Abstract
Molecular assays designed to provide bacterial identification and detection of resistance genes directly from positive blood cultures can significantly reduce the time to definitive results. This has the potential to improve patient management and antimicrobial stewardship. However, the extent of such an impact is yet to be fully assessed. We tested two such assays, the Verigene® System Bloodstream Infection Tests (Nanosphere, Inc., Northbrook, IL, USA) (both Gram-positive and Gram-negative cartridges) and the FilmArray® Blood Culture Identification Panel (BioFire® Diagnostics, Inc., Salt Lake City, UT, USA). We compared their accuracy and speed of organism and resistance gene identification to conventional culture-based methods for 173 positive blood cultures. We also retrospectively determined, for organisms deemed not to be contaminants, the potential impact on antimicrobial prescribing. Both the Verigene® and FilmArray® assays accurately identified organisms, on average, 27.95 and 29.17 h earlier than conventional methods, respectively. There were a significant number of false-positives for Pseudomonas aeruginosa with the FilmArray® assay, which may have been related to contamination of the bioMérieux BacT standard anaerobic blood culture bottles, which the manufacturer has acknowledged. Both panels provided results significantly faster than conventional methods. In our setting, the extent of the potential positive impact on antimicrobial prescribing was modest (9 out of 173 samples). However, this may be an underestimation, since probable contaminants were not included in this analysis. In conclusion, both panels gave accurate results with significantly improved turnaround times.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Despite advances in its management, sepsis remains a leading cause of morbidity and mortality. Between 32 and 47 thousand deaths per year are attributed to sepsis, accounting for up to 7.7 % of all deaths in England [1]. Twenty-seven percent of intensive care admissions in England and Wales are for severe sepsis and almost half of these patients die in hospital [2]. Outcomes can be improved by the early administration of effective antimicrobials [3, 4]. Increasing antibiotic resistance has lead to both a broadening of the spectrum of activity of agents chosen to empirically treat septic patients and a critical need for improved antimicrobial stewardship [3, 5]. The tension between these important goals is exacerbated by the slow speed with which culture-based techniques provide diagnostic information (12–72 h) [6].
Conventional microbiological methods for identification of micro-organisms from blood cultures (such as agar-based culture techniques) are slow and sometimes insensitive. The introduction of automated techniques and, more recently, mass spectrometry have improved accuracy and turnaround times [7, 8]. Further reductions in the time to identification of organisms and provision of antimicrobial susceptibility data are now being explored using molecular techniques and automated technologies [9–23].
We assessed two automated molecular assays; the Verigene® Blood Culture Gram-Positive (BC-GP) and Gram-Negative (BC-GN) tests (Nanosphere, Inc., Northbrook, IL, USA) (cartridges) and the FilmArray® Blood Culture Identification Panel (BioFire® Diagnostics, Inc., Salt Lake City, UT, USA). Both assays are designed to provide bacterial identification and detection of resistance genes directly from pre-incubated positive blood cultures. Our aims were to compare the diagnostic accuracy of each assay against conventional culture-based laboratory methods and estimate the average time to definitive results. Although results of the molecular panels were not disclosed to physicians, we retrospectively reviewed antimicrobial prescribing data to determine whether the earlier provision of information could have impacted upon clinical management.
Materials and methods
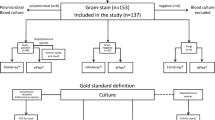
The study was conducted in a large tertiary academic hospital in central London. Clinicians routinely investigate all patients with fever or suspected systemic infection by obtaining blood cultures (up to 10 mL each taken into separate bioMérieux BacT/ALERT® standard aerobic and anaerobic bottles; paediatric patients have only aerobic samples taken). Aliquots of all positive blood cultures between 18th June and 22nd August 2013 were removed from blood culture bottles as close to the time of identification of positive growth as possible (within 10 h) and stored frozen at −20 °C until they could be tested (within 7 days). After testing by conventional methods, frozen aliquots were thawed and tested using each of the molecular platforms (see below for details). Neither manufacturer recommends freezing samples before testing, but this allowed more convenient retrospective testing. Only residual anonymised specimens collected for standard-of-care purposes were used in the analysis, samples were included as a performance evaluation and patient consent was not sought.
