Abstract
We used transcriptome sequencing to investigate the hepatic postprandial responses of Rachycentron canadum (cobia), an important commercial fish species. In total, 150 cobia juveniles (50 per tank, triplicate) were fed ad libitum with a commercial diet for 7 days, fasted for 24 h, and fed for 10 min. The liver was sampled 10 min prior to feeding and 30 min, 1, 2, 4, 8, 12, and 24 h after the feeding event. Each sample was evaluated in terms of liver fatty acid profile and gene expression. Differential gene expressions were evaluated, focusing on fatty acid synthesis and oxidation pathways. In general, the liver fatty acid profile reflected diet composition. Docosahexaenoic acid (DHA), eicosapentaenoic acid (EPA), and n-3 long-chain polyunsaturated fatty acids (n-3 LC-PUFA) levels increased at 8 to 12 h but decreased at 24 h after the feeding event. A high number of differentially expressed genes (DEGs) were observed comparing fish that fasted for 8 h with those fasted for 30 min and 24 h, while a reduced number of DEGs was observed comparing individuals who fasted for 30 min compared with those who fasted for 24 h. Similarly, the main differences in the expression of genes related to the fatty acid biosynthesis and oxidation pathways were noticed in individuals who fasted for 8 h compared with those who fasted for 30 min and 24 h. The results suggested that the adequate time to sample the individuals ranged between 8 and 12 h after the meal since, apparently, after 24 h, differential gene expression was not necessarily influenced by food intake.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The main aim of performing nutritional trials with commercial finfish species is to define adequate levels of nutrients (primarily lipids and proteins) to promote optimum performance and health (Glencross 2009; Wade et al. 2014). Essential to animals, lipids and proteins are related to several functions, mainly energy production, cellular structuring, fat-soluble vitamin transport, and eicosanoid synthesis (Higgs and Dong 2000; Turchini et al. 2009). Thus, knowledge about the metabolic pathways that modulate lipid synthesis and oxidation processes is crucial in defining specific nutritional strategies based on the physiological characteristics of the target species (Glencross 2009; Tocher et al. 2003; Wade et al. 2014; Araújo et al. 2016). For most fish species, the liver is the primary lipogenic and lipolytic tissue, and it is related to the modulation of lipid metabolism, including fatty acid synthesis and oxidation processes (Tocher 2003). Thus, to understand the dynamics of the lipid metabolism pathways influenced by diet composition in fish, firstly, it is necessary to investigate the hepatic molecular responses after a single meal (Leaver et al. 2008; Wade et al. 2014; Araújo et al. 2016).
Molecular tools like transcriptome sequencing have been practical in different applications, such as identifying molecular markers and discovering and describing new genes and pathways, and differential analyses of gene expression. Such tools have been applied to several commercial aquaculture fish species, such as rainbow trout (Oncorhynchus mykiss, Rivas-Aravera et al. 2019), Atlantic salmon (Salmo salar, Glencross et al. 2015), barramundi (Lates calcarifer, Wade et al. 2014), dusky grouper (Epinephelus marginatus, Araújo et al. 2018), sea bass (Dicentrarchus labrax, Magnanou et al. 2014), sea bream (Sparus aurata, Mininni et al. 2014), and tilapia (Oreochromis niloticus, Zhao et al. 2020). Additionally, the development of transcriptome sequence technology and the advance of bioinformatics tools help in providing a broad overview of the post-feeding effects on the expression of genes related to the growth, immunological system, and energetic metabolism (e.g., synthesis, oxidation, and deposition of specific fatty acids) in finfish species (Leaver et al. 2008;Calduch-Giner et al. 2013; Qian et al. 2014; Glencross et al. 2015). Using information generated by barramundi liver transcriptome sequencing, Wade et al. (2014) mapped out, identified, and quantified the expression of several metabolic-relevant genes (mainly those related to carbohydrates, lipids, and proteins metabolism), improving the knowledge about the physiology of this species and, consequently, contributing to the development of further research with that species.
The knowledge regarding the postprandial effects on the expression of metabolic-relevant genes is essential to precisely investigate the physiological effects of the nutrients in diets. However, such understanding is only achievable if the adequate moment to sample the animals is known. Aiming to avoid stress by handling and poor water quality, several previous nutritional studies investigate the influence of diet composition on gene transcription, fasted the animals for 24 h before sampling. However, previous studies performed with finfish species such as Atlantic salmon (Valente et al. 2012), barramundi (Wade et al. 2014; Araújo et al. 2016; Poppi et al. 2019), and rainbow trout (Mente et al. 2017) showed that metabolic-relevant genes, especially those related to the modulation of fatty acid syntheses and oxidation pathways, such as fatty acid synthase (fas), stearoyl-CoA desaturase (scd), ATP citrate lyase (acyl), carnitine palmitoyltransferase (cpt), and 3-hydroxybutyrate dehydrogenase (bdh2), were drastically down-regulated between 8 and 12 h after the last meal, presenting consistent expression compared to the moment when these animals were fed. Therefore, based on that, it is assumed that in 24 h of fasting, the expression of protein- and lipid-relevant genes reached basal levels, probably not integrally reflecting the influence of the diet on their expression. However, it is worth mentioning that these responses will be modulated by fasting and interact with several other parameters, such as health condition, life stage, diet composition, and temperature (Leaver et al. 2008; Wade et al. 2014). Thus, the characteristics of every single nutritional trial are unique, and the time to sample the individuals/tissues after the last meal needs to be always considered to understand the animal’s physiological response properly.
