Abstract
Staphylococcus argenteus is a novel Staphylococcus species closely related to Staphylococcus aureus that has been recently described. In this study, we investigated the proportion and the characteristics of S. argenteus recovered from humans in Belgium. S. aureus. human isolates collected in Belgium from 2006 to 2015 (n = 1,903) were retrospectively characterised via the presence of non-pigmented colonies on chocolate agar, spa typing and rpoB sequencing to determine if some of them were in fact S. argenteus. Out of 73 strains non-pigmented on chocolate plates, 3 isolates (0.16 %) showed rpoB sequences, in addition to spa and sequence types (ST2250/t5787, ST2250/t6675, ST3240/t6675), related to S. argenteus. Two of them were methicillin-resistant, harbouring a SCCmec type IV. The three S. argenteus isolates carried genes (sak, scn) of the immune evasion cluster. This first Belgian nationwide analysis showed a low occurrence of S. argenteus. Further studies should be conducted to identify the distribution range and the clinical impact of this new species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Staphylococcus argenteus is a novel Staphylococcus species that has recently been described [1]. Staphylococcus argenteus isolates have been ascribed to phylogenetically distinct Staphylococcus aureus lineages (clonal complex [CC] 75, CC2198, CC2483, CC1594) [2–5]. This species is characterised by a lack of the genes that produce the carotenoid pigment staphyloxanthin [6] and therefore shows a non-pigmented phenotype on chocolate agar plates [1, 6]. S. argenteus displays a reduced virulence compared with S. aureus [7], but isolates carrying Panton–Valentine leukocidin (PVL) have been described [8, 9]. Although this new species may be widely distributed, its geographical distribution range remains unknown. Technical difficulties in performing multilocus sequence typing (MLST) using conventional primers on these isolates may contribute to underreporting [3]. However, other phylogenetic analyses such as the rpoB, gap, sodA, tuf and hsp60 sequence identification are able to successfully identify S. argenteus [3]. In this work, we described the first Belgian nationwide analysis on human S. argenteus occurrence.
Materials and methods
Strain collection
Two collections of S. aureus strains (n = 1,903) were retrospectively analysed for the presence of S. argenteus. Most isolates (n = 1,650) were sent by Belgian microbiology laboratories from 2012 to 2015 to the National Reference Centre—Staphylococcus aureus (NRC), Brussels. Belgian microbiology laboratories were invited to refer S. aureus isolates to the NRC in the following cases:
-
1.
Diagnostic problems including identification and susceptibility testing
-
2.
Exotoxin gene detection
-
3.
Outbreak investigation
The remaining isolates (n = 253) corresponded to nasopharyngeal samples recovered from healthy children attending kindergartens from Brussels between 2006 and 2008 [10].
Identification and molecular typing
All isolates were analysed by 16S/mecA/nuc, 16S/mecC PCR and spa typing, as previously described [10, 11]. The spa types were determined with Ridom StaphType software (www.ridom.de/staphtype) and analysed by the based upon repeat pattern (BURP) algorithm using default parameters (types shorter than five repeats were excluded, and spa types were grouped into the same group if cost was less or equal to four) and non-restrictive conditions (types shorter than five repeats were included, and spa types were grouped into the same group if cost was less or equal to six). All isolates were inoculated onto Chocolate agar PolyViteX (bioMérieux, France) for 48 h at 35 °C. Isolates with non-pigmented (white) phenotype on chocolate agar plates were considered to be possible S. argenteus [6] and were further analysed for the presence of the dehydrosqualene synthase gene crtM involved in staphyloxanthin production (Table S1) and subjected to rpoB typing using primers and conditions previously described [12]. rpoB sequences were analysed with the Bionumerics 6.5 software (Applied Mathematics, Belgium). A similarity dendrogram was constructed using the multiple sequence alignment and the unweighted pair group method with arithmetic averages (UPGMA). rpoB sequences from S. argenteus, S. aureus and other staphylococci (http://www.ncbi.nlm.nih.gov/) were used as controls. MLST of S. argenteus isolates was performed, as previously described [13].
