Abstract
Currently, diagnosis of legionellosis relies mainly on urinary antigen testing (UAT) for Legionella pneumophila serogroup 1 (Lp1). However, this test has several limitations, particularly missing non-Lp1 infections. The purpose of this large multicenter study was to investigate the risk of missing legionellosis relying on UAT solely. Molecular results of Legionella detection as part of a first-line (syndromic) testing algorithm for severe respiratory tract infections were investigated retrospectively and compared with UAT results in 14 Belgian laboratories. Overall, 44.4% (20/45) UAT results appeared false negative and were reclassified as legionellosis based on PCR findings [Legionnaires’ disease, 37.5% (15/40); Pontiac fever, 100% (5/5)]. A total of 39.4% (26/66) diagnosis probably would have been missed or delayed without a syndromic approach, as UAT or specific molecular testing for Legionella was not requested by the clinician. Furthermore, we confirmed the higher sensitivity of molecular Legionella detection in lower respiratory tract compared with upper respiratory tract specimens (p = 0.010).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Legionella species are intracellular, Gram-negative, rod-shaped bacteria with strict growth requirements existing ubiquitously in aqueous environments. Currently, the genus knows more than 50 species and 70 serogroups. Almost half of the species have been associated with human disease, with Legionella pneumophila serogroup 1 (Lp1) being the most virulent and most common cause [1,2,3].
Legionella infection can cause two main distinct clinical presentations: Pontiac fever (PF) and Legionnaires’ disease (LD). PF is a mild upper respiratory infection with non-specific influenza-like illness (ILI), mostly self-limiting. LD is an atypical pneumonia with symptoms ranging from mild disease to severe pneumonia with need for oxygenation and a high mortality rate [1, 4]. Host factors associated with increased susceptibility for legionellosis include immunosuppression, hepatic or renal failure, chronic obstructive lung disease, smoking history, patient age over 50 years, and male sex (at least partially attributed to the fact that more adult men than women are smoking) [1, 4, 5]. One of the most important determinants of outcome is a rapid diagnosis to start accurate antibiotic treatment [4, 6, 7]. However, both PF and LD are underdiagnosed because of the large list of pathogens causing similar symptoms [8].
Although culture of respiratory samples is still considered the gold standard for diagnosis of legionellosis, it is a very demanding test necessitating incubation for several days on complex media and expertise [4]. Because of the ease of specimen collection, simplicity of analysis, rapid (same day) result, and relatively low cost, diagnosis mostly relies on commercial urinary antigen tests (UAT) [8, 9]. The currently available rapid UATs are based on monoclonal antibodies targeting Lp1 soluble antigen in urine [8, 9]. The antigen can become detectable 48–72 h after onset of clinical symptoms, and the test may remain positive for several weeks or months [1, 2]. However, Lp1 causes 79% of culture-confirmed cases in Europe according to the Epidemiological Report for 2017 of the ECDC (European Centre for Disease Prevention and Control) [4, 10]; so theoretically, at least as many as 21% cases remain undiagnosed if relying on UAT solely. Other serogroups of L. pneumophila (especially serogroups 4 and 6) and non-pneumophila species (especially L. micdadei, L. bozemanii, and L. longbeachae) are increasingly recognized as causes of severe lower respiratory tract (LRT) infection [2, 3, 9]. Due to overreliance on UAT testing, a diagnostic gap for LD caused by non-serogroup 1 L. pneumophila and other species has been created [2, 4, 9]. UAT is estimated to represent as much as 82% to more than 90% of diagnostic tools used for LD confirmation in Europe [1, 4]. Subsequently, Lp1 may be overestimated in current estimates of LD. Although some manufacturers are claiming confidentially that other serogroups can be detected due to cross-reactivity, sensitivities for non-serogroup 1 L. pneumophila are highly variable and generally much lower than those for Lp1-associated infections [1, 4]. Another limitation of the test is that it only can be used for diagnosis of LD; the antigen is not detectable in the urine of patients with PF [9]. About 15 years ago, commercial and in-house developed nucleic acid amplification tests (NAATs) were introduced for diagnosis of legionellosis (isothermal amplification, conventional PCR, real-time PCR (single and multiplex)) [4]. As NAAT still necessitates trained personnel and sophisticated machines, it is not yet universally deployed, but it is becoming increasingly accessible to a larger number of laboratories on a moderate budget and often in a “syndromic approach” setting for community-acquired pneumonia (CAP) [3, 4)]. An important advantage of this diagnostic method is that NAATs can be developed to target Lp1 as well as non-serogroup 1 L. pneumophila and other species [8]. NAATs have a significant lower turnaround time than culture, with specificities close to 100% and sensitivities often reported to be better than UAT and equal to or greater than culture [1, 2, 4, 8]. For both, culture and PCR, optimal sensitivity is obtained using specimens of the LRT [2, 3, 8]. Besides specimen type, sensitivity of both methods is positively correlated with disease severity but NAATs can be considered superior in diagnosing mild LD cases compared with culture that is experiencing a greater decrease in sensitivity over the disease course [4, 8]. Nevertheless, sample culture of a respiratory specimen and subsequent serotyping and molecular typing is recommended as it can play an important role in identifying sources of infection through comparison of strains from clinical and environmental sources [1, 11]. If no isolate is cultured, direct molecular typing on the sample should be tried. Thus, all samples should be referred to a reference laboratory for epidemiological purposes.
