Abstract
The purpose of this investigation was to determine the prevalence of plasmid-mediated AmpC (pAmpC) and carbapenemases in Enterobacteriaceae collected from 35 hospitals in Spain and to establish their epidemiological relationships. We conducted a prospective multi-centre study on pAmpC- or carbapenemase-producing Enterobacteriaceae isolates from clinical samples collected from February to July 2009. The strains suspected to carry pAmpC were resistant or showed intermediate susceptibility to co-amoxiclav and second- or third-generation cephalosporins. Strains suspected to carry a carbapenemase were selected because they showed a minimum inhibitory concentration (MIC) to imipenem >1 mg/L. Polymerase chain reaction (PCR) and a sequencing strategy were used to characterise the enzymes. The clonal relationships between isolates was analysed by pulsed field gel electrophoresis (PFGE). Among 100,132 Enterobacteriaceae isolates collected, 1,654 were compatible with the production of pAmpC or carbapenemases. We found a prevalence of 0.64 % of pAmpC (n = 635) and 0.04 % of carbapenemases (n = 43). The most prevalent pAmpC enzymes were CMY-type (78.3 %), DHA-type (19.5 %), ACC-type (1.6 %) and FOX-type (0.6 %). The CMY-type was the most frequent in Escherichia coli and Proteus mirabilis species, whereas the DHA-type was mainly found in Klebsiella spp. The enzymes involved in carbapenem resistance were VIM-1, IMP-22 and the new IMP-28. Nine new bla genes were described: bla CMY-54, bla CMY-55, bla CMY-56, bla CMY-57, bla CMY-96, bla DHA-6, bla DHA-7, bla FOX-8 and bla IMP-28. The prevalence of pAmpC or carbapenemases found is not negligible. The CMY-types were the predominant pAmpC, whereas the VIM or IMP enzymes were the predominant carbapenemases. Furthermore, we observed a great genetic diversity among pAmpC-producing strains and a close clonal relationship between carbapenemase-producing strains.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
AmpC β-lactamases (AmpC) are clinically relevant cephalosporinases encoded on the chromosome of many Enterobacteriaceae. Some of them are also described on plasmids, usually in species that do not naturally produce AmpC, such as Klebsiella spp., Salmonella enterica or Proteus mirabilis.
Plasmid AmpC (pAmpC) derive from chromosomal ampC genes of the families Enterobacteriaceae and Aeromonadaceae. First characterised in 1988, pAmpC described to date can be grouped into seven families: ACC, CMY, DHA, FOX, MIR, ACT and MOX [1]. These enzymes have been identified throughout the world, with CMY-2 being the most prevalent. A previous study performed in Barcelona (Spain) showed a significant increase of pAmpC in Enterobacteriaceae from 0.06 % in 1999 to 1.3 % in 2007 [2], in agreement with other authors (0.13 % to 3 %) [3–6].
Classes A, B and D of acquired carbapenemases are also increasing in Enterobacteriaceae. Often involved in nosocomial outbreaks [7–12], these emerging enzymes are worrisome because their broad activity profiles encompass all β-lactam antibiotics. Additionally, carbapenemases are difficult to detect using phenotypical methods because their resistance level to carbapenems is frequently below the established clinical breakpoints [13, 14].
The prevalence of pAmpC and carbapenemases could be underestimated because of the lack of consensus on the methodology to detect them [15]. This study aimed to determine the prevalence of these enzymes in Enterobacteriaceae isolates from clinical samples collected in Spain over a six-month period in 2009.
Materials and methods
Study design, bacterial strains and susceptibility tests
A prospective multi-centre study was undertaken in Spain to collect pAmpC- or carbapenemase-producing Enterobacteriaceae isolates from clinical samples from February to July 2009. Thirty-five Spanish hospitals participated, six of which acted as co-ordinating centres. These hospitals were distributed throughout 14 of the 18 Autonomous Communities in Spain.