Conventional methods
During ‘working hours’ (between 0900 h and 1600 h Monday to Friday and 0900 and 1400 on Saturday and Sunday), positive blood culture bottles were processed immediately. Samples were inoculated directly onto appropriate agar plates determined by the Gram stain result. Outside of working hours, positive bottles were removed in batches at midnight and 0400 h, inoculated directly onto blood, chocolate and fastidious anaerobic agar plates (Oxoid, Basingstoke, UK), and a Gram stain prepared.
For identification, at 0900 h each day, all agar plates from the previous working day and from subculture at midnight and 0400 h were examined for growth. All morphologically distinct colonies were identified to the species level using matrix-assisted laser desorption ionisation time-of-flight (MALDI-TOF) (Bruker Daltonics, Bremen, Germany) and, where necessary, biochemical identification (basic bench-side biochemical tests and/or VITEK 2, bioMérieux, Marcy l’Etoile, France). Antimicrobial sensitivity testing was performed by automated microdilution using the appropriate testing card for VITEK 2.
Molecular methods
Separate aliquots of positive blood culture samples were tested using Verigene® Blood Culture Gram-positive and/or Gram-negative cartridges (according to the preliminary Gram stain results) and the FilmArray® Blood Culture Identification Panel. All assays were performed according to the manufacturers’ instructions.
The Verigene® Gram-positive assay detects nine species (Staphylococcus aureus, Staphylococcus epidermidis, Staphylococcus lugdunensis, Streptococcus anginosus group, Streptococcus agalactiae, Streptococcus pneumoniae, Streptococcus pyogenes, Enterococcus faecalis, Enterococcus faecium) and four genera (Staphylococcus spp., Streptococcus spp., Micrococcus spp., Listeria spp.), along with three antibiotic resistance genes (mecA for meticillin, vanA and vanB for vancomycin) in approximately 2.5 h. The Verigene® Gram-negative assay detects five species (Escherichia coli, Klebsiella pneumoniae, Klebsiella oxytoca, Pseudomonas aeruginosa, Serratia marcescens) and four genera (Acinetobacter spp., Citrobacter spp., Enterobacter spp., Proteus spp.), along with six antibiotic resistance genes (CTX-M for the detection of extended-spectrum beta-lactamases, IMP, KPC, NDM, OXA and VIM for the detection of carbapenemases) in approximately 2 h. Both assays run on the Verigene® Processor and Reader platforms, which extract and purify nucleic acids, followed by hybridisation to specific oligonucleotide-labelled gold nanospheres on a microarray.
The FilmArray® assay detects 24 pathogens (Enterococcus spp., Listeria monocytogenes, Staphylococcus spp., S. aureus, Streptococcus spp., S. agalactiae, S. pneumoniae, S. pyogenes, Acinetobacter baumannii, Haemophilus influenzae, Neisseria meningitidis, P. aeruginosa, Enterobacteriaceae, Enterobacter cloacae complex, E. coli, K. oxytoca, K. pneumoniae, Proteus spp., S. marcescens, Candida albicans, Candida glabrata, Candida krusei, Candida parapsilosis, Candida tropicalis) and three antibiotic resistance genes (mecA for meticillin, vanA/B for vancomycin and KPC for carbapenem resistance) in approximately 1 h. The system performs extraction and purification, nested polymerase chain reaction (PCR) and detection using end-point melt curve data.
Analysis
Duplicate blood cultures were excluded from analysis. For each sample, the transportation time (from sample collection to loading onto the automated blood culture system), bottle culture time (from loading onto the automated blood culture system to identified positive growth), Gram stain time (from identified positive growth to available Gram stain result), species-level identification time (from available Gram stain result to available species identification) and antimicrobial susceptibility testing time (from available Gram stain result to available antimicrobial susceptibility results) for conventional methods were calculated. For molecular techniques, the total turnaround time was calculated by adding the time taken to complete the molecular assay to the sum of the transportation, bottle culture and Gram stain times. Whilst the FilmArray® assay does not require a Gram stain, this time was included as it largely represented the standard workflow in our laboratory.
The conventional method for species identification and antimicrobial sensitivity testing was considered to be the gold standard against which both molecular methods were compared. Physicians were informed of all Gram stain results from positive blood cultures by a microbiology or infectious diseases physician. Advice given, including currently prescribed antimicrobials and any recommended changes, was documented in the electronic patient record. These data were used to determine the antimicrobials each patient was prescribed at the time the Gram stain result was known and to estimate the frequency with which the molecular techniques would have facilitated an earlier change to therapy. Samples with organisms that were considered to be contaminants were not included in the clinical utility analysis.