The purpose of this large multicenter study was to investigate the risk of missing legionellosis relying on UAT solely. Routinely obtained results from 14 Belgian laboratories that introduced an in-house developed or commercial real-time PCR method as part of a first-line (syndromic) testing algorithm for severe respiratory tract infections were investigated retrospectively and compared with UAT results.
Materials and methods
Patients and samples
Fourteen Belgian Hospitals participated in this laboratory-based retrospective study (AZ Sint-Jan Brugge-Oostende AV, UZ Leuven, OLVZ Aalst-Asse-Ninove, ZNA-Ziekenhuizen, AZ Zeno, AZ Alma, AZ Sint-Lucas Brugge, Sint-Andriesziekenhuis Tielt, Jan Yperman Ziekenhuis, AZ Delta, AZ Damiaan, AZ West, AZ Groeninge, AZ Sint-Rembert). Patients were included if Legionella DNA was detected in a respiratory sample. Subsequently, the laboratory information system was searched for results of Legionella UAT from these patients within the same episode of illness. Medical records of the patients were reviewed for relevant clinical and epidemiological factors associated with legionellosis. Patients were categorized as suffering from LD if the clinical and radiographic picture were compatible with pneumonia, not attributed to another infectious trigger. If there were no clinical or radiographic signs of pneumonia, but there was a respiratory disease (ILI) not attributed to another (infectious) trigger, the diagnosis of PF was retained.
Furthermore, a preliminary “reverse” study was performed in the laboratory of the AZ Sint-Jan Brugge-Oostende hospital between January 2013 and June 2018, including all patients for whom a positive result was observed with Legionella UAT, and Legionella PCR was performed on both an upper and lower respiratory tract sample within the same episode.
Ethical approval was obtained from the Ethics Committee of the AZ Sint-Jan Brugge-Oostende Hospital.
Urinary antigen test
A variety of commercial UATs were used in the different laboratories, according to the manufacturers’ instructions: Alere BinaxNOW® Legionella Urinary Antigen Card (n = 12) (Alere Health, Ghent, Belgium), TRU Legionella® (n = 1) (Meridian Bioscience, Braine-l’Alleud, Belgium), and Legionella V-TesT (n = 1) (Coris BioConcept, Gembloux, Belgium). Sensitivities of these similar-format UATs are considered comparable regardless of the manufacturer [1, 2, 4, 8].
Molecular method
The molecular analyses were performed in four of the 14 participating clinical laboratories: AZ Sint-Jan Brugge-Oostende (also testing the samples routinely sent by the other ten hospitals not performing molecular analysis for Legionella spp.), UZ Leuven, OLVZ Aalst-Asse-Ninove, and ZNA-Ziekenhuizen (Table 1). Quality and competence of these laboratories were assured by accreditation for the international standard ISO15189 by the Belgian Accreditation organization (BELAC) and/or by participation in internal and external quality control (eQC) programs (Table 1). There is a considerable risk of contamination in molecular assays for Legionella spp. given the environmental habitat of this microorganism [12]. Nevertheless, L. pneumophila species were correctly identified with each of the included assays in the yearly eQC evaluations and there was no report of false positive results nor cross-reaction with non-pneumophila species. No contamination of negative controls was mentioned in the runs included in this study for the assays used in UZ Leuven, OLVZ Aalst, and ZNA-Ziekenhuizen (no negative controls routinely tested in AZ Sint-Jan Brugge-Oostende). Furthermore, each case of legionellosis detected by the molecular method was clinically confirmed, and negative samples can be considered negative controls for the extraction and amplification processes. Over a period of almost 3 years (January 2017–October 2019), following positivity rates of the molecular Legionella assays were observed: AZ Sint-Jan Brugge-Oostende, 0.19% (41/21,134); UZ Leuven, 0.58% (56/9664); OLVZ Aalst, in-house syndromic panel 0.14% (23/16,580), specific Legionella spp. assay 13.42% (31/231); ZNA-Ziekenhuizen, in-house syndromic panel 0.16% (4/2572), commercial syndromic panel 0.24% (3/1255).