Only one isolate per patient was considered for analysis. Isolates were identified in each hospital, using standard methods. Disc-diffusion and microdilution susceptibility tests were performed according to Clinical and Laboratory Standards Institute (CLSI) guidelines [16]. As some pAmpC do not confer resistance to cefoxitin, each hospital collected all the strains showing resistance or intermediate susceptibility to both co-amoxiclav (<18 mm; >8/4 mg/L) and any of the following: cefotaxime (<23 mm; >8 mg/L), ceftazidime (<18 mm; >8 mg/L) or aztreonam (<21 mm; >8 mg/L).
All extended-spectrum β-lactamase (ESBL)-producing isolates were excluded, except those that were resistant to or had intermediate susceptibility to cefoxitin (<15 mm; >8 mg/L) or co-amoxiclav. ESBL production was studied using the CLSI confirmatory test [16], the double-disc synergy test [14] and/or Etest ESBL (AB Biodisk, Solna, Sweden). Enterobacteriaceae with inducible chromosomal AmpC β-lactamase were also excluded from this part of the study.
Isolates suspected of carrying a carbapenemase were first selected because they showed a minimum inhibitory concentration (MIC) to imipenem >1 mg/L. The modified Hodge test [17] and Etest MBL IP/IPI (AB bioMérieux, Solna, Sweden) were used to confirm carbapenemase activity.
Susceptibility to aminoglycosides, quinolones or co-trimoxazole was determined by broth microdilution (MicroScan, Siemens Healthcare Diagnostics, Deerfield, IL, USA).
Characterisation of pAmpC and carbapenemases
The detection and characterisation of their genes were performed using the specific primers listed in Table S1 (supplementary data) or as previously described [1, 7, 18–22]. All amplicons obtained were purified and sequenced as previously described [7].
PFGE
For clonal studies, a group of strains were selected according to the following criteria: (i) all carbapenemase-producing isolates, (ii) all pAmpC-producing isolates from the co-ordinating centres and (iii) two pAmpC-producing isolates per species and enzyme (one at the start and one at the end of the study period) from the remaining participant hospitals. The genetic relationship among isolates of the same species was determined by pulsed field gel electrophoresis (PFGE) using a CHEF-DRII device (Bio-Rad, Hercules, CA, USA) after total chromosomal DNA digestion with XbaI (Escherichia coli, Klebsiella pneumoniae, K. oxytoca, Enterobacter cloacae and Citrobacter koseri) [23] or NotI (P. mirabilis) [24]. DNA relatedness was calculated based on the criteria of Tenover et al. [25].
Nucleotide sequence accession numbers
The new β-lactamase gene sequences were submitted to GenBank under accession numbers HM544039 (bla CMY-54), HM544040 (bla CMY-55), HQ322613 (bla CMY-56), HQ285243 (bla CMY-57), HQ267531 (bla CMY-96), HQ322612 (bla DHA-6), HQ456945 (bla DHA-7), HM565917 (bla FOX-8) and HQ263342 (bla IMP-28).
Results and discussion
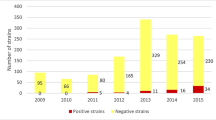
Our study showed that the prevalence of pAmpC and carbapenemases in Spain in 2009 was 0.64 % and 0.04 %, respectively. These data were obtained from 100,132 isolates of Enterobacteriaceae, of which 1,654 showed a susceptibility phenotype compatible with the production of pAmpC or carbapenemases. These results were similar to the prevalences found around the world [2, 4–6, 26].
Genes coding for pAmpC or carbapenemase were detected in 674 isolates (Table 1). Using phenotypic methods, we found that those isolates with negative polymerase chain reaction (PCR) results were ESBL-producers, serine class A-producers (Klebsiella spp.) or hyperproducers of chromosomal AmpC (E. coli). However, non-enzymatic resistance mechanisms such as altered permeability may have also contributed to resistance in these isolates [14].