Rachycentron canadum, popularly known as cobia, is an important worldwide commercial species and a targeted species for research to inform the aquaculture industry. However, no studies have been performed on the modulation of energetic metabolism during the postprandial period. Against this background, this study aimed to assess the molecular responses on the hepatic tissue of cobia juveniles after a single feed event. Additionally, this study is part of a project that aimed to investigate and understand the lipid metabolism of this important commercial species. The results provide insights into the physiological mechanisms modulated by the influence of the postprandial period, generating essential knowledge about hepatic lipid metabolism during fasting, which is essential to adequately design further trials focusing on lipid nutrition aiming to improve the performance and composition of this important commercial marine fish species.
Material and Methods
Experimental Design
All procedures performed in this study followed the Mogi das Cruzes University Institutional Animal Care and Use Ethics Committee (approval number: #008/2017). In total, 150 cobia juveniles (90.85 ± 11.75 g, mean ± SD) were acquired by Redemar Alevinos (Ilhabela, SP, Brazil), acclimated during 15 days in a 10,000 L tank (temperature average of 28 ± 0.4 ℃; dissolved oxygen 6.57 ± 0.7 mg L−1; and total ammonia nitrogen (TAN) ˂0.05 mg L−1, mean ± SD), and fed twice a day until apparent satiation with a commercial fish diet (Guabipirá, Guabi Nutrição e Saúde Animal S.A., S.P., Brazil). After acclimatization, the animals were transferred to three 1,000 L tanks (50 animals per tank, triplicate) and fed twice a day for 7 days with the same commercial diet. At the end of the seventh day, the animals were fasted for 24 h. After the fasting period, an uninterrupted single feed event (ad libitum) was carried out for 10 min. Ten minutes before and after 30 min, 1, 2, 4, 8, 12, and 24 h of the feed event, three fish from each tank (a total of 9 individuals) were carefully removed at the same time using a small net (aiming to avoid stress in the remained stock), immediately anesthetized and euthanized, and aliquots of the liver were frozen in liquid nitrogen and then transferred to the ultra-freezer (− 80 ℃) until the analysis.
Diet and Liver Fatty Acid Profile
The total lipids of the diet and liver were extracted using a chloroform/methanol/water mix (2:1:0.5) (modified from Folch et al. 1957). The lipid extracts were methylated using 5% HCl methanol prepared by dissolving 10% vol acetyl chloride in methanol, according to Christie (2003). The fatty acid analysis was performed using a gas chromatograph (GC, model 3900, CA, USA), a flame ionization detector (FID), and a CP-8410 autosampler. The FAME analysis used a capillary column (CP-Wax 52 CB, 0.25 μm thick, 0.25 mm inner diameter, and 30 m length). Hydrogen was used as a carrier gas at a linear velocity of 22 cm/s. The column was programmed to start at 170 °C for 1 min, followed by a 2.5 °C/min ramp to 240 °C and a final hold time of 5 min. The injector and FID temperature were kept at 250 and 260 °C, respectively. The FAMEs were identified by comparing their retention times to those obtained with commercial standards (Supelco, 37 components; Sigma-Aldrich; mixture Me93, Larodan, and Qualimix; PUFA fish M, menhaden Oil, Larodan).
RNA Isolation, Library Preparation, and Sequencing
Total RNA from the liver was extracted using an RNeasy Lipid Tissue kit (Qiagen, Germantown, MD, USA) following the manufacturer’s instructions. The quantity of extracted RNA was assessed by NanoDrop™ Spectrophotometer (Thermo Fisher Scientific, USA), and the quality was assessed by Agilent 2100 Bioanalyzer (Agilent Technologies, Germany). The RIN values of the RNA used were all above 7.5.
RNA from the liver of the three animals of each tank was used in the library construction using the TruSeq RNA Sample Preparation kit (Illumina Inc., USA) according to the manufacturer’s specifications. Transcriptome sequencing was performed with the Illumina NextSeq® platform (Illumina Inc., San Diego, CA, USA) using a paired-end sequencing strategy (2 × 75 bp).
De Novo Assembly, Functional Annotation, and Differential Expression
The detailed methodology, statistics, and summary of the de novo transcriptome assembled of cobia liver tissue were previously described by Aciole Barbosa et al. (2021). Due to the massive dataset generated by the transcriptome sequencing, the differential expression and quantitative real-time PCR (qRT-PCR) validation were restricted to three comparisons (30 min vs. 8 h, 30 min vs. 24 h, and 8 h vs. 24 h) based on the main differences observed in the liver fatty acid profile. In a UseGalaxy environment (Galaxy Community 2022), raw libraries were submitted to quality control checking using FastQC (Andrews 2010), and Fastp was used to remove low-quality (Q < 30) reads, adapters, and other contaminant sequences. To remove rRNA reads, the high-quality reads were aligned to sequences in the SILVA ribosomal RNA (rRNA) gene database (http://www.arbsilva.de/) using SortMeRNA (Kopylova et al. 2012). The National Center for Biotechnology Information (NCBI) UniVec database (https://www.ncbi.nlm.nih.gov/tools/vecscreen/univec/) was used to remove contaminants from libraries. Quality-filtered reads were then mapped to the R. canadum transcriptome available and described by Aciole Barbosa et al. (2021) using HISAT2 (Kim et al. 2015) with default parameters. StringTie (Pertea et al. 2015) was then used to assemble the mapped reads into transcripts, using the de novo transcriptome reconstruction method, allowing the identification of all transcripts present in each sample (including currently annotated genes, as well as newly identified elements and isomorphs). StringTie Merge combined redundant transcription structures, providing a non-redundant reference transcriptome with unique identifiers. Cufflinks (Trapnell et al. 2012) were next used to estimate expression values (FPKM) for each element in the StringTie-generated reference transcriptome. Transcriptome completeness was assessed using the Benchmarking Universal Single-Copy Orthologs (BUSCO) software, version 3.0 (Simão et al. 2015). Finally, the reads and the merged assembly were used to calculate expression levels and the effect size and statistical significance of observed differences using Cuffdiff with its default options (false discovery rate = 0.05) to obtain significant differentially expressed genes (DEGs) (Trapnell et al. 2010). Heatmaps based on expression patterns were built with a Complex Heatmap package (Gu et al. 2016) in a custom R script. We applied Eukaryotic Non-Model Transcriptome Annotation Pipeline (EnTAP) (Hart et al. 2020) on the query transcripts (blastx, e-value ≤ e−5) for homology against the National Center for Biotechnology Information non-redundant protein database (NCBI nr), NCBI proteins reference database (RefSeq), the curated Swiss-Prot database from UniProt Knowledgebase (UniProtKB) (UniProt Consortium 2023), and the EggNOG (Hernández-Plaza et al. 2023) protein database. The EggNOG hits also helped to assign the biological function to the genes, identifying GO (Gene Ontology) (Gene Ontology Consortium 2023) and KEGG (Kyoto Encyclopedia of Genes and Genomes) (Kanehisa et al. 2023) terms. EnTAP functional annotation process was carried out using a Dugong container environment (Menegidio et al. 2018) in BioPortainer Workbench (Menegidio et al. 2019).