PCR was considered the reference method based on its ability to detect all L. pneumophila serogroups as well as on previous publications comparing the performance characteristics of Legionella UAT versus PCR [2, 8].
Data analysis
Sensitivity is reported, including the 95% confidence interval. A two-sample permutation test was used to compare proportions. Statistical tests were performed two sided at a significance level of 0.05 using exact p values. Microsoft® Excel (Microsoft Corp., Redmond, WA, USA) was used for descriptive statistics. Statistical analyses were performed using R version 3.3.2 [13].
Results
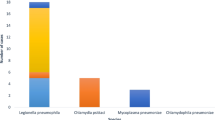
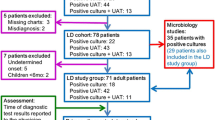
In our retrospective multicenter study, L. pneumophila was detected by PCR in the respiratory samples of 71 patients (Table 2). Fifty-eight of them were suffering from LD, mostly being men (ratio men/women, 41/17), with a mean and median patient age of 64 and 65 years respectively (range, 11–90). Thirteen patients presented with PF, mostly being women (ratio men/women, 5/8), with a mean and median patient age of 46 and 60 years respectively (range, 0–79).
In 45 cases both diagnostic methods, UAT and PCR, were requested. In 15/40 (37.5%) patients with a positive Legionella PCR result on a respiratory sample and a clinical and radiographic picture compatible with pneumonia, the diagnosis of LD was missed by UAT not being able to detect the antigen in urine. In patients presenting with PF, a negative UAT result was observed for all analyzed samples (5/5), in contrast to the positive PCR result. Sensitivity of UAT compared with PCR was 62.5% (45.8–77.3) for LD and 0% (0.00–52.2) for PF. The sensitivity of UAT was significantly better (p = 0.013) in patients suffering from LD compared with those suffering from PF.
Sixty-six of the included positive PCR results (n = 71) were obtained by a syndromic approach PCR for diagnosis of respiratory infection or atypical pneumonia targeting multiple microorganisms simultaneously; the other five cases were found by a molecular assay only targeting L. pneumophila (Table 1). When considering all Legionella-positive respiratory samples analyzed with a syndromic approach PCR method, only PCR analysis was requested in 39.4% (26/66) without a request for UAT by the clinician. It concerned 18 patients suffering from LD and 8 patients suffering from PF (Table 2).
In the reverse single-center study, five patients were included with a positive UAT for whom PCR was performed on an upper respiratory tract (URT) and LRT sample within the same disease episode during a study period of over 5 years. All included patients were suffering from LD, consequently no PF cases were detected by UAT. All five URT samples (nasopharyngeal swabs) were negative by PCR in contrast to the positive UAT result on urine. However, a positive result was found when PCR was repeated on a LRT sample (bronchoalveolar lavage, BAL) from these LD cases. PCR for Legionella spp. was significantly (p = 0.010) more often positive when performed on a LRT specimen (100% (47.8–100)) compared with a URT specimen (0% (0–52.2)) in patients with a positive UAT result.
Discussion
In this retrospective Belgian (Flemish) study, we evaluated the added value of PCR on a respiratory specimen compared with UAT on urine for diagnosis of legionellosis. Our study population of 71 Legionella PCR positive patients was predominantly male (64.6%), with a mean age of 61 years and a median of 64 years. This is in concordance with the risk factors described in literature and data from the World Health Organization with 75–80% of reported legionelloses being in patients over 50 years and 60–70% being male [2, 8, 14].