Results
Between 18th June and 22nd August 2013, there were 191 positive blood cultures. Eighteen of these samples were excluded from analysis, as they were duplicate samples. This left 173 samples from 159 patients with a median age of 53 years (interquartile range 21 to 70 years), 67 of whom were female. Ninety-seven (56 %) of the 173 samples were taken within 48 h of the patient’s admission to hospital and 25 (15.7 %) patients died within 30 days of the blood culture being taken (all-cause mortality).
Turnaround times
The mean time taken for each step of the laboratory process is shown in Table 1. Since the molecular assays were performed retrospectively at convenience, we assumed that these assays were run immediately following the Gram stain result. In reality, the total turnaround time may be considerably longer than this, depending on when the samples are actually processed. The mean total turnaround time from sample collection to species-level identification for conventional culture-based methods was 57.29 h. Both molecular panels were significantly faster, with mean turnaround times of 41.44 and 40.22 h for the Verigene® and FilmArray® assays, respectively. The conventional culture-based method took an additional mean of 12.1 h to obtain antimicrobial susceptibility results. The Verigene® assay produced six failed tests, which were repeated with success. No failed tests were reported by the FilmArray® assay.
Identification of micro-organisms
A total of 180 organisms were identified by conventional culture-based methods, including those where two or more pathogens were identified in the same sample. These are shown in Table 2. The most common pathogens identified were S. epidermidis (25.6 %), E. coli (16.8 %), coagulase-negative staphylococci (CNS) (15 %) and S. aureus (9.8 %). The molecular panels were not able to identify all organisms isolated by conventional techniques. There were 17 organisms belonging to genera not featured on the Verigene® panel and which produced a ‘not detected’ result. Eleven of these were considered to be contaminants (Propionibacterium spp., 5; Corynebacterium spp., 4; Paenibacillus macerans, 1; and Brevibacterium casei, 1) and six were significant isolates (Haemophilus parainfluenzae, 2; Bacteroides fragilis, 1; Fusobacterium necrophorum, 1; Prevotella denticola, 1; and Aeromonas spp., 1).
There were 23 organisms belonging to genera not featured on the FilmArray® panel and which produced a ‘not detected’ result. Seventeen of these were considered to be contaminants (as above, with the addition of six samples with Micrococcus spp.) and six significant isolates, identical to those for the Verigene® assay. In total, the Verigene® and FilmArray® assays detected 90.6 % and 87.2 %, respectively, of all organisms detected by conventional techniques.
Organism-specific sensitivity, specificity, and positive and negative predictive values for the molecular assays are shown in Tables 3 (Verigene®) and 4 (FilmArray®). These should be interpreted with caution, as there were small numbers for any individual pathogen, resulting in large confidence intervals.
The Verigene® assay produced occasional false-negative results (three for Streptococcus spp. and one each for Staphylococcus species, Micrococcus spp., Enterococcus faecalis and Acinetobacter spp.). There were also misidentifications for Streptococcus pneumoniae (two cases where Streptococcus viridans was identified by conventional methods), Staphylococcus epidermidis (one case where Staphylococcus hominis was identified by conventional methods), Enterococcus faecium (one case where Enterococcus avium was identified by conventional methods), Streptococcus agalactiae (one case where S. aureus was identified by conventional methods but was also detected by Verigene®) and Citrobacter spp. (one case where Enterococcus faecalis was identified by conventional methods).
The FilmArray® assay produced only one false-negative result, for Klebsiella oxytoca. There were 25 presumed false-positive results for Pseudomonas aeruginosa; these were repeated but produced the same result. In all cases, conventional methods failed to yield any Pseudomonas species, but did isolate an alternative organism. Additionally, testing by the Verigene® assay did not produce any positive results for Pseudomonas for any of these samples. In 22 of the samples with presumed false-positive results, the correct organism was also identified by the FilmArray® assay. The remaining three samples produced positive results for Pseudomonas in addition to the correctly identified organism (positive for more than one target). The details of all samples with discrepant results are shown in Table 5.
There were seven samples where conventional techniques identified two pathogens. The Verigene® assay failed to detect one of the pathogens in two samples (a Group C/G Streptococcus and an E. faecalis) and the FilmArray® assay failed to detect a Klebsiella in one sample. Full details of results for the seven samples that had two pathogens detected are shown in Table 6.