KEGG terms were annotated and organized in Pathway Level 2 classes from KEGG Pathway Maps (https://www.genome.jp/brite/htext=br08901.keg). KEGG Automatic Annotation Server (KAAS) (Moriya et al. 2007) was used to annotate the transcript sequences in KEGG Orthology (KO) terms with the bi-directional best hit (BBH) method. KOs were utilized as input in the KEGG Mapper Color tool (Kanehisa et al. 2022) to map the differentially expressed transcripts based on their modulation; up-regulated elements were assigned as green, while down-regulated ones received the color red. Barramundi (Lates calcarifer) was set as the background because it is a species closely related to cobia and contained in KEGG.
Validation of DEGs
According to the manufacturer’s instructions, total RNA from liver samples was purified using PureLink™ RNA Mini Kit (12183018A Invitrogen™). To eliminate the potential contamination by genomic DNA, samples were treated with DNase using TURBO DNA-free™ Kit (AM1907 Invitrogen™); for this, previously, the RNA samples were diluted to 100 ng/µL and, after treatment, were stored at − 80 ℃. A total of 500 ng of total RNA was used as a template to synthesize cDNA using SuperScript™ III Reverse Transcriptase (18,080,093 Invitrogen™), and random primers were used according to the manufacturer’s instructions. Negative controls were also performed without reverse transcriptase, and the cDNA was stored at − 20 ℃.
A total of 10 differentially expressed unigenes (DEGs) from the liver were selected to verify the RNA-seq data’s reliability. The primers were designed using the PrimerQuest tools from IDT (https://www.idtdna.com/pages/tools/primerquest). The elongation factor 1 alpha (ef1a) and β-actn were used as endogenous reference genes. The oligonucleotides used as primers are described in Table 1. The sequences used in this procedure were obtained from the transcriptome sequencing of the liver from fish used in this same trial (Aciole Barbosa et al. 2021, BioProject PRJNA675281). They presented minimum efficiency of 95% and R2 of 0.96. All primers were synthesized by Thermo Fisher (Invitrogen™).
Statistical Analysis
The liver fatty acid profile data is presented as mean ± SD (Table 3). For the comparisons between the hepatic fatty acid composition of different sample times, normality and homogeneity of variance were tested using Levene’s test, and when the data allowed a parametric analysis, it was performed using one-way analysis of variance (ANOVA), followed by Tukey’s HSD test, using the software SIGMASTAT for Windows version 3.5 (SigmaStat Software, CA, USA). A significance level of P < 0.05 was used for all statistical tests.
Results
Liver Fatty Acid Profile
The fatty acid profile of the diet used in the trial is presented in Table 2. Several significant differences were observed in the hepatic fatty acid profile of the experimental animals sampled at different postprandial periods (Table 3). The most relevant was observed in the n-3 and n-6 PUFAs. Lower levels of DHA, EPA, 22:5n-3, 20:4n-6, 22:4n-6, and 22:5n-6 were observed in animals before the feed event and 30 min after feeding, especially compared with animals sampled at 8 and 12 h, and the percentage of these same fatty acids was significantly reduced in 24 h after the meal. These changes resulted in the same profile of total n-3 polyunsaturated fatty acids (n-3 PUFA) and, consequently, total long-chain polyunsaturated fatty acids (LC-PUFA).
DEG Analysis
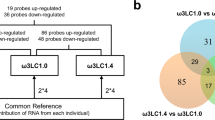
Figure 1A presents the DEGs in three comparison pairs for the different postprandial periods. The results showed 2,507 DEGs between animals sampled at 30 min vs. 8 h, with 1,591 transcripts up- and 916 down-regulated in fishes sampled at 30 min, compared with those sampled at 8 h after feeding. Comparing individuals sampled at 30 min vs. 24 h, a reduced number of 448 DEGs was observed, with 220 up- and 228 down-regulated in fish sampled at 30 min compared to those sampled at 24 h after a meal. Finally, 2,401 DEGs were observed comparing fish sampled at 8 h vs. 24 h, with 1,071 up- and 1,330 down-regulated in individuals sampled at 8 h compared to those sampled at 24 h after a meal. The heatmap based on the DEGs generated by the transcriptome sequencing corroborated the same expression profile observed in Fig. 1A, with a higher number of DEGs and an opposite profile between 30 min vs. 8 h and 8 h vs. 24 h, and a reduced number of DEGs at 30 min vs. 24 h (Fig. 1B).
