Abstract
The aim of this study was to investigate the molecular features and the antibiotic resistance profile of 98 clinical Staphylococcus aureus isolates collected during 6 months in two hospitals of Kabul, Afghanistan. For all isolates, antimicrobial resistance patterns were determined by the disc diffusion method (including methicillin resistance which was detected using cefoxitin). The presence of the mecA/mecC genes was detected by PCR. Strains were then extensively characterized using microarray analysis. Of the 98 S. aureus isolates, methicillin-resistant S. aureus (MRSA) prevalence was high at 66.3%. Antibiotic susceptibility testing also revealed a high resistance rate to penicillin (100%), erythromycin (66.3%), ciprofloxacin (55.1%), and cotrimoxazole (40.8%). Resistance to tobramycin was detected in 25.5%, to gentamicin in 16.3%, to chloramphenicol in 34.7%, and to doxycycline in 23.5% of the isolates. All the MRSA isolates were mecA-positive and none of them harbored mecC. Isolates were grouped into twelve clonal complexes and twenty-seven distinct clones. The most frequently detected clones were the Southwest Pacific clone (CC30-MRSA-IV PVL+) (21/65 MRSA, 32.3%), the CC22-MRSA-IV TSST-1+ clone (11/65 MRSA, 16.9%), and the Bengal Bay clone (ST772-MRSA-V PVL+) (11/65 MRSA, 16.9%). The PVL genes were found in 59.2% (46/65 MRSA and 12/33 methicillin-susceptible S. aureus, MSSA) and tst1 gene in 16.3% of isolates. This molecular study highlights the high prevalence of MRSA and the large genetic diversity of the S. aureus isolates circulating and detected in two hospitals of Kabul, with the presence of multiple virulence and antibiotic resistance genes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Staphylococcus aureus is a major human pathogen responsible for serious infections in both community and hospital settings [1]. S. aureus can cause a wide range of infections, from minor skin infections, such as carbuncles, folliculitis, and impetigo, to deep-seated (abscesses or cellulitis) or life-threatening infections such as pneumonia, osteomyelitis, bacteremia, toxic shock syndrome, or endocarditis. The global emergence and spread of methicillin-resistant S. aureus (MRSA) in hospitals since the 1960s are a public health concern and limit the therapeutic arsenal for severe staphylococcal infections. Indeed, healthcare-associated (HA-) MRSA are usually resistant to most antimicrobial agents. In the case of severe infections due to these multi-resistant strains, glycopeptides are often the drugs of last resort; however, isolates with reduced susceptibility to vancomycin are frequently reported [2], and some vancomycin-resistant S. aureus (VRSA) strains harboring the vanA gene from enterococci have also been described but are very rare [3]. On the other hand, the emergence and spread of community-acquired (CA-) MRSA clones expressing the Panton-Valentine leukocidin (PVL) in the last two decades are an additional cause for concern and have changed the epidemiology of MRSA [4, 5]. These strains are generally associated with recurrent skin and soft tissue infections (SSTIs) but can also cause necrotizing pneumonia or destructive bone and joint infection [6]. Interestingly, some CA-MRSA clones predominate in geographically restricted areas while others have achieved a pandemic level.
In central Asia, including Afghanistan, studies about MRSA have mainly focused on the prevalence of MRSA, but few data are available about the circulating clones. In this part of the world, S. aureus infections in healthcare settings show a high prevalence of MRSA [7]. For example, in Pakistan, a multicentric study in four hospitals showed a MRSA rate of 41.9% in 2006–2008 [8]. In India, two multicentric studies described comparable MRSA rates of 41% and 45% in 2008–2009 and 2011, respectively [9, 10]. In Iran, a systematic review reported a mean MRSA rate of 43.0% between 2000 and 2016 [11] and several VRSA isolates were described [12]. More specifically, in Afghanistan, a recent study concerning S. aureus infections diagnosed in Kabul hospitals highlighted a high rate of methicillin resistance (56.2%) and an increase in the prevalence of multidrug-resistant (MDR)-MRSA compared to the neighboring countries [13]. In parallel, an overuse of antimicrobials was described during the last decade in primary healthcare clinics of five major provinces of Afghanistan, including Kabul [14] and, in 2014, some authors raised the question of over prescription of antibiotics in a district hospital in Kabul where more than 50% of out-patients were prescribed at least one antibiotic [15]. This widespread use of antimicrobial therapies could have contributed to the increase of MRSA prevalence in Afghanistan and the selection of MDR clones.