Detection of resistance markers
The results for the detection of antimicrobial resistance genes are listed in Table 7. On most occasions where an mecA gene was identified, it was in a coagulase-negative Staphylococcus. All four isolates of S. aureus that were phenotypically resistant to meticillin were identified as having an mecA gene by both the Verigene® and FilmArray® assays. One S. aureus isolate was identified by both Verigene® and FilmArray® assays as having an mecA gene, but was phenotypically meticillin-sensitive; however, no further testing was undertaken to resolve this result. This particular patient was known to be colonised at several sites with meticillin-resistant S. aureus (MRSA).
For the remaining CNS samples, the Verigene® assay produced two false-negatives and three false-positives, and the FilmArray® assay produced nine false-negatives and five false-positives. The Verigene® assay only tests for mecA in S. aureus and S. epidermidis, so the total number of evaluable samples was lower.
vanA/B was detected in only one sample (an Enterococcus faecium) by both Verigene® and FilmArray® assays, with compatible phenotypic results. CTX-M was detected in five samples by the Verigene® assay (four cases with E. coli and one case with Enterobacter cloacae). In all cases, the phenotypic susceptibility testing was compatible, showing resistance to amoxicillin, co-amoxiclav, cefuroxime, ceftazidime, cefpodoxime, cefotaxime and cephalothin. There were no samples testing positive for any of the carbapenemase genes. The results for resistance markers are shown in Table 7.
Clinical utility
We assessed the potential impact that earlier species and resistance mechanism identification could have had on patient management and antimicrobial stewardship, had the tests been performed prospectively and the results released to physicians. The difference in the mean total time to the final identification of species and antibiotic resistance between standard and molecular methods was 27.95 h for the Verigene® assay and 29.17 h for the FilmArray® assay. This time period represented the window of opportunity in which earlier identification by the molecular panels could potentially benefit patient management.
Organisms were separated into those where the species would have indicated a need to broaden the spectrum of antimicrobial coverage (Enterococcus faecium, Pseudomonas aeruginosa and Enterobacteriaceae with predictable chromosomal resistance to beta-lactams), those where the combination of organism and resistance gene indicated a need to broaden antimicrobial therapy, and those where the species would indicate a change from empirical therapy (usually co-amoxiclav) to a more targeted antimicrobial was possible (e.g. S. aureus and flucloxacillin, E. faecalis and amoxicillin, and streptococci and penicillin). For each sample in these categories, the antimicrobial that the patient was receiving when the Gram stain was performed was noted and an assessment made as to whether knowledge of the species or resistance gene could have changed this choice.
There were 13 (7.5 %) samples where species identification suggested a need to broaden the spectrum of antimicrobial coverage. Of these, two patients were not prescribed antimicrobials that would have covered the identified organism at the point of Gram stain. The other 11 patients were prescribed antimicrobials that deviated from hospital guidelines due to previous isolation of resistant organisms or their clinical condition (unwell on the intensive care unit or neutropaenic).
There were 11 (6.4 %) samples which had an identified resistance gene and, of these, nine had already received appropriate antimicrobials by the time the blood culture Gram stain was available and one had died prior to the blood culture becoming positive. Again, the reason for deviation from hospital antimicrobial guidelines was due to prior knowledge of resistant organisms isolated from other sites on the same patient.
Thirty-one (17.9 %) samples isolated a Streptococcus, meticillin-sensitive S. aureus (MSSA) or E. faecalis. When clinical details were reviewed, six patients could have had their antimicrobials changed sooner if the molecular result had been available. The other patients were either already on an appropriate antimicrobial, were suspected to have a mixed infection or were thought to have a contaminant in the blood culture.
Discussion
A total of 173 bacteraemic episodes were assessed in 159 patients, covering a broad age range and divided approximately equally between community- and hospital-acquired infection. Reflecting the high morbidity associated with bacteraemia, 16 % of patients died within 30 days of microbiological confirmation of bloodstream infection.
Both molecular assays correctly identified targeted organisms with high sensitivity and specificity. Samples were frozen, which is not recommended by either manufacturer and we are unable to assess the effect this might have had on the performance characteristics. The FilmArray® assay identified P. aeruginosa in 25 samples where this organism had not been found by conventional testing or using the Verigene® panel. On one occasion, where the true organism was Fusobacterium necrophorum, this may have resulted in the use of inappropriate antimicrobials had the physician acted on the result.