Enrichment Analysis of Gene Ontology (GO) and KEGG Pathways of the DEGs
The GO annotation was used to classify the DEGs of each comparison by three categories, biological process (BP), cellular component (CC), and molecular function (MF) (Fig. 2). In general, the top 10 GO annotations were similar between different comparisons (with a few exceptions mainly in 30 min vs. 24 h). A higher number of transcripts were observed in all three categories for 30 min vs. 24 h and 8 h vs. 24 h, while fewer transcripts were observed at 30 min vs. 24 h.
The top 30 KEGG pathways of the Tier 2 classification were utilized to detect the main pathways for the DEGs (Fig. 3). The profile of KEGG pathway classification was consistent between different comparisons. The “signal transduction,” “cell growth and death,” and “carbohydrate metabolism” were the most prominent pathways observed in 30 min vs. 8 h, 30 min vs. 24 h, and 8 h vs. 24 h, respectively. The “lipid metabolism,” which is the primary metabolic pathway explored in this study, was also represented by a high number of transcripts in the three comparisons, with a higher number of DEGs noticed in 30 min vs. 8 h, followed by 8 h vs. 24 h and 30 min vs. 24 h (Fig. 3).
To further explore the physiological response profiles induced by fasting at specific pathways (fatty acid synthesis and oxidation), KEGG mapper analysis was performed to identify the DEGs (Fig. 4). Several differences were observed in the expression of genes related to the fatty acid biosynthesis and degradation pathways (Fig. 4A and B, respectively). The expression of fatty acid synthase (fasn – 2.3.1.85), acetyl CoA-carboxylase (acc—6.4.1.2.), and medium-chain acyl dehydrolase (mch—3.1.2.21) was up-regulated in animals sampled at 30 min compared to those sampled at 8 h after the meal. On the other hand, animals fasted by 8 h showed lower expression of these genes compared with those fasted by 24 h. The long-chain acyl-CoA synthetase (acsl – 6.2.1.3) expression was up-regulated in animals fasted by 30 min compared to those fasted by 8 h, while 3-oxoacyl-synthase II (fabf – 2.3.1.179) was down-regulated in fish fasted by 8 h compared with those fasted by 24 h (Fig. 4A). Regarding the fatty acid catabolism pathway, fish fasted by 30 min showed higher expression of long-chain acyl-CoA synthetase (acsl – 6.2.1.3) and butyryl-CoA dehydrogenase (acads – 1.3.8.1) compared to individuals sampled at 8 h. The acyl-CoA oxidase (acox – 1.3.3.6) expression was down-regulated, and carnitine palmitoyltransferase 1 (cpt1 – 2.3.1.21) was up-regulated in fish sampled at 30 min and 24 h compared to those sampled at 8 h after last meal, while acyl-CoA dehydrogenase (acadm – 1.3.8.7) was up-regulated in fish sampled at 30 min compared to those sampled at 8 and 24 h. Finally, the expression of alcohol dehydrogenase 1/7 (adh1_7 – 1.1.1.1) and aldehyde dehydrogenase (aldh – 1.2.1.3) was up-regulated in animals fasted by 30 min and 24 h compared to those fasted by 8 h (Fig. 4B).
Lipid-relevant DEGs identified by KEGG in the A fatty acid biosynthesis and B degradation pathways. The genes highlighted in yellow were significantly expressed in at least one of the three comparisons. Red squares represent a comparison between 30 min vs. 24 h, blue squares between 30 min vs. 8 h, and purple squares between 8 h vs. 24 h. The arrows in the colorful squares represent (↑) up-regulation of the specific transcript in the lower fasting time compared to the higher fasting time in each of the three comparisons and (↓) down-regulation of the specific transcript in the lower fasting time compared to the higher fasting time in each of the three comparisons. Arrow absence indicates no significant difference in the specific comparison
Verification and Validation of DEGs by qRT-PCR
To verify the accuracy of the RNA-seq data, we screened ten DEGs, analyzed them with qRT-PCR, and compared the differences. The expression profile of these genes (comparing different sample periods) was consistent with those observed in the RNA-seq analysis (Fig. 5). Therefore, the qRT-PCR results confirmed the reliability and accuracy of the RNA-seq data. The Pearson correlation between DEGs and qRT-PCR results is presented in the supplementary material.
Comparison of gene expression patterns obtained using comparative transcriptome analysis and qRT-PCR. A elovl6-like (elongase 6-like), B elovl6 (elongase 6), C mcm3 (minichromosome maintenance complex component 3), D hspa1s (heat shock 70 kDa protein 1), E acads (acyl-CoA dehydrogenase short chain), F acsl (long-chain acyl-CoA synthetases), G cpt (carnitine palmitoyltransferase), H fas (fatty acid synthase), I pdk (pyruvate dehydrogenase kinase), and J scd (stearoyl-CoA desaturase). The transcript expression levels of the target genes were normalized by the β-actin expression
Discussion
In order to understand the nutritional requirements of finfish species, it is crucial to investigate the molecular responses that occur each time the animals receive food since several metabolic pathways are differentially modulated during the postprandial period (Valente et al. 2012; Wade et al. 2014). However, studies focusing on the lipid metabolism of finfish species after feeding are limited/scarce. Despite the commercial relevance, cobia have not ever been used cobia as a model. Thus, the data generated in this study are essential in providing bases for further physiological and nutritional studies using this important marine commercial species.