Overall 44.4% (20/45) of the UAT results appeared false negative; consequently, diagnosis of the corresponding patients was reclassified as legionellosis based on PCR findings. All five PF cases diagnosed by PCR were not detected by UAT; what is in line with the expectations as UAT is not approved for diagnosis of PF. More alarming is the number of missed LD cases with UAT, as most laboratories worldwide still are relying on this single test for diagnosis [4]. In total, 37.5% (15/40) of the Legionella pneumonia diagnoses would have been missed if UAT would have been performed as the only diagnostic test. This implicates an increased risk of fatal outcome as one of the most important determinants for cure is rapid diagnosis followed by early initiation of adequate antibiotic therapy [4, 6, 7]. In a previously performed study by Peci et al., similar results were found with 25.3% (19/75) of all patients with a positive PCR result being not identified by UAT. They further investigated the responsible Legionella spp. by culture, giving following results: Lp1 (n = 2), Lp6 (n = 1), L. pneumophila without serogroup testing (n = 6), and L. non-pneumophila (n = 10) [8]. Those data are confirming that not only L. non-pneumophila infections are missed by UAT but also infections with L. pneumophila serogroups other than serogroup 1 (despite the claimed cross-reaction phenomenon by some manufacturers) and even Lp1. They reported an overall sensitivity of 74.7% and a specificity of 98.3% for UAT compared with PCR with a positive predictive value of 77.7% and a negative predictive value of 98.1% [8]. In a study conducted by Chen et al., only 2.7% (1/37) of cases being positive by PCR were found negative by UAT [2].
We observed a significant (p = 0.013) higher sensitivity of UAT in patients suffering from LD (62.5%) compared with those suffering from PF (0%). The reported sensitivities of UATs in previous studies are ranging from 56 to 99% with lower sensitivities for non-Lp1 infection [1, 2, 4, 8]. The urinary analysis is highly specific for L. pneumophila (95–100%) [4, 8]. For molecular testing on LRT secretions, better sensitivities of 80–100% are reported, with specificities of 93–100% [1, 2]. However, it is of course necessary to use a well-validated PCR test and to integrate appropriate quality assurance measures to guarantee a sustained high analytical quality [3].
Important to consider when applying molecular analysis for diagnosis is the type of respiratory sample used. Historically, BAL fluid is considered the optimal respiratory specimen for identification of Legionella spp., partially because few LD patients can effectively produce sputa. Data of our reverse study (n = 5) confirm the higher sensitivity (p = 0.010) of detecting Legionella by PCR in LRT specimens (BAL) in comparison with URT specimens, as all LD cases were missed by PCR when performed on a nasopharyngeal swab, yet detected on BAL fluid of the same patients within the same disease episode. Some previous studies were performed concerning the preferred respiratory specimen source for Legionella diagnosis, with divergent conclusions. Sensitivities of less than 10 to 80% have been reported for URT specimens in diagnosis of legionellosis [2, 3, 8]. In a study performed by Peci et al., no significant difference in sensitivity of PCR was observed between BAL fluid (12.8%) and induced sputum (10.3%) when different specimen sources from the same patient within the same episode were compared [8]. In a systematic review conducted by Avni et al., equally high sensitivity values for all respiratory samples (including BAL fluid, sputum, pharyngeal swab, tissue biopsy) with near perfect specificity values were reported [15]. Findings of Diederen et al. on the other hand indicate that oropharyngeal swabs are not reliable for Legionella PCR, as only 27% of confirmed LD cases were detected [12].
UAT is only performed when specifically requested by the clinician, so clinical suspicion of LD is necessary [3]. However, this is often not the case in the initial differential diagnosis as there is no pathognomonic sign differentiating the clinical picture from other causes of CAP. Therefore, the use of a syndromic approach molecular diagnosis for respiratory infections can have an added value [2]. This approach enables early detection and rapid response for suspected as well as unexpected cases that otherwise may have gone undiagnosed [3]. Syndromic molecular methods (in-house developed as well as commercial assays) are becoming more widespread accessible for clinical laboratories [4, 16]. In 39.4% (26/66) of all legionelloses in our study tested by syndromic approach, diagnosis was probably made, thanks to a syndromic respiratory molecular testing protocol. In those cases, there was no specific UAT requested by the clinician suggesting that diagnosis eventually would have been missed or delayed without the syndromic approach.