Nevertheless, although molecular typing data for S. aureus and MRSA are abundantly available for Western Europe, North America, or Australia, few studies have been conducting to describe S. aureus epidemiology in the rest of the world, including Central Asia and more specifically, Afghanistan. In this context, the aim of this work was to investigate the molecular features of S. aureus strains isolated from clinical samples in two hospitals of Kabul, as well as their antibiotic resistance profile.
Materials and methods
Strains collection
From January to June 2017, all S. aureus isolates cultured from in- and out-patient samples in two hospitals located in the center of Kabul (Maiwand and Ibn-Sina hospitals) were collected. During the 6-month study period, a total of 98 isolates of S. aureus obtained from 98 individual patients were characterized. The strains were isolated from various types of clinical samples: pus (skin and soft tissue infections; 65.3%), ear pus (18.4%), blood (7.1%), urine (3.1%), vaginal swab (3.1%), tracheal aspirations (2%), and semen (1%). Out of the 98 patients, 33 (33.7%) were women and 65 (66.3%) were men. The mean age was 19 years ([1–75] years, median 12 years).
Phenotypic assays
Standard microbiological procedures were conducted on clinical samples with minimum delay for culture, confirmatory tests, and antibiotic susceptibility testing (AST). Presumptive S. aureus isolates were subcultured for 18–24 h at 37 °C onto blood agar base medium (Oxoid, Basingstoke, UK) supplemented with 5% sheep blood. Confirmatory tests were carried out for the identification of S. aureus isolates, by performing Pastorex Staph Plus (Bio-Rad, Marnes-la-Coquette, France) and coagulase assay. AST was performed on Mueller-Hinton agar (Oxoid) by Kirby Bauer disc diffusion method according to the 2017 guidelines of the European Committee on Antimicrobial Susceptibility Testing (EUCAST) [16]. The following panel of antibiotics was tested: penicillin G, cefoxitin (used to detect methicillin/oxacillin resistance), tobramycin, gentamicin, erythromycin, clindamycin, ciprofloxacin, chloramphenicol, doxycycline, rifampicin, and cotrimoxazole.
Detection of mecA/mecC genes and agr typing by multiplex PCR
Cellular DNA was obtained from S. aureus colonies grown overnight on blood agar plates using DNA Extraction Kit (Promega, USA) in accordance with the manufacturer’s instructions. Detection of the mecA and mecC genes and agr typing were performed by multiplex PCR using primers already published [17,18,19].
Molecular characterization of strains by DNA microarray
DNA was extracted and purified using commercial extraction kits (DNeasy kit and Promega instrument; Qiagen, Hilden, Germany), according to the manufacturer’s protocol. The DNA microarray Identibac S. aureus genotyping® (Alere Technologies, Jena, Germany) was used as previously described [20]. This microarray allows the detection of 336 different target sequences corresponding to 185 genes and their allelic variants. The assignation of isolates to clonal complexes (CCs) was determined by comparison of the hybridization profiles to previously typed multi-locus sequence typing reference strains [20].
Statistical analysis
Statistical analysis was done using SPSS 21 (IBM Inc., Chicago). Binary logistic regression was used to determine the association between MRSA or MSSA status, gender, and age. Chi-square test was used to compare the resistance profiles and the presence of virulence genes between MRSA and MSSA (methicillin-susceptible S. aureus). A p value less than 0.05 was considered statistically significant.
Results
Antibiotic susceptibility testing
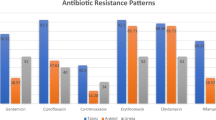
Out of the 98 S. aureus isolates, all were resistant to penicillin and 65 (66.3%) were methicillin-resistant (Table 1). Twenty-five (25.5%) were resistant to tobramycin, 16 (16.3%) to gentamicin, 65 (66.3%) to erythromycin, 11 (11.2%) to clindamycin, 54 (55.1%) to ciprofloxacin, 34 (34.7%) to chloramphenicol, 23 (23.5%) to doxycycline, 3 (3.1%) to rifampicin, and 40 (40.8%) to cotrimoxazole. MRSA isolates were statistically more frequently resistant to gentamicin (p = 0.002), erythromycin (p < 0.001), ciprofloxacin (p < 0.001), and cotrimoxazole (p = 0.017) than MSSA (Table 1). The difference of MRSA distribution was not statistically significant according to gender (p = 0.39) and age (p = 0.25).