As observed in previous studies performed with finfish species (Rombenso et al. 2016; Araújo et al. 2018; Gou et al. 2020; Fisher et al. 2022) and specifically with cobia (Araújo et al. 2021; Marques et al. 2021), with a few exceptions, the liver fatty acid profile reflected the composition of the diet. However, a lower percentage of important LC-PUFA, such as DHA and EPA, was observed in the liver compared to the diet, reflecting in the same profile as the total n-3 PUFA. The LC-PUFAs, mainly DHA and EPA, play several essential physiological functions in marine fish, such as cell membrane structuring and eicosanoid synthesis. The animals commonly retain it in the same proportion as those in the diet (Bell and Sargent 2003;Tocher 2003; Glencross 2009). Therefore, this paradigm can be changed by the influence of several other variables, especially tissue specificity, diet composition, and temperature (Tocher 2003). Based on the profile observed in the liver fatty acid composition, there are several statistical differences in the n-3 and n-6 PUFA levels, especially between 8 and 12 h compared to 30 min and 24 h after the feed event. These corroborate previous results from studies that also investigated the molecular responses of finfish species during the postprandial period (Valente et al. 2012; Wade et al. 2014; Araújo et al. 2016; Mente et al. 2017). Thus, as previously mentioned, the molecular results were focused on three main comparisons: (1) 30 min vs. 8 h, (2) 30 min vs. 24 h, and (3) 8 h vs. 24 h after feeding.
A reduced number of DEGs comparing individuals sampled at 30 min with those sampled at 24 h (448 transcripts in total, 220 up-regulated and 228 down-regulated at 30 min) after feeding were observed, while animals sampled at 8 h showed many DEGs compared to those sampled at 30 min (2,507 transcripts in total, 1,591 up-regulated and 916 down-regulated at 8 h) and 24 h (2,401 transcripts in total, 1,071 up-regulated and 1,330 down-regulated at 8 h) after feeding (see Fig. 1A). These results (DEGs and qRT-PCR) indicate an opposite profile in the expression of metabolic-relevant genes at 8 h compared with 30 min and 24 h (which showed consistent expression) after the feed event (Fig. 1B). Previous studies performed with barramundi juveniles (Wade et al. 2014; Araújo et al. 2016; Poppi et al. 2019) showed a differential expression of several metabolic-relevant genes after 8 h and 12 h compared with 1 h and 24 h after feeding. Additionally, Mente et al. (2017) observed significant changes in the expression of several protein-relevant genes in the liver of rainbow trout 6 to 12 h after the last meal, while at 24 h, the expression levels of these same genes were consistent to the moment after the feed event. Similarly, in Atlantic salmon juveniles, growth-relevant genes such as insulin-like growth factor (igf) and ubiquitin ligase MAFbx/atrogin-1 were initially down-regulated but restored to the basal expression levels after 12 h of fasting (Valente et al. 2012). Aiming to avoid stress by handling and poor water quality (due to the possibility of fish regurgitating during handling), previous studies performed with finfish species standardized a period of 24 h of fasting to sample the experimental animals (Zheng et al. 2004; Manor et al. 2015; Jin et al. 2017; Zhang et al. 2019; Gou et al. 2020). However, based on our results with cobia and previous results with other finfish species (Valente et al. 2012; Wade et al. 2014; Araújo et al. 2016; Diez et al. 2007; Mente et al. 2017; Poppi et al. 2019), after 24 h of the last meal, the expression of metabolic-relevant genes reaches basal levels, not necessarily reflecting the influence of the feed intake/composition.
Lipids are the primary source of energy for finfish, and the liver is considered the central lipolytic tissue (especially under fasting) and notably an essential fat deposit for energy in several species (Tocher 2003; Turchini et al. 2009). Thus, previous knowledge regarding the hepatic fatty acid synthesis and catabolism pathways is essential for designing physiological and nutritional trials to define finfish species’ nutrient requirements. Specifically related to the lipid-relevant pathways (biosynthesis and β-oxidation), a similar profile was observed in the expression of the total transcripts, with only one DEG (acadm) comparing fish fasted by 30 min vs. 24 h. In contrast, all the other DEGs were observed comparing fish fasted by 8 h vs. 30 min and 24 h. These results suggest that after 24 h of a single meal, the expression of lipid-relevant genes (such as the expression of other metabolic genes) reached basal levels (30 min after meal). Most lipogenic genes (mainly fas, acc, and acsl) were significantly down-regulated in individuals sampled at 8 h after a meal compared with those sampled at 30 m or 24 h (or both periods). Generally, the expression of fatty acid synthesis-relevant genes is strictly related to the tissue and diet fatty acid composition (Leaver et al. 2008; Torstensen et al. 2009). Previous studies in vivo (Alvarez et al. 2000; Datsomor et al. 2019) and in vitro (Menoyo et al. 2003) showed that higher levels of hepatic EPA and DHA inhibited lipogenesis in salmonids, corroborating the results observed herein since it was observed higher levels of fatty acids and total n-3 PUFAs in fish sampled at 8 h compared to those sampled at 30 min and 24 h after a meal. Acetyl CoA is activated to malonyl-CoA by ACC and iteratively assembled in the cytosol by FAS to form 16- and 18-carbon saturated fatty acids. Thus, consistent expression levels between both genes are expected. Consistent expression of fas and acc was also observed in zebrafish (Danio rerio) under fasting (Seiliez et al. 2013). However, differently than observed in cobia, zebrafish juveniles showed an up-regulation of these genes (and other lipid-relevant genes) after 6 h of the last meal.
In comparison, at 24 h, the expression levels of these genes reached basal levels. Similarly, Wade et al. (2014) observed that fatty acids synthesis-relevant genes (e.g., fas and scd) were up-regulated in barramundi sampled at 12 h compared with those fish sampled at 1 h and 24 h after feeding. These contradictory results in the expression of lipogenic genes during a postprandial period between different finfish species corroborate the hypothesis that lipogenesis is highly modulated by the interaction of several variables, primarily species specificity (capacity to tolerate fasting), fasting period, diet composition, and culture conditions (Tocher 2003; Leaver et al. 2008).