False-positive tests resulting in pseudo-outbreaks due to contamination of blood culture bottles have previously been described [24, 25], as has contamination of molecular reagents [26]. Evaluation by other authors of the FilmArray® Blood Culture Identification Panel have not identified this issue [27]. However, in May 2014, BioFire® released an advisory note which detailed the risk of false-positive results for P. aeruginosa and Enterococcus spp. when using bioMérieux BacT standard anaerobic bottles. This is thought to be related to the presence of nucleic acid from non-viable organisms in these bottles, and the company advises that any positive results for these organisms should be confirmed by another method prior to reporting the test results.
The Verigene® assay produced occasional false-positive results (6/173); however, two of these were S. pneumoniae, which has previously been noted to give false-positives for this organism [15, 16]. It is difficult to comment on the presumed false-positive results, since we performed no further analysis of these samples (e.g. by sequencing). The clinical significance of these findings are unknown; however, in all cases, an alternative pathogen was identified by both conventional techniques and the molecular assays.
The average total time to organism identification for both molecular assays was significantly shorter than for the conventional methods. Had these assays been used to inform clinical practice, then the final results (including antimicrobial susceptibility data) would, on average, have been available 27.95 h earlier for the Verigene® assay and 29.17 h earlier for the FilmArray® assay. This is broadly similar to previously published studies [13, 17, 18]. The potential for an earlier result to positively impact upon clinical management in our setting was relatively modest (9 out of 173 samples, 5.2 %); however, we did not include samples that were thought to have contaminants only in the analysis. Three patients could have benefited from an earlier change from ineffective to effective antimicrobials. Antimicrobial stewardship could have been improved in a further six patients whose antimicrobial spectrum could have been narrowed at an earlier stage. Previous studies have examined the use of the Verigene® assay for improving antimicrobial prescribing for S. aureus and Enterococcus spp. [20, 21]. These studies found a greater potential benefit, but had much higher frequencies of resistant organisms.
Most patients are prescribed antimicrobials because the attending physician is concerned that they have a bacterial infection. Knowledge of a contaminant in the blood culture might not be an indication for changing or stopping antimicrobial therapy. We excluded samples that isolated contaminant organisms only from the prescribed antimicrobial analysis; however, this knowledge could feasibly affect antimicrobial prescribing behaviour and would have likely led to an underestimation of the clinical utility of these tests. On these occasions, earlier knowledge of species would expedite cessation of antimicrobials.
Although the molecular assays are rapid and generally performed well, they have yet to establish a clear role in day-to-day clinical practice. They are a tool to augment rather than replace conventional methods, as samples still require an initial blood culture incubation step and the assays do not detect all organisms and resistance mechanisms. Consequently, they are likely to result in a significant additional cost for laboratories. In our setting, where extensive data were available for local resistance rates and previous microbiology results were frequently available, the impact of earlier organism identification on antimicrobial therapy was limited. The low rates of antimicrobial resistance locally also reduced the opportunity for the assays to identify occasions where broadening or narrowing initial empirical antimicrobials was appropriate. A final consideration is that each molecular assay can process only one sample at a time. In order to gain maximum benefit from timely results, samples should be processed in real time using multiple platforms in parallel, providing staff are available to operate them.
There were a number of limitations to our study. The use of frozen samples (outside of the manufacturers’ recommended protocol) may have resulted in degradation and decreased the sensitivity of the molecular assays. The number of samples tested, although comparable to previous studies [12, 20], was relatively small. This resulted in large confidence intervals around performance characteristics for less frequently detected organisms and resistance mechanisms. We did not review the full antimicrobial prescribing data for patients; instead, we determined which antimicrobials were prescribed at the time clinicians were notified of the Gram stain, together with the initial microbiology advice given. It is reasonable to assume that these are the antimicrobials and decisions the molecular assays could have an impact upon; however, it is an incomplete analysis and excluded contaminants such as CNS. The study was performed retrospectively, introducing potential bias, as the assessor was not blinded to both conventional and molecular assay results.