Molecular characterization of S. aureus isolates
Various resistance genes were detected in the 98 collected isolates: blaZ in almost all isolates (97/98, 99%), msrA and mphC in 43/98 (43.9%), ermC in 16/98 (16.3%), aphA3 in 44/98 (44.9%), aadD in 22/98 (22.4%), aacA-aphD in 16/98 (16.3%), and tetK in 39/98 isolates (39.8%) (Table 2). No strain harbored the resistance genes vanA, qacA, qacC, fusB, mupR, and cfr. The presence of msrA, mphC, aacA-aphD, aphA3, sat, and dfrA was higher in MRSA than in MSSA whereas the presence of lnuA, tetK, and tetEfflux was higher in MSSA. Out of the 65 isolates phenotypically resistant to methicillin, all harbored the mecA gene and no isolate was positive for mecC. Concerning the staphylococcal chromosome cassette mec (SCCmec) element of these MRSA isolates, 47 (72.3%) harbored SCCmec type IV, 17 (26.2%) harbored SCCmec type V, and one (1.5%) harbored SCCmec type III.
Regarding the molecular typing of the isolates by microarray, a wide clonal diversity was observed. The 98 isolates belonged to twelve different clonal complexes (CCs) and were assigned to 27 distinct clones (Tables 3 and 4). Three CCs (CC1, CC22, and CC30) dominated and accounted for 65.3% of the isolates; four other CCs (CC8, CC15, CC121, and CC398) accounted for 24.5%.
The most frequent detected clone was the Southwest Pacific clone (CC30-MRSA-IV PVL+), which accounted for 21.4% (21/98) of all isolates and 32.3% (21/65) of MRSA. It was mostly isolated from skin and soft tissue infections. Isolates of this clone were resistant to multiple antibiotics, particularly oxacillin (100%), erythromycin (95.2%), ciprofloxacin (95.2%), and cotrimoxazole (57.1%). Various resistance genes were detected in these isolates: blaZ operon, msrA, mphC, fosB and tetEfflux in all isolates; aphA3 and sat in 19/21 (90.5%) isolates; aadD and tetK in 1/21 (4.8%). The enteroxin gene cluster egc (seg, sei, sem, sen, seo, and seu) was detected in all these isolates.
The second most frequent detected clone was the Bengal Bay clone (ST772-MRSA-V PVL+), which accounted for 11.2% (11/98) of all isolates and 16.9% (11/65) of MRSA. Isolates of this clone were also resistant to multiple antibiotics, particularly oxacillin (100%), tobramycin (63.6%), gentamicin (54.6%), erythromycin (100%), ciprofloxacin (100%), chloramphenicol (63.6%), and doxycycline (63.6%). Various resistance genes were detected in these isolates: blaZ operon, msrA, mphC, fosB, and tetEfflux in all isolates; tetK in 6/11 (54.6%); and aacA-aphD in 5/11 (45.5%). Nine enterotoxin genes (sea, sec, sel, and the egc) were detected in almost all of these isolates (Table 3). Another clone, the CC22-MRSA-IV TSST1+ (toxic shock syndrome toxin-1) clone, accounted for 11/98 (11.2%) of all isolates and 11/65 (16.9%) of MRSA isolates. Isolates of this clone were less resistant to antibiotics: oxacillin (100%), erythromycin (27.3%), doxycycline (27.3%), rifampicin (18.2%), and cotrimoxazole (36.4%). Various resistance genes were detected in these isolates: blaZ operon in 10/11 (90.9%), ermC in 2/11 (18.2%), dfrA in 11/11 (100%), and tetK in 3/11 (27.3%). The egc cluster was detected in all of these isolates. All other clones of MRSA were detected in 6 or less isolates (< 7%) and presented a high diversity (Table 3).
Out of the 33 MSSA isolates, 6 (18.2%) belonged to the CC15 and did not harbor genes for PVL, TSST-1, and enterotoxins. Five MSSA isolates (15.2%) belonged to the CC121-MSSA PVL+ clone and harbored the resistance genes blaZ, fosB, tetK, and tetEfflux, and virulence genes encoding PVL and enterotoxins (seb, and egc). Five MSSA isolates (15.2%) belonged to the ST291/813 and four (12.1%) belonged to the CC30-MSSA PVL+ clone. All other clones of MSSA were detected in three or less isolates (4% of all strains) but showed high diversity (Table 4).
