inhibitors and chemotherapy and the gut microbiota and oral microbiota in irAEs, and researchers from around the world are working on a number of hot topics.
The analysis of intestinal microflora, including the study of stool transplantation, will become increasingly important with respect to the development of ICI treatment in the future.
References
Cani PD (2017) Gut microbiota-at the intersection of everything? Nat Rev Gastroenterol Hepatol 14:321–322. https://doi.org/10.1038/nrgastro.2017.54
Arboleya S et al (2012) Establishment and development of intestinal microbiota in preterm neonates. FEMS Microbiol Ecol 79:763–772. https://doi.org/10.1111/j.1574-6941.2011.01261.x
Backhed F et al (2015) Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17:852. https://doi.org/10.1016/j.chom.2015.05.012
Gopalakrishnan V et al (2018) Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359:97–103. https://doi.org/10.1126/science.aan4236
Routy B et al (2018) Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359:91–97. https://doi.org/10.1126/science.aan3706
Gevers D et al (2014) The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 15:382–392. https://doi.org/10.1016/j.chom.2014.02.005
Ahn J et al (2013) Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst 105:1907–1911. https://doi.org/10.1093/jnci/djt300
Lepage P et al (2011) Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology 141:227–236. https://doi.org/10.1053/j.gastro.2011.04.011
Matson V et al (2018) The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 359:104–108. https://doi.org/10.1126/science.aao3290
Dubin K et al (2016) Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat Commun 7:10391. https://doi.org/10.1038/ncomms10391
Mao J, Wang D, Long J et al (2021) Gut microbiome is associated with the clinical response to anti-PD-1 based immunotherapy in hepatobiliary cancers. J Immunother Cancer. https://doi.org/10.1136/jitc-2021-003334
Ribas A (2012) Tumor immunotherapy directed at PD-1. N Engl J Med 366:2517–2519. https://doi.org/10.1056/NEJMe1205943
Qureshi OS et al (2011) Trans-endocytosis of CD80 and CD86: a molecular basis for the cell-extrinsic function of CTLA-4. Science 332:600–603. https://doi.org/10.1126/science.1202947
Soskic B, Qureshi OS, Hou T et al (2014) A transendocytosis perspective on the CD28/CTLA-4 pathway. Adv Immunol 124:95–136. https://doi.org/10.1016/B978-0-12-800147-9.00004-2
Vetizou M et al (2015) Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 350:1079–1084. https://doi.org/10.1126/science.aad1329
Sivan A et al (2015) Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 350:1084–1089. https://doi.org/10.1126/science.aac4255
Wang Y et al (2018) Fecal microbiota transplantation for refractory immune checkpoint inhibitor-associated colitis. Nat Med 24:1804–1808. https://doi.org/10.1038/s41591-018-0238-9
Schwabe RF, Jobin C (2013) The microbiome and cancer. Nat Rev Cancer 13:800–812. https://doi.org/10.1038/nrc3610
Derosa L et al (2018) Negative association of antibiotics on clinical activity of immune checkpoint inhibitors in patients with advanced renal cell and non-small-cell lung cancer. Ann Oncol 29:1437–1444. https://doi.org/10.1093/annonc/mdy103
Hamada K et al (2021) Antibiotic usage reduced overall survival by over 70% in non-small cell lung cancer patients on anti-PD-1 immunotherapy. Anticancer Res 41:4985–4993. https://doi.org/10.21873/anticanres.15312
Sun M, Wu W, Liu Z et al (2017) Microbiota metabolite short chain fatty acids, GPCR, and inflammatory bowel diseases. J Gastroenterol 52:1–8. https://doi.org/10.1007/s00535-016-1242-9
Skelly AN, Sato Y, Kearney S et al (2019) Mining the microbiota for microbial and metabolite-based immunotherapies. Nat Rev Immunol 19:305–323. https://doi.org/10.1038/s41577-019-0144-5
Coutzac C et al (2020) Systemic short chain fatty acids limit antitumor effect of CTLA-4 blockade in hosts with cancer. Nat Commun 11:2168. https://doi.org/10.1038/s41467-020-16079-x
Reck M, Ciuleanu TE, Cobo M et al (2021) First-line nivolumab plus ipilimumab with two cycles of chemotherapy versus chemotherapy alone (four cycles) in advanced non-small-cell lung cancer: CheckMate 9LA 2 year update. ESMO Open 6:100273
Sakai K, Sakurai T, De Velasco MA et al (2021) Intestinal microbiota and gene expression reveal similarity and dissimilarity between immune-mediated colitis and ulcerative colitis. Front Oncol 11:763468
Davar D et al (2021) Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science 371:595–602. https://doi.org/10.1126/science.abf3363
Baruch et al (2021) Fecal microbiota transplant promotes response in immunotherapy refractory melanoma patients. Science 371:602–609
Acknowledgements
We would like to thank Editage (www.editage.com) for English language editing.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
About this article
Cite this article
Yoshimura, K. Association of microbiota with cancer treatment. Int J Clin Oncol 28, 341–346 (2023). https://doi.org/10.1007/s10147-023-02302-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10147-023-02302-4