In conclusion, both the Verigene® and FilmArray® molecular assays for positive blood cultures provided accurate results more rapidly than conventional culture-based methods. However, in our setting, where antimicrobial resistance is low and there is good knowledge of resistance patterns at both population and individual levels, the impact of earlier results on antimicrobial choices was limited. These assays are likely to have the greatest impact in settings with higher levels of antimicrobial resistance and where there are no prior microbiological samples available for patients.
References
McPherson D, Griffiths C, Williams M, Baker A, Klodawski E, Jacobson B, Donaldson L (2013) Sepsis-associated mortality in England: an analysis of multiple cause of death data from 2001 to 2010. BMJ Open. doi:10.1136/bmjopen-2013-002586
Padkin A, Goldfrad C, Brady AR, Young D, Black N, Rowan K (2003) Epidemiology of severe sepsis occurring in the first 24 hrs in intensive care units in England, Wales, and Northern Ireland. Crit Care Med 31:2332–2338
Dellinger RP, Levy MM, Rhodes A, Annane D, Gerlach H, Opal SM, Sevransky JE, Sprung CL, Douglas IS, Jaeschke R, Osborn TM, Nunnally ME, Townsend SR, Reinhart K, Kleinpell RM, Angus DC, Deutschman CS, Machado FR, Rubenfeld GD, Webb SA, Beale RJ, Vincent J-L, Moreno R; Surviving Sepsis Campaign Guidelines Committee including the Pediatric Subgroup (2013) Surviving sepsis campaign: international guidelines for management of severe sepsis and septic shock: 2012. Crit Care Med 41:580–637
Kumar A, Roberts D, Wood KE, Light B, Parrillo JE, Sharma S, Suppes R, Feinstein D, Zanotti S, Taiberg L, Gurka D, Kumar A, Cheang M (2006) Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med 34:1589–1596
Davies SC (2013) Annual report of the Chief Medical Officer 2011: volume two, 2011. Infections and the rise of antimicrobial resistance. Department of Health, London
Stoneking LR, Patanwala AE, Winkler JP, Fiorello AB, Lee ES, Olson DP, Wolk DM (2013) Would earlier microbe identification alter antibiotic therapy in bacteremic emergency department patients? J Emerg Med 44:1–8
Pérez-Vázquez M, Oliver A, Sánchez del Saz B, Loza E, Baquero F, Cantón R (2001) Performance of the VITEK2 system for identification and susceptibility testing of routine Enterobacteriaceae clinical isolates. Int J Antimicrob Agents 17:371–376
Fournier P-E, Drancourt M, Colson P, Rolain J-M, La Scola B, Raoult D (2013) Modern clinical microbiology: new challenges and solutions. Nat Rev Microbiol 11:574–585
Peters RPH, van Agtmael MA, Danner SA, Savelkoul PHM, Vandenbroucke-Grauls CMJE (2004) New developments in the diagnosis of bloodstream infections. Lancet Infect Dis 4:751–760
La Scola B (2011) Intact cell MALDI-TOF mass spectrometry-based approaches for the diagnosis of bloodstream infections. Expert Rev Mol Diagn 11:287–298
Chang S-S, Hsieh W-H, Liu TS, Lee S-H, Wang C-H, Chou H-C, Yeo YH, Tseng C-P, Lee C-C (2013) Multiplex PCR system for rapid detection of pathogens in patients with presumed sepsis—a systemic review and meta-analysis. PLoS One 8:e62323
Samuel LP, Tibbetts RJ, Agotesku A, Fey M, Hensley R, Meier FA (2013) Evaluation of a microarray-based assay for rapid identification of Gram-positive organisms and resistance markers in positive blood cultures. J Clin Microbiol 51:1188–1192
Wojewoda CM, Sercia L, Navas M, Tuohy M, Wilson D, Hall GS, Procop GW, Richter SS (2013) Evaluation of the Verigene Gram-positive blood culture nucleic acid test for rapid detection of bacteria and resistance determinants. J Clin Microbiol 51:2072–2076
Buchan BW, Ginocchio CC, Manii R, Cavagnolo R, Pancholi P, Swyers L, Thomson RB Jr, Anderson C, Kaul K, Ledeboer NA (2013) Multiplex identification of gram-positive bacteria and resistance determinants directly from positive blood culture broths: evaluation of an automated microarray-based nucleic acid test. PLoS Med 10:e1001478
Sullivan KV, Turner NN, Roundtree SS, Young S, Brock-Haag CA, Lacey D, Abuzaid S, Blecker-Shelly DL, Doern CD (2013) Rapid detection of Gram-positive organisms by use of the Verigene Gram-positive blood culture nucleic acid test and the BacT/Alert Pediatric FAN system in a multicenter pediatric evaluation. J Clin Microbiol 51:3579–3584