The highest prevalence of pAmpC in Spain was found in the Autonomous Community of Catalonia (0.92 %), followed by Asturias (0.85 %). The lowest prevalence was observed in the Balearic Islands (0.35 %). No cases of pAmpC were detected in Castilla y León (Figure S1, supplementary data). This geographical variability was probably due to both the number of centres and the different kind of healthcare institutions included per autonomous community.
The most prevalent pAmpC enzymes found were CMY (78.3 %), followed by DHA (19.5 %), ACC (1.6 %) and FOX (0.6 %) (Table 2). CMY-2 remained the most widely distributed, especially in E. coli (87.1 %) and P. mirabilis (88.8 %) species, whereas DHA-1 enzyme was the most predominant in Klebsiella spp. isolates. Remarkably, both these enzymes were also found in other Enterobacteriaceae species, such as C. koseri, P. penneri and S. enterica (Table 2, Figure S1, supplementary data). FOX-type enzymes were found exclusively in E. coli. Nucleotide sequence analysis of all bla genes identified eight new pAmpC (bla CMY-54, bla CMY-55, bla CMY-56, bla CMY-57, bla CMY-96, bla DHA-6, bla DHA-7 and bla FOX-8).
Table 3 shows the correlation between resistance to non-β-lactam antibiotics and the most frequent species/pAmpC enzymes. The resistance of pAmpC-producers to non-β-lactam antibiotics was high: 70.2 % to ciprofloxacin, 65.9 % to co-trimoxazole, 44.8 % to tobramycin, 35.8 % to gentamicin and 7.4 % to amikacin. Co-resistance to ciprofloxacin, co-trimoxazole, gentamicin and tobramycin was detected in 23.4 % of the isolates. Regarding resistance to non-β-lactams, we did not find any statistically significant differences between CMY-2- and DHA-1-producing isolates.
VIM-1, VIM-2 and IMP-1 are the most frequent carbapenemases reported in Enterobacteriaceae around the world [27]. In Spain, the first VIM-1 enzyme was described in 2003 in K. pneumoniae and E. coli [7, 28]. Since then, several outbreaks involving K. pneumoniae and E. cloacae strains have been published [7, 28, 29]. In the present study, we found that the VIM-1 enzyme was the most frequent carbapenemase. We also detected IMP-22-producers and the new IMP-28-producing strain that was recently published [30]. As Gaibani et al. [31], we did not find any KPC, NDM or OXA carbepenemases. Our study, therefore, preceded the emergence of carbapenemases such as OXA-48 and NDM-1 that were recently described in Spain [32, 33]. The resistance of carbapenemase-producers to non-β-lactam antibiotics was high and similar to the pAmpC-producers: 60 % to ciprofloxacin, 80 % to co-trimoxazole, 33 % to tobramycin, 27.5 % to gentamicin and 7.5 % to amikacin. Co-resistance to ciprofloxacin, co-trimoxazole, gentamicin and tobramycin was detected in 17.5 % of the isolates.
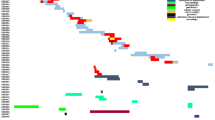
A total of 324 non-duplicate isolates were selected to determine their molecular typing. PFGE analysis showed a great genetic diversity: 172 different PFGE patterns in 179 E. coli isolates, 51 in 67 K. pneumoniae isolates, 45 in 48 P. mirabilis isolates, 10 in 15 K. oxytoca isolates, 11 in 11 E. cloacae isolates and 3 in 3 C. koseri isolates. The PFGE results revealed four well-defined clusters: (i) all nine IMP-22-producing K. pneumoniae isolates from Hospital Puerta del Mar (Cadiz), (ii) five VIM-1-producing K. oxytoca from Hospital Ramón y Cajal (Madrid), (iii) three CMY-2-producing E. coli from Hospital Virgen del Rocío (Seville) and (iv) three CMY-2-producing P. mirabilis from Hospital Sant Pau (Barcelona). We also detected 15 pairs of identical isolates: seven pairs of CMY-2-producing isolates (four of E. coli and three of K. pneumoniae), three of VIM-1-producing isolates (two K. pneumoniae and one K. oxytoca), two of DHA-1-producing K. pneumoniae, one of AAC-1-producing K. pneumoniae, one of FOX-8-producing E. coli and one of DHA-7- and VIM-1-producing E. cloacae. All but one of these strain pairs consisted of isolates from the same hospital. The highest number of strain pairs was detected in Hospital Vall d’Hebron (Barcelona) and corresponded to three CMY-2-producing isolates (two of E. coli and one of K. pneumoniae) and one pair of E. cloacae isolates co-producing DHA-7 and VIM-1. Finally, one pair of VIM-1-producing K. oxytoca strains comprised of isolates from Hospital Vall d’Hebron and from Laboratori de Referència de Catalunya (both in Barcelona), but no epidemiologic relationship between the patients carrying these isolates could be demonstrated.
