Abstract
Even if Panton–Valentine leukocidin (PVL), toxic shock syndrome toxin-1 (TSST-1), staphylococcal enterotoxins (SEB and SEC), and exfoliative toxins (ETA and ETB) may be associated with severe infections, the clinical significance of their presence in clinical isolates of Staphylococcus aureus remains poorly documented. In this study, we evaluated the prevalence of toxin genes and the relationship between their presence and the severity of infection. We screened for the presence of these six toxin genes among 186 consecutive S. aureus clinical isolates (resistant or not to methicillin) during a two-month period. We compared the toxin gene profile between strains recovered from patients presenting uncomplicated infections (n = 151) and from patients suffering from severe infections (n = 35). At least one toxin gene was detected in 55 (29.6%) isolates as follows: pvl (n = 1), tst + sec (n = 5), seb (n = 19), seb + sec (n = 1), sec (n = 28), and eta (n = 1). The proportion of toxin-producing strains among patients with uncomplicated infections (27.8%) and patients with severe infections (37.1%) was not statistically different (p = 0.3044), even if the severity of infection tended to be associated with the presence of sec (p = 0.0655). Although the prevalence of toxin genes was relatively high herein, no statistically significant association between the severity of infection and the presence of toxin genes was observed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Staphylococcus aureus is a major human pathogen that causes both suppurative and toxin-mediated nosocomial and community-acquired infections worldwide [1]. Numerous toxins have been identified in S. aureus, including some of them that have been potentially associated with specific infections or syndromes [2].
Among these virulence factors, Panton–Valentine leukocidin (PVL) is a pore-forming toxin generally associated with complicated skin and skin structure infections, extensive cellulitis, necrotizing pneumonia, and acute osteomyelitis in children [3–5]. In particular, the leukotoxic action of PVL is responsible for the high mortality rate (up to 75%) reported in necrotizing pneumonia [4].
Several toxins have superantigenic properties, such as toxic shock syndrome toxin-1 (TSST-1) and staphylococcal enterotoxins B (SEB) and C (SEC). These superantigens directly activate T-cells through bypassing normal antigen presentation. They bind to the specific variable region of the T-cell antigen receptor-β-chain with strong affinity [2, 6]. Uncontrolled release of Th1 pro-inflammatory cytokines mediated by massive T-cell proliferation may be responsible for the symptomatology of staphylococcal toxic shock syndrome (STSS) [2]. Clinically, STSS combines rapid onset of fever, diffuse macular rash, hypotension, and multiple organ failure, with a mortality rate ranging from 4 to 22% [6, 7].
Exfoliative toxins A (ETA) and B (ETB) are epidermolytic proteases mainly responsible for staphylococcal scalded skin syndrome (SSSS), which typically occurs in neonates and infants, but can also affect predisposed adults. The syndrome is characterized by a fever and a rapid generalized desquamation without mucosal damage. The mortality rate is low in infants but can reach 67% in adults [8]. The strains producing these toxins can also cause localized forms (namely, bullous impetigo), where skin lesions are restricted to the site of infection [8].
Even if these toxins have been related to severe infections, the clinical significance of their presence in S. aureus isolates has been poorly investigated, and only among collections of non-consecutive clinical isolates [9–13]. The aim of this study was, then, to: (i) screen consecutive S. aureus clinical isolates (resistant or not to methicillin) for the presence of these six major toxins and (ii) assess the clinical significance of these toxin-producing strains. The proportion of virulence genes was compared between the isolates groups associated with uncomplicated infection and severe infection.
Materials and methods
Bacterial isolates
From May to June 2009, all non-duplicate S. aureus clinical isolates recovered from patients hospitalized at the university hospital of Caen (ca. 2,000-bed) were studied. They were identified by using conventional phenotypic methods, such as Gram staining, catalase activity, and latex agglutination test (Pastorex Staph-Plus®; Bio-Rad, Marnes-la-Coquette, France). In doubtful cases, an identification by using the ID-GP card on the Vitek 2 system (bioMérieux, Marcy-l’Etoile, France) was performed. Antimicrobial susceptibility was determined by disc diffusion on Mueller–Hinton agar according to the Antibiogram Committee of the French Society for Microbiology guidelines (http://www.sfm.asso.fr).
Patient characteristics and definitions
Demographic and clinical data were retrospectively recorded for each patient regarding age, sex, underlying conditions (including heart disease, hemodialysis, diabetes mellitus, hematological disorder, HIV infection, current malignancy, immunosuppressive therapy), site of isolation, clinical presentation, hospital ward, treatment, and clinical outcome.
Sepsis, severe sepsis, septic shock, and multiple organ dysfunction syndrome were defined according to the criteria of the American College of Chest Physicians and Society of Critical Care Medicine Conference [14], while STSS was defined as described elsewhere [6]. Hospital-acquired infections were defined according to the CDC definitions [15] (http://www.cdc.gov/ncidod/dhqp/pdf/nnis/NosInfDefinitions.pdf). All other cases were considered as community-acquired infections.