A different profile in the expression of genes related to fatty acid catabolism was observed in cobia sampled at different postprandial periods. While acox expression was significantly up-regulated, cpt1 was significantly down-regulated in fish sampled at 8 h compared with those sampled at 30 min and 24 h after the meal. Fatty acid catabolism can occur in two organelles: mitochondria and peroxisomes (Leaver et al. 2008). Acox catalyzes the rate-limiting step in the peroxisomal β-oxidation. At the same time, CPT1 is related to the fatty acid transport over the inner mitochondrial membrane to be β-oxidized (mitochondrial oxidation) (Leaver et al. 2008). This opposite profile in the acox and cpt1 expressions suggests that fish sampled at 8 h catabolized hepatic fatty acids preferentially by peroxisomal β-oxidation, while those sampled at 30 min and 24 h after feeding preferentially catabolized by mitochondrial β-oxidation. According to Leaver et al. (2008), typically in fish liver, peroxisomal β-oxidation contributes more significantly to the total β-oxidation. However, the capacity to catabolize fatty acids depends on the interaction of several biotic and abiotic factors, mainly temperature, fish size, life stage, and diet composition, that together will differently modulate fatty acid catabolism processes in fish.
Additionally, malonyl-CoA produced from ACC activity inhibits CPT1, justifying an opposite profile in the expression of both genes. Similar to those found in cobia, Diez et al. (2007) observed higher peroxisomal β-oxidation (ACOX activity) in sea bream juveniles after 6 h of the last meal, accompanied by a decrease in a mitochondrial β-oxidation (L-3-hydroxyacyl-CoA dehydrogenase—L3HOAD activity). However, interestingly, these effects were not observed at 24 h after feeding. Unfortunately, studies investigating fatty acid catabolism of finfish species during the postprandial period are scarce in the literature. However, the results obtained in this study and those found in sea bream by Diez et al. (2007) and barramundi by Wade et al. (2014) reinforce the importance of evaluating the expression of fatty acid catabolism-relevant genes during the postprandial period in finfish species.
Conclusion
This study performed a transcriptional profiling analysis of fasting in cobia juveniles to identify metabolism-related genes and pathways, focusing on lipid metabolism. The results showed the main differences in liver fatty acid profile and molecular responses between 8 and 12 h after the last meal. In comparison, after 24 h, the n-3 LC-PUFAs, global transcription, and differential expression of lipid-relevant genes reached basal levels (30 min after the last meal). Our results, differently than previous studies performed with other teleost species, suggest an adequate time to sample the animals/tissues ranging between 8 and 12 h after the last meal, since apparently, after 24 h, gene expression is not more influenced by feed intake/composition. However, these molecular responses should be evaluated for every single species/trial since the interaction of several biotic and abiotic variables, such as species specificity, life stage, diet composition, and temperature, can directly influence the hepatic molecular responses in finfish species, especially post-feeding.
Data Availability
Sequencing raw data were deposited in the Sequence Read Archive (SRA) repository of the National Center for Biotechnology Information (NCBI), under accession numbers SRR13009897, SRR13009896, SRR13009895, SRR13009894, SRR13009893, SRR13009892, SRR13009891, SRR13009890, SRR13009889, SRR13009888, SRR13009887, and SRR13009886, associated to the BioProject number PRJNA675281 and BioSamples numbers SAMN16708758, SAMN16708759, SAMN16708760, SAMN16708761, SAMN16708762, SAMN16708763, SAMN16708764, SAMN16708765, SAMN16708766, SAMN16708767, SAMN16708768, and SAMN16708769. The Transcriptome Shotgun Assembly (TSA) project has been deposited at DDBJ/EMBL/GenBank under accession number GIWT00000000. Intermediate data and high resolution figures can be found in the Open Science Framework (https://osf.io/ut5sn/).
References
Alvarez MJ, Diez A, Lopez-Bote C, Gallego M, Bautista JM (2000) Short-term modulation of lipogenesis by macronutrients in rainbow trout (Oncorhynchus mykiss) hepatocytes. Br J Nutr 84:619–628
Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc
Araújo B, Salini M, Glencross B, Wade N (2016) The influence of dietary fatty acid and fasting on the hepatic lipid metabolism of barramundi (Lates calcarifer). Aquac Res 7:3879–3893
Araújo B, Wade MW, de Mello PH, Rodrigues-Filho JA, Garcia CEO, de Campos MF, Botwright N, Hashimoto DT, Moreira RG (2018) Characterization of lipid metabolism genes and the influence of fatty acid supplementation in the hepatic lipid metabolism of dusk grouper (Epinephelus marginatus). Comp Biochem Physis Part A 219:1–9
Araújo B, Rodriguez M, Honji RM, Rombenso AN, Rio-Zaragoza OB, Cano A, Tinajero A, Mata-Sotres JA, Viana MT (2021) Arachidonic acid modulated lipid metabolism and improved the productive performance of striped bass (Morone saxatilis) juveniles under sub- to optimal temperatures. Aquaculture 530:735939
Aciole Barbosa D, Araujo BC, Branco GS, Simeone AS, Hilsdorf AWS, Jabes DL, Nunes LR, Moreira RG, Menegidio FB (2021) Transcriptome profiling and microsatellite identification in cobia (Rachycentron canadum), using high-throughput RNA sequencing. Mar Biotechnol 24:255–262