pAmpC-producing strains cause nosocomial, healthcare-associated and community infections mainly in predisposed patients. The clinical data of the patients included in our study have already been published, showing that invasive infections were associated with high mortality, which might be partly related to inappropriate empirical therapy [22].
In spite of the predominance of CMY-2- and DHA-1-producing E. coli, K. pneumoniae and P. mirabilis, we found a great genetic diversity among these isolates, higher in E. coli and P. mirabilis than in K. pneumoniae. This finding supports the theory that the spread of these antibiotic resistance mechanisms is mainly due to the dissemination of mobile genetic elements among a polyclonal population. Previous studies have demonstrated the polyclonal dissemination of CMY-2-producing E. coli isolates [23, 34, 35]. However, nosocomial outbreaks due to DHA-1- or ACC-1-producing K. pneumoniae or CMY-2-producing P. mirabilis have been only sporadically described [36, 37]. In this study, we describe two small nosocomial outbreaks due to CMY-2-producing E. coli and P. mirabilis, respectively.
MBL-producing clinical Enterobacteriaceae isolates have been reported all over the world [7, 38, 39]. In this study, we describe two nosocomial outbreaks in two hospitals in Spain, one due to VIM-1-producing K. oxytoca in Hospital del Mar (Barcelona) and the other due to IMP-22-producing K. pneumoniae in Hospital Puerta del Mar (Cadiz). Conejo et al. [9] described a nosocomial outbreak of IMP-8-producing K. oxytoca strain in the same period and in the same area. In contrast to the wide spread of the KPC-producing ST258 K. pneumoniae strain [10], the dissemination of VIM and IMP carbapenemases seems to be associated to both clonal intra-hospital spread of local strains [7, 8, 31] and to non-clonal dissemination [7, 29, 38].
The detection and surveillance of pAmpC- and carbapenemase-producing strains is needed in order to implement appropriate therapeutic options and appropriate infection control measures [13]. However, the lack of standardised phenotypic methods has become evident in this study, as only 63 % of all evaluated isolates (n = 1,654) were pAmpC- or carbapenemase-producers.
In conclusion, the prevalence of pAmpC and carbapenemases found is not negligible. The CMY-type enzymes were the predominant pAmpC found, whereas the VIM or IMP enzymes were the most frequent carbapenemases. A great genetic diversity among pAmpC-producing strains and a closer clonal relationship between carbapenemase-producing strains was found.