The overall rate of PVL-positive isolates was 59.2% (58/98), these isolates belonging to 8 MRSA and 5 MSSA clones. The prevalence of PVL genes was higher in MRSA than in MSSA (70.8% vs 36.4%, p = 0.001). The arginine catabolic mobile element (ACME) locus was present in only one strain of MRSA belonging to the USA300 clone (ST8-MRSA-IV PVL+), isolated from an ear pus. Contrary to PVL, few isolates harbored the TSST-1 (16/98, 16.3%). Table 5 shows the prevalence data for virulence genes in the S. aureus isolates collected.
Discussion
This epidemiological study investigated the antibiotic resistance profile and performed the molecular characterization of MRSA and MSSA clinical isolates at two main health facilities in Kabul. The overall prevalence of MRSA was high at 66.3% (65/98). A very high genetic diversity was detected using microarray analysis, with 27 distinct clones among the 98 isolates collected during a 6-month period.
This study is the first molecular study of S. aureus clinical isolates in Afghanistan and reports the various clones circulating in its capital. The prevalence of MRSA in this study was higher compared to findings of similar studies conducted in neighboring countries such as Pakistan [21], Iran [22], Saudi Arabia [23], and Turkey [24]. In this study, a wide clonal diversity was detected with twelve different clonal complexes, along with 13 distinct MRSA and 14 MSSA clones. The typing results revealed a predominance of three MRSA clones, representing 44% of the isolates: Southwest Pacific (CC30-MRSA-IV PVL+), Bengal Bay (ST772-MRSA-V PVL+), and CC22-MRSA-IV TSST-1+ clones. The 24 other clones were present in one to 6 isolates.
The main limitation of this study is that only S. aureus isolates collected in two hospitals of Kabul were included. Therefore, even if these hospitals perform most of the bacteriological analyses in Kabul, the results do not reflect the global epidemiology in Afghanistan and are only representative of S. aureus epidemiology for infections diagnosed in hospitals. Nevertheless, the huge diversity of strains highlighted by the typing results illustrates the position of Kabul as a center of intense international exchanges.
Three different MRSA and two different MSSA clones were identified within CC30. The Southwest Pacific clone (CC30-MRSA-IV PVL+, USA1100) was the most prevalent clone, detected in 32.3% of MRSA isolates. This MDR and PVL+ clone is widely distributed in the Pacific islands and was first identified in New Zealand among the Samoan immigrant population [25]. It is a widespread CA-MRSA clone in Australia and has been found in several European countries as well as in Hong Kong, Taiwan, the USA, Saudi Arabia, the United Arab Emirates, and Iran [25,26,27]. The major occurrence of this clone in Afghanistan could possibly be linked to the large migratory movements of Afghans to Australia, Saudi Arabia, Iran, European countries, and some of the southwest Pacific countries, especially Indonesia, and Malaysia. The WA MRSA-124 clone (CC30-MRSA-V/VT PVL+), detected in 9.2% of MRSA isolates, is another virulent and MDR clone and was isolated mainly from SSTIs. The same lineage has already been described in Egypt [28] and in a patient living in the German/Polish border region and presenting a SSTI [4].
Two MSSA and two MRSA clones were identified within the CC1. Within this clonal complex, the Bengal Bay clone (ST772-MRSA-V PVL+), detected in 19.6% of MRSA isolates, is nowadays the dominant clone in India [29, 30]. As the Southwest Pacific clone, this one is also PVL+ and MDR. Studies from Pakistan, Nepal, and Australia also reported the predominance of this clone [31,32,33] which has become endemic in these regions. The presence of this clone in Kabul may illustrate an epidemiological link between Afghanistan and the neighboring countries, especially India and Pakistan where many Afghan families travel for medical treatment, and many young students go to pursue their higher education. This clone appears to be an increasing public health threat in several countries in the region, as it can be encountered in hospital as well as in community settings [31]. One PVL+ isolate, collected in Kabul from a SSTI, belonged to the USA400 pulsotype, one of the first-described PVL+ CA-MRSA. This one was reported to cause fatal infections in healthy children in the late 1990s in the USA [34] and then sporadically described in Australia, Germany, and the UK. Prior to this study, USA400 had not been reported in the neighboring countries such as Pakistan and Iran.