Antimicrobial resistance
Antimicrobial susceptibility of S. argenteus isolates was determined using the disk diffusion method and interpreted according to CLSI breakpoints for S. aureus [14] for the following antibiotics: penicillin, oxacillin, cefoxitin, gentamicin, kanamycin, tobramycin, fusidic acid, trimethoprim/sulfamethoxazole, ciprofloxacin, minocycline, tetracycline, rifampicin, erythromycin, clindamycin, mupirocin, chloramphenicol and linezolid. Vancomycin susceptibility was determined using the E-test (bioMérieux) and interpreted according to CLSI [14].
Antimicrobial resistance and detection of virulence determinants
S. argenteus isolates were tested for the presence of genes encoding β-lactamase (blaZ), PVL, toxic shock syndrome toxin 1 (TSST-1), exfoliatins (eta, etb), staphylokinase (sak), staphylococcal complement inhibitor (scn), chemotaxis inhibitory protein (chp) and enterotoxins (sea to see, seg to selu; Table S1). Staphylococcal chromosomal cassette mec (SCCmec) typing was determined for methicillin-resistant S. argenteus (MRSArg) isolates (Table S1).
Results
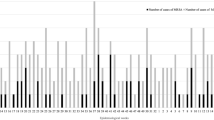
All 1,903 isolates collected at the NCR were positive for nuc and 16S genes, 48.0 % (n = 913) were mecA-positive and 0.3 % (n = 6) were mecC-positive. The 1,903 isolates belonged to 445 different spa types. The BURP analysis using default parameters (Table 1) grouped the 1,903 isolates into 20 groups, whereas 124 isolates were excluded or designated as singletons. Most groups could be related to typical S. aureus lineages (Table 1). The BURP analysis using relaxed grouping parameters (Table 1) allowed the isolates to be clustered into three groups, including a cluster grouping of spa types related to S. argenteus CC2483 (n = 3).
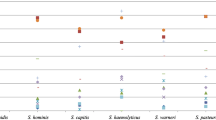
Most isolates coloured grey to yellow on chocolate agar plates (n = 1,830), and only 73 (3.8 %) isolates were non-pigmented. Twenty-nine out of the 73 non-pigmented isolates (28.8 %) carried crtM (Fig. 1). Non-pigmented isolates were subjected to rpoB typing. Three crtM-negative isolates with spa types t5787 or t6675 that grouped in the spa-CC related to S. argenteus CC2483 showed rpoB sequences that grouped at 99.5 % similarity with sequences of S. argenteus control strains. The remaining non-pigmented isolates (n = 70) carried diverse spa types (n = 26), and their rpoB sequences grouped at 99.2 % similarity with sequences of S. aureus control isolates. The clusters grouping S. argenteus and S. aureus rpoB sequences were associated at 93.2 % similarity. Other staphylococci rpoB sequences used as controls showed less similarity than S. argenteus and S. aureus sequences (<90.9 %).
Unweighted pair group method with arithmetic averages (UPGMA) tree based on partial RNA polymerase B (rpoB) gene sequences showing the phylogenetic relationship among different species and subspecies of the genus Staphylococcus (n = 59) and National Reference Centre—Staphylococcus aureus (NRC) and healthy children (HC) collection isolates, which showed a non-pigmented phenotype on agar chocolate plates (n = 73). The partial rpoB gene sequences corresponded to a 485-bp fragment between nucleotides 1444 and 1928, as previously described [12]. The scale indicates the similarity on the basis of the multiple sequence alignment. S. argenteus control isolates were sequenced by Tong et al. [1], whereas the remaining control isolates corresponded to sequences of strains deposited from diverse culture collections. spa types, and the presence (+) or absence (−) of the mecA and crtM genes, are indicated at the right of the NRC and HC collection isolates. The asterisk indicates that the strain NCR-2013S018 carried the mecC gene. ATCC American Type Culture Collection, Manassas, USA; DSM Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany; FRI Food Research Institute, University of Wisconsin, Madison, USA; CCM Česká Sbírka Mikroorganismů, Brno, Czech Republic
Overall, only three isolates (0.16 %) corresponded to S. argenteus isolates by rpoB typing. The three isolates were collected from two hospitals and one kindergarten located in different Belgian cities (Table 2). The three S. argenteus isolates belonged to CC2883 (two isolates were ST2250, and the third was ST3240). All isolates showed resistance to penicillin and carried the blaZ gene. The two hospital-associated isolates were MRSArg-SCCmec IV. The three isolates were fully susceptible to the remaining antimicrobials tested. Regarding virulence factors, one isolate carried enterotoxin-like genes (selk, selq), and all carried the immune evasion cluster (IEC) sak and scn genes.