References
Pérez-Pérez FJ, Hanson ND (2002) Detection of plasmid-mediated AmpC β-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol 40:2153–2162
Mata C, Miró E, Rivera A et al (2010) Prevalence of acquired AmpC β-lactamases in Enterobacteriaceae lacking inducible chromosomal ampC genes at a Spanish hospital from 1999 to 2007. Clin Microbiol Infect 16:472–476
Navarro F, Pérez-Trallero E, Marimón JM et al (2001) CMY-2-producing Salmonella enterica, Klebsiella pneumoniae, Klebsiella oxytoca, Proteus mirabilis and Escherichia coli strains isolated in Spain (October 1999–December 2000). J Antimicrob Chemother 48:383–389
Yamasaki K, Komatsu M, Abe N et al (2010) Laboratory surveillance for prospective plasmid-mediated AmpC β-lactamases in the Kinki region of Japan. J Clin Microbiol 48:3267–3273
Song W, Kim JS, Kim HS et al (2006) Increasing trend in the prevalence of plasmid-mediated AmpC β-lactamases in Enterobacteriaceae lacking chromosomal ampC gene at a Korean university hospital from 2002 to 2004. Diagn Microbiol Infect Dis 55:219–224
Empel J, Baraniak A, Literacka E et al (2008) Molecular survey of β-lactamases conferring resistance to newer β-lactams in Enterobacteriaceae isolates from Polish hospitals. Antimicrob Agents Chemother 52:2449–2454
Miró E, Segura C, Navarro F et al (2010) Spread of plasmids containing the bla VIM-1 and bla CTX-M genes and the qnr determinant in Enterobacter cloacae, Klebsiella pneumoniae and Klebsiella oxytoca isolates. J Antimicrob Chemother 65:661–665
Oteo J, Hernández-Almaraz JL, Gil-Antón J et al (2010) Outbreak of VIM-1-carbapenemase-producing Enterobacter cloacae in a pediatric intensive care unit. Pediatr Infect Dis J 29:1144–1146
Conejo MC, Domínguez MC, López-Cerero L et al (2010) Isolation of multidrug-resistant Klebsiella oxytoca carrying bla IMP-8, associated with OXY hyperproduction, in the intensive care unit of a community hospital in Spain. J Antimicrob Chemother 65:1071–1073
Giakoupi P, Maltezou H, Polemis M et al; Greek System for the Surveillance of Antimicrobial Resistance (2009) KPC-2-producing Klebsiella pneumoniae infections in Greek hospitals are mainly due to a hyperepidemic clone. Euro Surveill 14. pii: 19218
Cuzon G, Ouanich J, Gondret R et al (2011) Outbreak of OXA-48-positive carbapenem-resistant Klebsiella pneumoniae isolates in France. Antimicrob Agents Chemother 55:2420–2423
Struelens MJ, Monnet DL, Magiorakos AP et al; European NDM-1 Survey Participants (2010) New Delhi metallo-β-lactamase 1-producing Enterobacteriaceae: emergence and response in Europe. Euro Surveill 15. pii: 19716
Miriagou V, Cornaglia G, Edelstein M et al (2010) Acquired carbapenemases in Gram-negative bacterial pathogens: detection and surveillance issues. Clin Microbiol Infect 16:112–22
Navarro F, Calvo J, Cantón R et al (2011) Detection of resistance phenotypes in gram-negative bacteria. Enferm Infecc Microbiol Clin 29:524–534
Walther-Rasmussen J, Høiby N (2007) Class A carbapenemases. J Antimicrob Chemother 60:470–482
Clinical and Laboratory Standards Institute (CLSI) (2005) Performance standards for antimicrobial susceptibility testing; fourteenth informational supplement, M100-S14. CLSI, Wayne, PA, USA
Pasteran F, Mendez T, Guerriero L et al (2009) Sensitive screening tests for suspected class A carbapenemase production in species of Enterobacteriaceae. J Clin Microbiol 47:1631–1639
Dubois V, Poirel L, Marie C et al (2002) Molecular characterization of a novel class 1 integron containing bla GES-1 and a fused product of aac(3)-Ib/aac(6″)-Ib” gene cassettes in Pseudomonas aeruginosa. Antimicrob Agents Chemother 46:638–645