Two MRSA and one MSSA clones were identified within the CC22. This is a common and widespread clonal complex, and different MRSA lineages have emerged from this genetic background. The CC22-MRSA-IV TSST-1+ clone, detected in 19.6% of all MRSA isolates, has been mostly reported in Saudi Arabia and the Middle East region [25]. Its presence in Afghanistan could be related to the large movements of Afghans to Saudi Arabia for pilgrimage each year. Seven MRSA isolates were assigned to the CC22-MRSA-IV PVL+. This clone has been described in Germany, Australia, England, Ireland, Hong Kong, and the United Arab Emirates [4, 35]. Both clones were reported in hospital-acquired infections from Pakistan as well [31].
Four different MRSA clones were identified within the CC8. Among them, the USA500 clone (CC8-MRSA-IV PVL-) was predominant. This clone has been identified in the USA, Australia, the UK, and Ireland, and sporadically in Germany [4, 36]. Interestingly, only one isolate was identified as the well-known USA300 (CC8-MRSA-IV PVL+) clone. This clone of CA-MRSA, mainly reported in the USA, was also reported in numerous countries in Europe or Asia such as Saudi Arabia, Qatar, and Pakistan [4, 25, 32, 37]. One isolate was also identified as belonging to the Vienna/Hungarian/Brazilian clone (ST239-MRSA-III). This HA-MRSA lineage is probably the oldest pandemic MRSA strain and has been reported in many European countries [4, 38], as well as in Asia including Saudi Arabia [25], Iran [27], and Pakistan [32].
The overall prevalence of PVL among S. aureus isolates collected in Kabul was 59.2%, these isolates belonging to 8 MRSA and 5 MSSA clones, with a higher prevalence in MRSA. This high prevalence and the diversity of PVL+ isolates must be highlighted and could be linked to the import of different strains via international exchanges. Contrary to other countries, we did not observe the predominance of one single CA-MRSA clone such as the USA300 in North America. In studies in neighboring countries, the prevalence of PVL was generally lower: from 0 to 12.7% in Turkey [39], from 7.4 to 55.6% in Iran according to the studies [40], and around 9% of S. aureus isolates responsible for community-acquired infections in West Bengal of India [41].
In conclusion, the prevalence of MRSA in S. aureus isolates circulating in Kabul was quite high (66.3%) when compared with the prevalence rates obtained from other similar studies conducted in neighboring countries. Most of S. aureus isolates, especially MRSA, were multidrug-resistant (generally resistant to beta-lactams, macrolides, quinolones, and cotrimoxazole and less often to aminoglycosides, chloramphenicol, and tetracyclines). This may be related to the overuse or the inappropriate use of antimicrobials which was recently reported in Afghanistan [14, 15]. A great diversity of S. aureus clones was brought to light using microarray analysis, with a predominance of the Southwest Pacific, the CC22-MRSA-IV TSST-1, and the Bengal Bay clones. This diversity and the clones identified reflect epidemiological links, including travels, migrations, commerce, and international military actions, with South West Pacific, Indian, and Middle Eastern/Arabian Gulf regions, as well as Western Europe, the USA, and Africa. In the future, major efforts have to be made on the monitoring, distribution, and spread of multidrug-resistant bacteria to control their diffusion and reduce the prevalence of MRSA in Afghanistan.
Data Availability
All relevant materials and data supporting the findings of this study are contained within the manuscript.
References
Rasigade J-P, Vandenesch F (2014) Staphylococcus aureus: a pathogen with still unresolved issues. Infect Genet Evol 21:510–514. https://doi.org/10.1016/j.meegid.2013.08.018
Zhang S, Sun X, Chang W, Dai Y, Ma X (2015) Systematic review and meta-analysis of the epidemiology of vancomycin-intermediate and heterogeneous vancomycin-intermediate Staphylococcus aureus isolates. PLoS One 10:e0136082. https://doi.org/10.1371/journal.pone.0136082