Bell JG, Sargent JR (2003) Arachidonic acid in aquaculture feeds: current status and future opportunities. Aquaculture 218:491–499
Calduch-Giner JCA, Bermejo-Nogales A, Benedito-Palos L, Estensoro I, Ballester-Lozano G, Sitjà Bobadilla A, Pérez-Sánchez J (2013) Deep sequencing for de novo construction of a marine fish (Sparus aurata) transcriptome database with a large coverage of protein coding transcripts. BMC Genomics 14:178
Christie WW (2003) Lipids analysis. The Oily Press, Bridgwater, p 298
Datsomor AK, Zic N, Li K, Olsen RE, Jin Y, Vik JO, Edvardsen RB, Grammes F, Wargelius A, Winge P (2019) CRISPR/Cas9-mediated ablation of elovl2 in Atlantic salmon (Salmo salar L.) inhibits elongation of polyunsaturated fatty acids and induces Srebp-1 and target genes. Sci Rep 7533
Diez A, Menoyo D, Perez-Benavente S, Calduch-Giner JA, Vega-Rubin de Celis S, Obach A, Favre-Krey L, Boukouvala E, Leaver MJ, Tocher DR, Perez-Sanchez J, Krey G, Bautista JM (2007) Conjugated linoleic acid affects lipid composition, metabolism, and gene expression in gilthead sea bream (Sparus aurata L). J Nutr 137:1363–1369
Fisher H, Romano N, Renukdas N, Kumar V, Sinha AK (2022) Comparing black soldier fly (Hermetia illucens) larvae versus prepupae in the diets of largemouth bass, Micropterus salmoides: effects on their growth, biochemical composition, histopathology, and gene expression. Aquaculture 546:737323
Folch J, Less M, Stanley GH (1957) A simple method for the isolation and purification of total lipids from animal tissues. J Biol Chem 226:497–503
Galaxy Community (2022) The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2022 update. Nucleic Acids Res 50:W345–W351. Advance online publication. https://doi.org/10.1093/nar/gkac247
Gene Ontology Consortium (2023) The Gene Ontology Knowledgebase in 2023 Genetics, iyad031. Advance Online Publication. https://doi.org/10.1093/genetics/iyad031
Glencross BD (2009) Exploring the nutritional demand for essential fatty acids by aquaculture species. Rev Aquac 1:71–124
Glencross BD, De Santis C, Bicskei B, Taggart JB, Bron JE, Betancor MB, Tocher DR (2015) A comparative analysis of the response of the hepatic transcriptome to dietary docosahexaenoic acid in Atlantic salmon (Salmo salar) post-smolts. BMC Genomics 16:684
Gou N, Ji H, Chang Z, Zhong M, Deng W (2020) Effects of dietary essential fatty acid requirements on growth performance, fatty acid composition, biochemical parameters, antioxidant response and lipid related genes expression in juvenile Onychostoma macrolepis. Aquaculture 528:735590
Gu Z, Eils R, Schlesner M (2016) Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32:2847–2849
Hart AJ, Ginzburg S, Xu M, Fisher CR, Rahmatpour N, Mitton JB, Paul R, Wegrzyn JL (2020) EnTAP: bringing faster and smarter functional annotation to non-model eukaryotic transcriptomes. Mol Ecol Resour 20:591–604
Hernández-Plaza A, Szklarczyk D, Botas J, Cantalapiedra CP, Giner-Lamia J, Mende DR, Kirsch R, Rattei T, Letunic I, Jensen LJ, Bork P, von Mering C, Huerta-Cepas J (2023) eggNOG 6.0: enabling comparative genomics across 12 535 organisms. Nucleic Acids Res 51:D389–D394
Higgs DA, Dong FM (2000) Lipids and fatty acids. In: Stickney RR (ed) Encyclopedia of Aquaculture. John Wiley & Sons Inc, New York, pp 476–496
Jin M, Lu Y, Yuan Y, Li Y, Qiu H, Sun P, Ma H, Ding L, Zhou Q (2017) Regulation of growth, antioxidant capacity, fatty acid profiles, hematological characteristics and expression of lipid related genes by different dietary n-3 highly unsaturated fatty acids in juvenile black seabream (Acanthopagrus schlegelii). Aquaculture 471:55–65
Kanehisa M, Furumichi M, Sato Y, Kawashima M, Ishiguro-Watanabe M (2023) KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res 51:D587–D592
Kanehisa M, Sato Y, Kawashima M (2022) KEGG mapping tools for uncovering hidden features in biological data. Protein Science: a Publication of the Protein Society 31:47–53
Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360
Kopylova E, Noé L, Touzet H (2012) SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28:3211–3217
Leaver MJ, Bautista JM, Björnsson LR, Jönsson E, Krey G, Tocher DR, Torstensen BE (2008) Towards fish lipid nutrigenomics: current state and prospects for fin-fish aquaculture. Rev Fish Sci 56:1064–1262
Magnanou E, Klopp C, Noirotm C, Besseau L, Falcon J (2014) Generation and characterization of the sea bass Dicentrarchus labrax brain and liver transcriptomes. Gene 544:56–66
Manor ML, Weber GM, Cleveland BM, Yao J, Kenney PB (2015) Expression of genes associated with fatty acid metabolism during maturation in diploid and triploid female rainbow trout. Aquaculture 435:178–186
Marques VH, Moreira RG, Branco GS, Honji RM, Rombenso AN, Viana MT, Mello PH, Mata-Sotres JA, Araujo BC (2021) Different saturated and monounsaturated fatty acids levels in fish oil-free diets to cobia (Rachycentron canadum) juveniles: effects in growth performance and lipid metabolism. Aquaculture 541:736843
Menegidio FB, Jabes DL, Costa de Oliveira R, Nunes LR (2018) Dugong: a Docker image, based on Ubuntu Linux, focused on reproducibility and replicability for bioinformatics analyses. Bioinformatics 34:514–515
Menegidio FB, Barbosa DA, Gonçalves RS, Nishime MM, Jabes DL, Oliveira RC, Nunes LR (2019) Bioportainer Workbench: a versatile and user-friendly system that integrates implementation, management, and use of bioinformatics resources in Docker environments. Giga Sci 8(4):giz041
Menoyo D, Lopez-Bote CJ, Bautista JM, Obach A (2003) Growth, digestibility, and fatty acid utilization in large Atlantic salmon (Salmo salar) fed varying levels of n-3 and saturated fatty acids. Aquaculture 225:295–307