Discussion
In this study, 0.16 % of the Belgian human S. aureus isolates investigated were identified as S. argenteus. This low occurrence should be considered carefully as two different populations (clinical samples and healthy carriers) were analysed. In this sense, only 0.12 % of the human clinical isolates investigated corresponded to S. argenteus. In Australia, S. argenteus has been reported to be the predominant community-acquired methicillin-resistant lineage in Aboriginal communities, with a prevalence of 71 % [2]. However, recent studies have reported S. argenteus at a low occurrence in hospital settings from Australia [8], Fiji [15], Thailand [5, 16] and Trinidad and Tobago [13]. S. argenteus carriage has also been reported in Cambodia [17] and French Guyana [4]. In Europe, S. argenteus studies are scarce, and this species has only been reported in two patients in France, who had an epidemiological link with Mayotte (Indian Ocean) [9]. In our study, one out of the three S. argenteus isolates had an epidemiological link with the Philippines (Table 2). The three Belgian isolates were related to carriage state (n = 2) or to soft-tissue infections (n = 1), like most S. argenteus isolates described so far [4, 15]. However, some S. argenteus isolates have been related to invasive infections [5, 16].
The screening of non-pigmented isolates on chocolate agar plates appeared to be a useful tool for the identification of S. argenteus isolates [6]. However, some S. aureus isolates were also non-pigmented. S. aureus defective in earlier enzymes of the staphyloxanthin pathway or with mutations in genes involved in regulation can show differences in the pigment production [18]. Therefore, the chocolate agar screening should be confirmed by other genetic analyses to accurately identify S. argenteus.
The three S. argenteus isolates described in this study carried spa types (t5078, t6675) that have previously been related to CC2483 (http://saureus.mlst.net). The spa types t127, t376 and t1635 have been associated with S. argenteus CC1594 in the literature [4]. In this study, we found some non-pigmented t127 isolates (n = 4), but they carried crtM, and were ascribed to S. aureus by using rpoB typing. Thus, spa typing does not seem to be a discriminatory typing technique for S. argenteus isolates. Moreover, non-typeable types have been reported in few S. argenteus isolates (http://saureus.mlst.net). Further revision of the spa typing scheme is needed to improve this technique for S. argenteus isolates.
In this study, the two CC2483 MRSArg isolates carried SCCmec type IV, which has previously been related to S. argenteus isolates of the CC75 [2]. The S. argenteus isolates described in this study carried IEC genes and/or enterotoxin-like genes (selk, selq), which have been related to hlb-converting prophages in S. aureus. These results, together with the recent characterisation of PVL-positive strains [8, 9, 16], underline the capacity of this new bacterium to acquire typical S. aureus virulence factors.
As far as we know, our study represents the first large study of this new species in a European country. Although its occurrence in Belgium seems low, it is notable that the first isolate was recovered in 2007. Further studies are needed to determine the geographical distribution of this new species.