Cong Y, Yang S, Rao X (2020) Vancomycin resistant Staphylococcus aureus infections: a review of case updating and clinical features. J Adv Res 21:169–176. https://doi.org/10.1016/j.jare.2019.10.005
Monecke S, Coombs G, Shore AC, Coleman DC, Akpaka P, Borg M, Chow H, Ip M, Jatzwauk L, Jonas D, Kadlec K, Kearns A, Laurent F, O’Brien FG, Pearson J, Ruppelt A, Schwarz S, Scicluna E, Slickers P, Tan H-L, Weber S, Ehricht R (2011) A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS One 6:e17936. https://doi.org/10.1371/journal.pone.0017936
Calfee DP (2017) Trends in community versus health care-acquired methicillin-resistant Staphylococcus aureus infections. Curr Infect Dis Rep 19:48. https://doi.org/10.1007/s11908-017-0605-6
Lina G, Piémont Y, Godail-Gamot F, Bes M, Peter MO, Gauduchon V, Vandenesch F, Etienne J (1999) Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis 29:1128–1132. https://doi.org/10.1086/313461
Chen C-J, Huang Y-C (2014) New epidemiology of Staphylococcus aureus infection in Asia. Clin Microbiol Infect 20:605–623. https://doi.org/10.1111/1469-0691.12705
Bukhari SZ, Ahmed S, Zia N (2011) Antimicrobial susceptibility pattern of Staphylococcus aureus on clinical isolates and efficacy of laboratory tests to diagnose MRSA: a multi-centre study. J Ayub Med Coll Abbottabad 23:139–142
Indian Network for Surveillance of Antimicrobial Resistance (INSAR) group, India (2013) Methicillin resistant Staphylococcus aureus (MRSA) in India: prevalence & susceptibility pattern. Indian J Med Res 137:363–369
Mendes RE, Mendoza M, Banga Singh KK, Castanheira M, Bell JM, Turnidge JD, Lin SSF, Jones RN (2013) Regional resistance surveillance program results for 12 Asia-Pacific nations (2011). Antimicrob Agents Chemother 57:5721–5726. https://doi.org/10.1128/AAC.01121-13
Dadashi M, Nasiri MJ, Fallah F, Owlia P, Hajikhani B, Emaneini M, Mirpour M (2018) Methicillin-resistant Staphylococcus aureus (MRSA) in Iran: a systematic review and meta-analysis. J Glob Antimicrob Resist 12:96–103. https://doi.org/10.1016/j.jgar.2017.09.006
Shekarabi M, Hajikhani B, Salimi Chirani A, Fazeli M, Goudarzi M (2017) Molecular characterization of vancomycin-resistant Staphylococcus aureus strains isolated from clinical samples: a three year study in Tehran, Iran. PLoS One 12:e0183607. https://doi.org/10.1371/journal.pone.0183607
Naimi HM, Rasekh H, Noori AZ, Bahaduri MA (2017) Determination of antimicrobial susceptibility patterns in Staphylococcus aureus strains recovered from patients at two main health facilities in Kabul, Afghanistan. BMC Infect Dis 17:737. https://doi.org/10.1186/s12879-017-2844-4
Green T, Zafar O, Zahir S, Jumahir A, Aisha N Afghanistan Medicine Use Study: a survey of 28 health facilities in 5 provinces | Management Sciences for Health. msh.org/resources/afghanistan-medicine-use-study-a-survey-of-28-health-facilities-in-5-provinces. Accessed 15 Jul 2020
Bajis S, Van den Bergh R, De Bruycker M, Mahama G, Van Overloop C, Satyanarayana S, Bernardo RS, Esmati S, Reid AJ (2014) Antibiotic use in a district hospital in Kabul, Afghanistan: are we overprescribing? Public Health Action 4:259–264. https://doi.org/10.5588/pha.14.0068
Société Française de Microbiologie (2017) Comité de l’Antibiogramme de la Société Française de Microbiologie CASFM / EUCAST
Murakami K, Minamide W, Wada K, Nakamura E, Teraoka H, Watanabe S (1991) Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J Clin Microbiol 29:2240–2244
Stegger M, Andersen PS, Kearns A, Pichon B, Holmes MA, Edwards G, Laurent F, Teale C, Skov R, Larsen AR (2012) Rapid detection, differentiation and typing of methicillin-resistant Staphylococcus aureus harbouring either mecA or the new mecA homologue mecA(LGA251). Clin Microbiol Infect 18:395–400. https://doi.org/10.1111/j.1469-0691.2011.03715.x
Lina G, Boutite F, Tristan A, Bes M, Etienne J, Vandenesch F (2003) Bacterial competition for human nasal cavity colonization: role of Staphylococcal agr alleles. Appl Environ Microbiol 69:18–23. https://doi.org/10.1128/aem.69.1.18-23.2003