According to national and international guidelines, it is recommended to associate a macrolide or fluoroquinolone with a β-lactam for empiric treatment of CAP in patients admitted to the intensive care unit (ICU) (CAP, subgroup IV) [17, 18]. Thus, in these patients, atypical pathogens including Legionella will be covered by initial therapy. However, for outpatients (CAP, subgroup I-II) and inpatients not admitted to the ICU (CAP, subgroup III), combination therapy with a macrolide or monotherapy with a fluoroquinolone (not for CAP III) is only recommended if there is no clinical improvement 3 days after initial antimicrobial monotherapy with a β-lactam according to the national guidelines [17]. Thus, especially in CAP subgroups I–III, a delay in diagnosis may contribute to an adverse outcome as Legionella is not covered by empiric treatment.
Despite the results of our study and earlier conducted studies showing the underdiagnosis of LD when relying on UAT solely because of a lower sensitivity compared with PCR methods, UAT remains the method of choice in national and international guidelines.
According to the criteria of the Flemish (Belgium) government, the diagnosis of a confirmed case can only be made by (i) UAT, by (ii) culture of L. pneumophila from respiratory secretions or any normally sterile site, or by (iii) seroconversion (a four-fold or greater rise in specific serum antibody titer to L. pneumophila on specimens collected 6 weeks apart). This implies that only UAT can be used for acute diagnosis since confirmation by culture and serology takes days to weeks. A positive PCR result for L. pneumophila can merely be used for diagnosis of a probable case. Furthermore, the Flemish guideline focuses only on L. pneumophila; the detection of other species unfortunately is not included in the criteria for diagnosis of legionellosis [19].
Similar criteria for confirmed and probable or suspected cases are found in the case definitions of the ECDC and CDC (Centers for Disease Control and Prevention), the public health institute of the USA. Though, they describe Legionella spp. in general instead of L. pneumophila for isolation by culture. And in the EU case definitions, the species in general are described for detection by PCR and serology too (probable cases in the case of non-Lp1) [20, 21].
In conclusion, there is an urgent need for updating national, European, and worldwide diagnostic guidelines. The criteria to confirm a LD case are obsolete, as they do not include analysis by PCR. Furthermore, non-Lp1 and non-pneumophila Legionella species should be clearly described additionally in the definition.
Moreover, the Belgian national reimbursement criteria for diagnostic analyses are quite limited; bacterial culture of sputa and other respiratory tract specimens is reimbursed since several decades but without specific attention for Legionella species; consequently, many laboratories do not use additional specific media for the growth of this bacterium; UAT is only reimbursed from September 2016 on; reimbursement for Legionella serology is stopped almost a decade ago, and Legionella PCR on BAL is refunded from the 1st of April 2019, but only in solid organ transplantation recipients.
Our study has some limitations. Firstly, no further typing was performed on the strains with discordant results between UAT and L. pneumophila PCR. Hence, we are not able to assess if the cases missed by UAT were mainly due to infection with L. pneumophila serogroups other than Lp1 or due to infection with Lp1 (assuming UAT has a lower sensitivity compared with PCR for Lp1 diagnosis).
Furthermore, the syndromic molecular assays in our study were targeting different serogroups of L. pneumophila, but no other Legionella species. Probably, the diagnostic rate can be increased further when other species are targeted by PCR too.
We can conclude that Legionella UAT is a valuable tool in diagnosis of LD as it is easy and rapid to perform on urine, a non-invasive specimen type. Nevertheless, clinicians should be aware of the limitations of the test, especially when interpreting a negative result. As shown in our study, reliance on UAT solely may result in serious underdiagnosis of legionellosis. Furthermore, we confirmed the higher sensitivity of molecular Legionella detection in LRT specimens in comparison with URT specimens.
The use of PCR as a first-line diagnostic screening method should be encouraged, as early diagnosis and accurate antibiotic treatment are important in reducing the high mortality risk. PCR is the only analysis method making it possible to search for Lp1 as well as non-serogroup 1 and, when targeted, non-pneumophila Legionella species within a clinically relevant and adequate time frame. A syndromic approach for diagnosis of CAP or invasive respiratory infections targeting multiple microorganisms simultaneously is recommended for this purpose.