References
Tong SY, Schaumburg F, Ellington MJ et al (2015) Novel staphylococcal species that form part of a Staphylococcus aureus-related complex: the non-pigmented Staphylococcus argenteus sp. nov. and the non-human primate-associated Staphylococcus schweitzeri sp. nov. Int J Syst Evol Microbiol 65:15–22
McDonald M, Dougall A, Holt D et al (2006) Use of a single-nucleotide polymorphism genotyping system to demonstrate the unique epidemiology of methicillin-resistant Staphylococcus aureus in remote aboriginal communities. J Clin Microbiol 44:3720–3727
Ng JW, Holt DC, Lilliebridge RA et al (2009) Phylogenetically distinct Staphylococcus aureus lineage prevalent among indigenous communities in northern Australia. J Clin Microbiol 47:2295–2300
Ruimy R, Angebault C, Djossou F et al (2010) Are host genetics the predominant determinant of persistent nasal Staphylococcus aureus carriage in humans? J Infect Dis 202:924–934
Thaipadungpanit J, Amornchai P, Nickerson EK et al (2015) Clinical and molecular epidemiology of Staphylococcus argenteus infections in Thailand. J Clin Microbiol 53:1005–1008
Holt DC, Holden MT, Tong SY et al (2011) A very early-branching Staphylococcus aureus lineage lacking the carotenoid pigment staphyloxanthin. Genome Biol Evol 3:881–895
Tong SY, Sharma-Kuinkel BK, Thaden JT et al (2013) Virulence of endemic nonpigmented northern Australian Staphylococcus aureus clone (clonal complex 75, S. argenteus) is not augmented by staphyloxanthin. J Infect Dis 208:520–527
Tong SY, Lilliebridge RA, Bishop EJ et al (2010) Clinical correlates of Panton-Valentine leukocidin (PVL), PVL isoforms, and clonal complex in the Staphylococcus aureus population of Northern Australia. J Infect Dis 202:760–769
Dupieux C, Blonde R, Bouchiat C et al (2015) Community-acquired infections due to Staphylococcus argenteus lineage isolates harbouring the Panton-Valentine leucocidin, France, 2014. Euro Surveill 20:21154
Blumental S, Deplano A, Jourdain S et al (2013) Dynamic pattern and genotypic diversity of Staphylococcus aureus nasopharyngeal carriage in healthy pre-school children. J Antimicrob Chemother 68:1517–1523
Deplano A, Vandendriessche S, Nonhoff C et al (2014) Genetic diversity among methicillin-resistant Staphylococcus aureus isolates carrying the mecC gene in Belgium. J Antimicrob Chemother 69:1457–1460
Mellman A, Becker K, von Eiff C et al (2006) Sequencing and staphylococci identification. Emerg Infect Dis 12:333–336
Monecke S, Stieber B, Roberts R et al (2014) Population structure of Staphylococcus aureus from Trinidad & Tobago. PLoS One 9:e89120
Clinical and Laboratory Standards Institute (2014) Performance standards for antimicrobial susceptibility testing: twenty-fourth informational supplement M100-S24. CLSI, Waye, Pennsylvania, USA
Jenney A, Holt D, Ritika R et al (2014) The clinical and molecular epidemiology of Staphylococcus aureus infections in Fiji. BMC Infect Dis 14:160
Chantratita N, Wikraiphat C, Tandhavanant S et al (2016) Comparison of community-onset Staphylococcus argenteus and Staphylococcus aureus sepsis in Thailand: a prospective multicentre observational study. Clin Microbiol Infect. doi:10.1016/j.cmi.2016.01.008
Ruimy R, Armand-Lefevre L, Barbier F et al (2009) Comparisons between geographically diverse samples of carried Staphylococcus aureus. J Bacteriol 191:5577–5583
Fey PD, Endres JL, Yajjala VK et al (2013) A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. MBio 4:e00537-12
Acknowledgements
We thank our microbiologist colleagues for sending their staphylococci strains to the NRC.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Nucleotide accession numbers
The rpoB sequences generated in this study were deposited in GenBank: accession numbers KU555518 to KU555590).
Funding
This study was supported by internal funding.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Ethical approval
For this type of study formal consent is not required.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 41 kb)
Rights and permissions
About this article
Cite this article
Argudín, M.A., Dodémont, M., Vandendriessche, S. et al. Low occurrence of the new species Staphylococcus argenteus in a Staphylococcus aureus collection of human isolates from Belgium. Eur J Clin Microbiol Infect Dis 35, 1017–1022 (2016). https://doi.org/10.1007/s10096-016-2632-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-016-2632-x