Monecke S, Slickers P, Ehricht R (2008) Assignment of Staphylococcus aureus isolates to clonal complexes based on microarray analysis and pattern recognition. FEMS Immunol Med Microbiol 53:237–251. https://doi.org/10.1111/j.1574-695X.2008.00426.x
Ullah A, Qasim M, Rahman H, Khan J, Haroon M, Muhammad N, Khan A, Muhammad N (2016) High frequency of methicillin-resistant Staphylococcus aureus in Peshawar Region of Pakistan. SpringerPlus 5:600. https://doi.org/10.1186/s40064-016-2277-3
Ghasemian A, Mirzaee M (2016) Methicillin-resistant Staphylococcus aureus (MRSA) strains and the staphylococcal cassette chromosome mec types in Iran. Infect Epidemiol Med 2:31–34 https://iem.modares.ac.ir/article-4-10977-en.pdf
Baddour MM, Abuelkheir MM, Fatani AJ (2006) Trends in antibiotic susceptibility patterns and epidemiology of MRSA isolates from several hospitals in Riyadh, Saudi Arabia. Ann Clin Microbiol Antimicrob 5:30. https://doi.org/10.1186/1476-0711-5-30
Eksi F, Gayyurhan ED, Bayram A, Karsligil T (2011) Determination of antimicrobial susceptibility patterns and inducible clindamycin resistance in Staphylococcus aureus strains recovered from southeastern Turkey. J Microbiol Immunol Infect 44:57–62. https://doi.org/10.1016/j.jmii.2011.01.011
Senok A, Ehricht R, Monecke S, Al-Saedan R, Somily A (2016) Molecular characterization of methicillin-resistant Staphylococcus aureus in nosocomial infections in a tertiary-care facility: emergence of new clonal complexes in Saudi Arabia. New Microbes New Infect 14:13–18. https://doi.org/10.1016/j.nmni.2016.07.009
Monecke S, Skakni L, Hasan R, Ruppelt A, Ghazal SS, Hakawi A, Slickers P, Ehricht R (2012) Characterisation of MRSA strains isolated from patients in a hospital in Riyadh, Kingdom of Saudi Arabia. BMC Microbiol 12:146. https://doi.org/10.1186/1471-2180-12-146
Goudarzi M, Bahramian M, Satarzadeh Tabrizi M, Udo EE, Figueiredo AMS, Fazeli M, Goudarzi H (2017) Genetic diversity of methicillin resistant Staphylococcus aureus strains isolated from burn patients in Iran: ST239-SCCmec III/t037 emerges as the major clone. Microb Pathog 105:1–7. https://doi.org/10.1016/j.micpath.2017.02.004
Enany S, Higuchi W, Okubo T, Takano T, Enany M, Yamamoto T (2007) Brain abscess caused by Panton-Valentine leukocidin-positive community-acquired methicillin-resistant Staphylococcus aureus in Egypt, April 2007. Euro Surveill 12:E070927.2. https://doi.org/10.2807/esw.12.39.03274-en
Afroz S, Kobayashi N, Nagashima S, Alam MM, Hossain ABMB, Rahman MA, Islam MR, Lutfor AB, Muazzam N, Khan MAH, Paul SK, Shamsuzzaman AKM, Mahmud MC, Musa AKM, Hossain MA (2008) Genetic characterization of Staphylococcus aureus isolates carrying Panton-Valentine leukocidin genes in Bangladesh. Jpn J Infect Dis 61:393–396
D’Souza N, Rodrigues C, Mehta A (2010) Molecular characterization of methicillin-resistant Staphylococcus aureus with emergence of epidemic clones of sequence type (ST) 22 and ST 772 in Mumbai, India. J Clin Microbiol 48:1806–1811. https://doi.org/10.1128/JCM.01867-09
Jamil B, Gawlik D, Syed MA, Shah AA, Abbasi SA, Müller E, Reißig A, Ehricht R, Monecke S (2018) Hospital-acquired methicillin-resistant Staphylococcus aureus (MRSA) from Pakistan: molecular characterisation by microarray technology. Eur J Clin Microbiol Infect Dis 37:691–700. https://doi.org/10.1007/s10096-017-3161-y
Madzgalla S, Syed MA, Khan MA, Rehman SS, Müller E, Reissig A, Ehricht R, Monecke S (2016) Molecular characterization of Staphylococcus aureus isolates causing skin and soft tissue infections in patients from Malakand, Pakistan. Eur J Clin Microbiol Infect Dis 35:1541–1547. https://doi.org/10.1007/s10096-016-2695-8
Pokhrel RH, Aung MS, Thapa B, Chaudhary R, Mishra SK, Kawaguchiya M, Urushibara N, Kobayashi N (2016) Detection of ST772 Panton-Valentine leukocidin-positive methicillin-resistant Staphylococcus aureus (Bengal Bay clone) and ST22 S. aureus isolates with a genetic variant of elastin binding protein in Nepal. New Microbes New Infect 11:20–27. https://doi.org/10.1016/j.nmni.2016.02.001