Mente E, Pierce GJ, Antonopoulou E, Stead D, Martin SAM (2017) Postprandial hepatic protein expression in trout Oncorhynchus mykiss a proteomics examination. Aquaculture 9:79–85
Mininni AN, Milan M, Ferrareso S, Petochi T, Marco PD, Marino G, Livi S, Romualdi C, Bargelloni L, Patarnello T (2014) Liver transcriptome analysis in gilthead sea bream upon exposure to low temperature. BMC Genomics 15:765
Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M (2007) KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res 35:W182–W185
Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL (2015) StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33:290–295
Poppi DA, Moore SS, Wade NM, Glencross BD (2019) Postprandial plasma free amino acid profile and hepatic gene expression in juvenile barramundi (Lates calcarifer) is more responsive to feed consumption than to dietary methionine inclusion. Aquaculture 501:345–358
Qian L, Qian W, Snowdon RJ (2014) Sub-genomic selection patterns as a signature of breeding in allopolyploid Brassica napus genome. BMC Genomics 15:1170
Rivas-Aravena A, Fuentes-Valenzuela M, Escobar -Aguirre S, Gallardo-Escarate C, Molina A, Valdés JA, (2019) Transcriptomic response of rainbow trout (Oncorhynchus mykiss) skeletal muscle to Flavobacterium psychrophilum. Comp Biochem Physiol Part D 31:100596
Rombenso AN, Trushenski JT, Jirsa D, Drawbridge M (2016) Docosahexaenoic acid (DHA) and arachidonic acid (ARA) are essential to meet LC-PUFA requirements of juvenile California yellowtail (Seriola dorsalis). Aquaculture 463:123–134
Seiliez I, Medale F, Aguirre P, Larquier M, Lanneretonne L, Alami-Durante H, Panserat S, Skiba-Cassy S (2013) Postprandial Regulation of Growth- and Metabolism-Related Factors in Zebrafish 10:237–248
Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM (2015) BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 19:3210–3212
Tocher DR (2003) Metabolism and functions of lipids and fatty acids in teleost fish. Rev Fish Sci 11:107–184
Torstensen BE, Nanton DA, Olsvik PA, Sundvold H, Stubhaug I (2009) Gene expression of fatty acid-binding proteins, fatty acid transport proteins (cd36 and FATP) and β-oxidation-related genes in Atlantic salmon (Salmo salar L.) fed fish oil or vegetable oil. Aquacult Nut 15:440–451
Trapnell C, Willians BA, Pertea G, Mortavazi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7:562–579
Turchini GM, Torstensen BE, Ng WK (2009) Fish oil replacement in finfish nutrition. Rev Aquac 1:10–57
UniProt Consortium (2023) UniProt: the Universal Protein Knowledgebase in 2023. Nucleic Acids Res 51:D523–D531
Valente LMP, Bower NI, Johnston IA (2012) Postprandial expression of growth-related genes in Atlantic salmon (Salmo salar L.) juveniles fasted for 1 week and fed a single meal to satiation. Br J Nutr 108:2148–2157
Wade NM, Skyba-Cassy S, Dias K, Glencross BD (2014) Postprandial molecular responses in the liver of the barramundi. Lates Calcarifer Fish Physiol Biochem 40:427–443
Zhang M, Chen C, You C, Chen B, Wang S, Li Y (2019) Effects of different dietary ratios of docosahexaenoic to eicosapentaenoic acid (DHA/EPA) on the growth, nonspecific immune indices, tissue fatty acid compositions and expression of genes related to LC-PUFA biosynthesis in juvenile golden pompano Trachinotus ovatus. Aquaculture 505:488–495
Zhao Y, Zhang C, Zhou H, Song L, Wang J, Zhao J (2020) Transcriptome changes for Nile tilapia (Oreochromis niloticus) in response to alkalinity stress. Comp Biochem Physiol 33:100651
Zheng XZ, Tocher DR, Dickson CA, Bell JB, Teale AJ (2004) Effects of diets containing vegetable oil on expression of genes involved in highly unsaturated fatty acid biosynthesis in liver of Atlantic salmon. Aquaculture 234:467–483
Funding
This study was supported by grants from the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP: 2019/26018–0). Bruno C. Araújo received a fellowship grant from São Paulo Research Foundation (FAPESP) (2016/12435–0). David Aciole Barbosa was a recipient of a scholarship grant from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES: 88882.365823/2019–01). A.W.S. Hilsdorf (309570/2021–2) and Renata G. Moreira (305493/2019–1) are recipients of CNPq productivity scholarships.
Author information
Authors and Affiliations
Contributions
Bruno C. Araújo, David Aciole Barbosa, Fabiano B. Menegidio, Giovana S. Branco, and Renato M. Honji: Conceptualization, data curation, formal analysis, investigation, writing, and editing. Victor H. Marques, Paulo H. de Mello, Marcelo V. Kitahara, and Artur N. Rombenso: Formal analysis, writing, review, and editing. Renata G. Moreira and Alexandre W.S. Hilsdorf: Conceptualization, methodology, resources, and review.
Corresponding author
Ethics declarations
Ethics Approval
This study was conducted according to the guidelines and approval of the Mogi das Cruzes University Institutional Animal Care and Use Ethics Committee (#008/2017).
Disclaimer
The authors alone are responsible for the content and the writing of the paper.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Araújo, B.C., Barbosa, D.A., Honji, R.M. et al. Post-feeding Molecular Responses of Cobia (Rachycentron canadum): RNA-Sequencing as a Tool to Evaluate Postprandial Effects in Hepatic Lipid Metabolism. Mar Biotechnol 25, 358–371 (2023). https://doi.org/10.1007/s10126-023-10209-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10126-023-10209-4