References
Cunha BA, Burillo A, Bouza E (2016) Legionnaires’ disease. Lancet 387(10016):376–385
Chen DJ, Procop GW, Vogel S, Yen-Lieberman B, Richter SS (2015) Utility of PCR, culture, and antigen detection methods for diagnosis of Legionellosis. J Clin Microbiol 53(11):3474–3477
Gadsby NJ, Helgason KO, Dickson EM, Mills JM, Lindsay DS, Edwards GF et al (2016) Molecular diagnosis of Legionella infections—clinical utility of front-line screening as part of a pneumonia diagnostic algorithm. J Inf Secur 72(2):161–170
Mercante JW, Winchell JM (2015) Current and emerging Legionella diagnostics for laboratory and outbreak investigations. Clin Microbiol Rev 28(1):95–133
World Health Organization. Facts on gender and tobacco [Internet]. 2010 [cited 1 November 2019]. Available from: https://www.who.int/gender/documents/10facts_gender_tobacco_en.pdf
Dirven K, Ieven M, Peeters MF, van der Zee A, De Schrijver K, Goossens H (2005) Comparison of three Legionella urinary antigen assays during an outbreak of legionellosis in Belgium. J Med Microbiol 54(Pt 12):1213–1216
Miyata J, Huh JY, Ito Y, Kobuchi T, Kusukawa K, Hayashi H (2017) Can we truly rely on the urinary antigen test for the diagnosis? Legionella case report. J Gen Fam Med 18(3):139–143
Peci A, Winter AL, Gubbay JB (2016) Evaluation and comparison of multiple test methods, including real-time PCR, for Legionella detection in clinical specimens. Front Public Health 4:175
Pierre DM, Baron J, Yu VL, Stout JE (2017) Diagnostic testing for Legionnaires’ disease. Ann Clin Microbiol Antimicrob 16(1):59
European Centre for Disease Prevention and Control (2019) Legionnaires’ disease. In: ECDC. Annual epidemiological report for 2017. ECDC, Stockholm
European Centre for Disease Prevention and Control (2017) European Legionnaires’ Disease Surveillance Network (ELDSNet) – Operating procedures for the surveillance of travel-associated Legionnaires’ disease in the EU/EEA. ECDC, Stockholm
Diederen BM, Peeters MF (2007) Are oropharyngeal swabs suitable as samples for Legionella-specific PCR testing? J Clin Microbiol 45(10):3482–3483
R Core Team (2016) R: A Language and Environment for Statistical Computing. Vienna, Austria. Available from: https://www.R-project.org
World Health Organization. Legionellosis [Internet]. February 2018 [cited 8 July 2019]. Available from: https://www.who.int/news-room/fact-sheets/detail/legionellosis
Avni T, Bieber A, Green H, Steinmetz T, Leibovici L, Paul M (2016) Diagnostic accuracy of PCR alone and compared to urinary antigen testing for detection of Legionella spp.: a systematic review. J Clin Microbiol 54(2):401–411
Steensels D, Reynders M, Descheemaeker P, Curran MD, Jacobs F, Denis O et al (2015) Clinical evaluation of a multi-parameter customized respiratory TaqMan(®) array card compared to conventional methods in immunocompromised patients. J Clin Virol 72:36–41
Belgische Vereniging voor Infectiologie en Klinische Microbiologie. IGGI Infectiologiegids - Guide d'Infectiologie [Internet]. October 2018 [cited 8 November 2019]. Available from: https://www.bvikm.org
Mandell LA, Wunderink RG, Anzueto A, Bartlett JG, Campbell GD, Dean NC et al (2007) Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin Infect Dis 44:S27–S72
Agentschap Zorg & Gezondheid. Legionellose [Internet]. April 2018 [cited 9 July 2019]. Available from: https://www.zorg-en-gezondheid.be/legionellose
European Centre for Disease Prevention and Control. EU case definitions [Internet]. July 2018 [cited 9 July 2019]. Available from: https://ecdc.europa.eu/en/surveillance-and-disease-data/eu-case-definitions
Centers for Disease Control and Prevention. Legionnaires Disease Case Definitions [Internet]. June 2010 [cited 9 July 2019]. Available from: https://www.cdc.gov/legionella/health-depts/surv-reporting/case-definitions.html
Acknowledgments
We would like to thank the participating laboratories for kindly providing the lab-technical and clinical data concerning their patients.
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Muyldermans, A., Descheemaeker, P., Boel, A. et al. What is the risk of missing legionellosis relying on urinary antigen testing solely? A retrospective Belgian multicenter study. Eur J Clin Microbiol Infect Dis 39, 729–734 (2020). https://doi.org/10.1007/s10096-019-03785-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-019-03785-8