Yigit H, Queenan AM, Anderson GJ et al (2001) Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother 45:1151–1161
Poirel L, Magalhaes M, Lopes M et al (2004) Molecular analysis of metallo-β-lactamase gene bla SPM-1-surrounding sequences from disseminated Pseudomonas aeruginosa isolates in Recife, Brazil. Antimicrob Agents Chemother 48:1406–1409
Poirel L, Lambert T, Türkoglü S et al (2001) Characterization of Class 1 integrons from Pseudomonas aeruginosa that contain the bla VIM-2 carbapenem-hydrolyzing β-lactamase gene and of two novel aminoglycoside resistance gene cassettes. Antimicrob Agents Chemother 45:546–552
Rodríguez-Baño J, Miró E, Villar M et al (2012) Colonisation and infection due to Enterobacteriaceae producing plasmid-mediated AmpC β-lactamases. J Infect 64:176–183
Oteo J, Navarro C, Cercenado E et al (2006) Spread of Escherichia coli strains with high-level cefotaxime and ceftazidime resistance between the community, long-term care facilities, and hospital institutions. J Clin Microbiol 44:2359–2366
Sabbuba NA, Mahenthiralingam E, Stickler DJ (2003) Molecular epidemiology of Proteus mirabilis infections of the catheterized urinary tract. J Clin Microbiol 41:4961–4965
Tenover FC, Arbeit RD, Goering RV et al (1995) Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 33:2233–2239
Ding H, Yang Y, Lu Q et al (2008) The prevalence of plasmid-mediated AmpC β-lactamases among clinical isolates of Escherichia coli and Klebsiella pneumoniae from five children’s hospitals in China. Eur J Clin Microbiol Infect Dis 27:915–921
Queenan AM, Bush K (2007) Carbapenemases: the versatile β-lactamases. Clin Microbiol Rev 20:440–458
Tórtola MT, Lavilla S, Miró E et al (2005) First detection of a carbapenem-hydrolyzing metalloenzyme in two Enterobacteriaceae isolates in Spain. Antimicrob Agents Chemother 49:3492–3494
Tato M, Coque TM, Ruíz-Garbajosa P et al (2007) Complex clonal and plasmid epidemiology in the first outbreak of Enterobacteriaceae infection involving VIM-1 metallo-β-lactamase in Spain: toward endemicity? Clin Infect Dis 45:1171–1178
Pérez-Llarena FJ, Fernández A, Zamorano L et al (2012) Characterization of a novel IMP-28 metallo-β-lactamase from a Spanish Klebsiella oxytoca clinical isolate. Antimicrob Agents Chemother 56:4540–4543
Gaibani P, Ambretti S, Berlingeri A et al (2011) Rapid increase of carbapenemase-producing Klebsiella pneumoniae strains in a large italian hospital: surveillance period 1 March–30 September 2010. Euro Surveill 16. pii: 19800
Solé M, Pitart C, Roca I et al (2011) First description of an Escherichia coli strain producing NDM-1 carbapenemase in Spain. Antimicrob Agents Chemother 55:4402–4404
Pitart C, Solé M, Roca I et al (2011) First outbreak of a plasmid-mediated carbapenem-hydrolyzing OXA-48 β-lactamase in Klebsiella pneumoniae in Spain. Antimicrob Agents Chemother 55:4398–4401
Oteo J, Cercenado E, Cuevas O et al (2010) AmpC β-lactamases in Escherichia coli: emergence of CMY-2-producing virulent phylogroup D isolates belonging mainly to STs 57, 115, 354, 393, and 420, and phylogroup B2 isolates belonging to the international clone O25b-ST131. Diagn Microbiol Infect Dis 67:270–276
Naseer U, Haldorsen B, Simonsen GS et al (2010) Sporadic occurrence of CMY-2-producing multidrug-resistant Escherichia coli of ST-complexes 38 and 448, and ST131 in Norway. Clin Microbiol Infect 16:171–178
Empel J, Hrabák J, Kozińska A et al (2010) DHA-1-producing Klebsiella pneumoniae in a teaching hospital in the Czech Republic. Microb Drug Resist 16:291–295