Alby K, Daniels LM, Weber DJ, Miller MB (2013) Development of a treatment algorithm for streptococci and enterococci from positive blood cultures identified with the Verigene Gram-positive blood culture assay. J Clin Microbiol 51:3869–3871
Mestas J, Polanco CM, Felsenstein S, Dien Bard J (2014) Performance of the Verigene Gram-positive blood culture assay for direct detection of Gram-positive organisms and resistance markers in a pediatric hospital. J Clin Microbiol 52:283–287
Mancini N, Infurnari L, Ghidoli N, Valzano G, Clementi N, Burioni R, Clementi M (2014) Potential Impact of a microarray-based nucleic acid assay for rapid detection of Gram-negative bacteria and resistance markers in positive blood cultures. J Clin Microbiol 52:1242–1245
Altun O, Almuhayawi M, Ullberg M, Ozenci V (2013) Clinical evaluation of the FilmArray blood culture identification panel in identification of bacteria and yeasts from positive blood culture bottles. J Clin Microbiol 51:4130–4136
Beal SG, Ciurca J, Smith G, John J, Lee F, Doern CD, Gander RM (2013) Evaluation of the Nanosphere Verigene Gram-positive blood culture assay with the VersaTREK blood culture system and assessment of possible impact on selected patients. J Clin Microbiol 51:3988–3992
Sango A, McCarter YS, Johnson D, Ferreira J, Guzman N, Jankowski CA (2013) Stewardship approach for optimizing antimicrobial therapy through use of a rapid microarray assay on blood cultures positive for Enterococcus species. J Clin Microbiol 51:4008–4011
Scott LJ (2013) Verigene® gram-positive blood culture nucleic acid test. Mol Diagn Ther 17:117–122
Sullivan KV, DeBurger B, Roundtree SS, Ventrola CA, Blecker-Shelly DL, Mortensen JE (2014) Pediatric multicenter evaluation of the Verigene gram-negative blood culture test for rapid detection of inpatient bacteremia involving gram-negative organisms, extended-spectrum beta-lactamases, and carbapenemases. J Clin Microbiol 52:2416–2421. doi:10.1128/JCM.00737-14
McNeil MM, Davis BJ, Anderson RL, Martone WJ, Solomon SL (1985) Mechanism of cross-contamination of blood culture bottles in outbreaks of pseudobacteremia associated with nonsterile blood collection tubes. J Clin Microbiol 22:23–25
Noskin GA, Suriano T, Collins S, Sesler S, Peterson LR (2001) Paenibacillus macerans pseudobacteremia resulting from contaminated blood culture bottles in a neonatal intensive care unit. Am J Infect Control 29:126–129
Spangler R, Goddard NL, Thaler DS (2009) Optimizing Taq polymerase concentration for improved signal-to-noise in the broad range detection of low abundance bacteria. PLoS One 4:e7010
Blaschke AJ, Heyrend C, Byington CL, Fisher MA, Barker E, Garrone NF, Thatcher SA, Pavia AT, Barney T, Alger GD, Daly JA, Ririe KM, Ota I, Poritz MA (2012) Rapid identification of pathogens from positive blood cultures by multiplex polymerase chain reaction using the FilmArray system. Diagn Microbiol Infect Dis 74:349–355
Conflict of interest
CW reports receiving an educational grant from Nanosphere to attend ECCMID 2014. All other authors report no conflicts of interest relevant to this article.
Financial support
This work was supported by the NIHR Comprehensive Biomedical Research Centre award to Guy’s and St Thomas’ NHS Foundation Trust in partnership with King’s College, London. We are grateful to Una Health, BioFire® Diagnostics, Grifols and Nanosphere for providing consumables and loan of equipment free of charge. The manufacturers took part in neither the study design and execution nor the preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ward, C., Stocker, K., Begum, J. et al. Performance evaluation of the Verigene® (Nanosphere) and FilmArray® (BioFire®) molecular assays for identification of causative organisms in bacterial bloodstream infections. Eur J Clin Microbiol Infect Dis 34, 487–496 (2015). https://doi.org/10.1007/s10096-014-2252-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-014-2252-2