Centers for Disease Control and Prevention (1999) Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus - Minnesota and North Dakota, 1997-1999. JAMA 282:1123–1125
Bouchiat C, Curtis S, Spiliopoulou I, Bes M, Cocuzza C, Codita I, Dupieux C, Giormezis N, Kearns A, Laurent F, Molinos S, Musumeci R, Prat C, Saadatian-Elahi M, Tacconelli E, Tristan A, Schulte B, Vandenesch F, ESCMID Study Group on Staphylococci and Staphylococcal Infections (ESGS) (2017) MRSA infections among patients in the emergency department: a European multicentre study. J Antimicrob Chemother 72:372–375. https://doi.org/10.1093/jac/dkw431
Frisch MB, Castillo-Ramírez S, Petit RA, Farley MM, Ray SM, Albrecht VS, Limbago BM, Hernandez J, See I, Satola SW, Read TD (2018) Invasive methicillin-resistant Staphylococcus aureus USA500 strains from the U.S. Emerging Infections Program constitute three geographically distinct lineages. mSphere 3(3):e00571–e00517. https://doi.org/10.1128/mSphere.00571-17
Albrecht N, Jatzwauk L, Slickers P, Ehricht R, Monecke S (2011) Clonal replacement of epidemic methicillin-resistant Staphylococcus aureus strains in a German university hospital over a period of eleven years. PLoS One 6:e28189. https://doi.org/10.1371/journal.pone.0028189
Monecke S, Slickers P, Gawlik D, Müller E, Reissig A, Ruppelt-Lorz A, Akpaka PE, Bandt D, Bes M, Boswihi SS, Coleman DC, Coombs GW, Dorneanu OS, Gostev VV, Ip M, Jamil B, Jatzwauk L, Narvaez M, Roberts R, Senok A, Shore AC, Sidorenko SV, Skakni L, Somily AM, Syed MA, Thürmer A, Udo EE, Vremerǎ T, Zurita J, Ehricht R (2018) Molecular typing of ST239-MRSA-III from diverse geographic locations and the evolution of the SCCmec III element during its intercontinental spread. Front Microbiol 9:1436. https://doi.org/10.3389/fmicb.2018.01436
Oksuz L, Dupieux C, Tristan A, Bes M, Etienne J, Gurler N (2013) The high diversity of MRSA clones detected in a university hospital in Istanbul. Int J Med Sci 10:1740–1745. https://doi.org/10.7150/ijms.6438
Shahini Shams Abadi M, Nikokar I, Hoseini Alfatemi SM, Malekzadegan Y, Azizi A, Sedigh Ebrahim-Saraie H (2017) Epidemiology of Panton-Valentine leukocidin harbouring Staphylococcus aureus in cutaneous infections from Iran: a systematic review and meta-analysis. Infez Med 25:217–223
Karmakar A, Jana D, Dutta K, Dua P, Ghosh C (2018) Prevalence of Panton-Valentine leukocidin gene among community acquired Staphylococcus aureus: a real-time PCR study. J Pathog 2018:4518541. https://doi.org/10.1155/2018/4518541
Acknowledgments
The authors would like to thank Christine GARDON and all the technicians of the French National Reference Center for staphylococci for technical assistance for PCR and microarray.
Funding
This work was financially supported by Santé Publique France and the French Ministry of Foreign Affairs.
Author information
Authors and Affiliations
Contributions
Frédéric Laurent and François Vandenesch were involved in the conception of the study, and in analysis and interpretation of data. Haji Mohammad Naimi was involved in conducting the study, statistically analyzing the data, and drafting the manuscript. Camille André was involved in antibiotic susceptibility testing. Céline Dupieux was involved in interpretation of data and revising and editing the manuscript. Michèle Bes, Anne Tristan, and Claude-Alexandre Gustave were involved in the interpretation of DNA microarray. Qand Agha Nazari conducted preliminary identifications of strains. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethics approval
The approval of an ethics committee was not necessary, because the clinical samples of our study were taken as part of standard patient care.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Code availability
Not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Naimi, H.M., André, C., Bes, M. et al. Antibiotic resistance profile and molecular characterization of Staphylococcus aureus strains isolated in hospitals in Kabul, Afghanistan. Eur J Clin Microbiol Infect Dis 40, 1029–1038 (2021). https://doi.org/10.1007/s10096-020-04130-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-020-04130-0