Ohana S, Leflon V, Ronco E et al (2005) Spread of a Klebsiella pneumoniae strain producing a plasmid-mediated ACC-1 AmpC β-lactamase in a teaching hospital admitting disabled patients. Antimicrob Agents Chemother 49:2095–2097
Psichogiou M, Tassios PT, Avlamis A et al (2008) Ongoing epidemic of bla VIM-1-positive Klebsiella pneumoniae in Athens, Greece: a prospective survey. J Antimicrob Chemother 61:59–63
Cagnacci S, Gualco L, Roveta S et al (2008) Bloodstream infections caused by multidrug-resistant Klebsiella pneumoniae producing the carbapenem-hydrolysing VIM-1 metallo-β-lactamase: first Italian outbreak. J Antimicrob Chemother 61:296–300
Acknowledgements
To the members of the GEMARA and GEIH groups: Emilia Cercenado (Hospital Gregorio Marañon, Madrid), Beatriz Orden (CE Argüelles, CEP Madrid, Hospital Universitario Puerta del Hierro, Madrid), Andrea Mª Gonzalez and Alberto Delgado-Iribarren (Hospital Fundación de Alcorcón, Madrid), Rafael Cantón and M. Isabel Morosini (Hospital Ramon y Cajal, Madrid), Carmen Aspiroz and Blanca Fortuño (Hospital Royo Villanova, Zaragoza), Fe Tubau and Josefina Ayats (Hospital de Bellvitge, Barcelona), F. Javier Castillo and Cristina Seral (Hospital Clínico Universitario Lozano Blesa, Zaragoza), Margarita Salvador and Concepción Segura (Laboratori de Referencia, Barcelona), Cayo Sádaba and Marta Lamata (Fundación Hospital de Calahorra, La Rioja), Ana Granados and Dionisia Fontanals (Hospital Parc Taulí, Barcelona), Frederic Ballester and Oscar Villuenda (Hospital de Sant Joan de Reus, Reus), Frederic Gómez and Rafael Sánchez (Hospital Universitari Joan XXIII, Tarragona), Cristina Pitart and Francesc Marco (Hospital Clínic de Barcelona, Barcelona), Gloria Royo and Montserrat Ruíz (Hospital General Universitari d’Elx, Elx), Mª Luz Núñez and Antonio Altuna Cuesta (Hospital Reina Sofia de Murcia, Murcia), José Luis López and Miguel Salavert (Hospital Universitari La Fe, Valencia), Pilar Marín (Hospital Puerta del Mar, Cadiz), Pilar Teno (Hospital San Pedro de Alcántara, Cáceres), Patricia Iraurgui and Cecilia Martín (Hospital Virgen del Rocío, Sevilla), Gloria Esteban and Begoña Fernández (Complejo hospitalario de Ourense, Ourense), M. Isabel Fernández-Natal (Complejo Asistencial de León, León), Carlos Fuster (Hospital de El Bierzo, León), Fernando García-Garrote and Amparo Coira Nieto (Complejo Hospitalario Xeral-Calde de Lugo, Lugo), Ana Fleites and Marta Lantero (Hospital Universitario Central de Asturias, Oviedo), Andrés Canut (Hospital Santiago Apostol Osakidetza, Servicio Vasco de Salud, Basurto), Estibaliz Ugalde and Carmen Torres (Hospital San Pedro, Logroño), Mª Luz Cordón and Ainara Rodríguez (Hospital de Alto Deba, Mondragón), Mirian Alkorta and Ana María Iturzaeta (Hospital de Zumárraga, Zumárraga), Joxe Mari Manterola (Hospital de Mendaro, Mendaro).
We thank C. Newey for revising the English language in this paper.
Funding
This study was partially supported by the Ministry of Health and Consumer Affairs, Instituto de Salud Carlos III—FEDER, the Spanish Network for Research in Infectious Diseases (REIPI RD06/0008), by a grant from the Fondo de Investigación Sanitaria (PS09/00125) and by AstraZeneca Farmacéutica Spain and Wyeth (now Pfizer) pharmaceutical industries.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Miró, E., Agüero, J., Larrosa, M.N. et al. Prevalence and molecular epidemiology of acquired AmpC β-lactamases and carbapenemases in Enterobacteriaceae isolates from 35 hospitals in Spain. Eur J Clin Microbiol Infect Dis 32, 253–259 (2013). https://doi.org/10.1007/s10096-012-1737-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-012-1737-0