Multiplex real-time PCR
Bacterial genomic DNA was extracted by using the InstaGene Matrix® Kit (Bio-Rad) according to the manufacturer’s recommendations. We developed two triplex real-time polymerase chain reaction (PCR) assays for the detection of pvl, tst, and eta genes (PCR A) and etb, seb, and sec genes (PCR B). Primers and TaqMan™ LNA probes were designed and synthesized by Sigma-Aldrich France (Table 1). PCR A and PCR B experiments were performed by using the QuantiFast® Multiplex PCR Kit (Qiagen, Courtaboeuf, France) in a 25-μl reaction mixture with final concentrations of 0.5 μM for each primer and 0.2 μM for each LNA probe. PCR amplifications were performed using a Rotor-Gene Q instrument (Qiagen) as follows: (i) initial denaturation step of 5 min at 95°C and (ii) 40 cycles of PCR, with one cycle consisting of 45 s at 95°C and 45 s at 60°C. The presence of amplified DNA was detected by measuring the fluorescence emitted at 470 nm (for pvl and etb), 530 nm (for tst and seb), and 585 nm (for eta and sec).
PFGE
Fingerprinting by pulsed-field gel electrophoresis (PFGE) was performed on toxin gene-positive strains as previously described [16]. SmaI-macrorestricted patterns were obtained by using a contour-clamped homogeneous electric field DR-III apparatus (Bio-Rad) as follows: ramped pulse times of 5 s and 35 s at 6 V/cm for 20 h at 14°C. Fingerprint® II software (Bio-Rad) was used to analyze PFGE patterns. The calculation of similarity matrices and dendrograms were obtained with the unweighted pair group method with arithmetic averages (UPGMA). Strains were considered to be clonally-related if the Dice coefficients were greater than 85%.
Comparative analysis
In order to evaluate the role of toxins in the severity of infections, patients were classified into two groups: (i) uncomplicated infections (UI) defined as sepsis and (ii) severe infections (SI) defined as severe sepsis, septic shock, multiple organ dysfunction syndrome, or STSS. These two groups were compared regarding demographic and clinical characteristics, as well as toxin gene prevalence.
Statistical analysis
Statistical analysis was performed using the GraphPad Software (http://www.graphpad.com/quickcalcs). The χ2 test or Fisher’s exact test was used, as appropriate, to compare categorical variables, whereas Student’s t-test was used to compare continuous variables. Differences between groups were considered to be statistically significant for p-values <0.05.
Results
Clinical data
During the two-month period, 186 strains were recovered from 186 patients, including 75 isolates obtained from patients with superficial infections (68 skin and soft tissue infections, six ear, nose, throat [ENT] infections, and one genital infection) and 111 from patients with invasive infections (33 respiratory tract infections, 30 bacteremia, 21 urinary tract infections, 17 bone or joint infections, and 10 deep abscesses). Among the 186 patients enrolled in the study, 151 suffered from uncomplicated infections and 35 suffered from severe infections (18 severe sepsis, 15 septic shock, two multiple organ dysfunction syndromes). No patients met the diagnostic criteria for STSS. Most infections were hospital-acquired (99/186; 53.2%), while the prevalence of methicillin-resistant Staphylococcus aureus (MRSA) infection was 25.8%. Forty surgical site infections (21.5%) were reported.
The demographic and clinical characteristics of the patients are shown in Table 2. There were no significant differences between UI and SI groups regarding age, sex, underlying diseases, prior hospitalization, prior surgery, nosocomial infection, MRSA infection, and surgical therapy (Table 2). By contrast, there was a significant difference for hospitalization ward, type of infection (superficial or invasive), and antimicrobial therapy (Table 2). Unsurprisingly, the 28-day crude mortality was significantly higher among SI patients than among UI patients (34.1% vs. 6.6%; p = 0.0002).
Prevalence of toxin genes
Of the 186 isolates, 55 (29.6%) harbored at least one toxin gene (Table 3), comprising 13 of 35 strains (37.1%) recovered from SI patients and 42 of 151 strains (27.8%) recovered from UI patients. The most prevalent gene was sec (15.1%), followed by seb (10.2%). Five (2.7%) strains were positive for both tst and sec genes, while seb and sec genes were detected in a single isolate. Finally, the eta gene was detected in one strain, whereas no strain was positive for the etb gene. Noteworthy, one or more toxin genes were detected in 46 (33.3%) of 138 methicillin-susceptible S. aureus (MSSA) and in 9 (18.7%) of 48 MRSA.
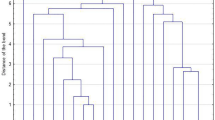
By using a cut-off value of 85% for the Dice coefficient, 16 and 10 different PFGE clusters were identified in the group of pvl-, tst- and sec-, seb-, seb- and sec-, and eta-positive strains (n = 27) and in the group of sec-positive strains (n = 28), respectively (Fig. 1). Noteworthy, three of the five tst-positive isolates were MRSA, while the single pvl-positive isolate was also MRSA (Fig. 1a). Concerning the 28 sec-positive strains, 13 (46.4%) clustered in a major group (Fig. 1b). Although they exhibited identical antimicrobial resistance profiles, they were recovered at different dates from patients hospitalized in different hospital wards (Fig. 1b).
Unweighted pair group method with arithmetic averages (UPGMA) dendrogram of pulsed-field gel electrophoresis (PFGE) results based on the Dice distance matrix of pvl-, tst and sec-, seb-, seb and sec-, and eta-positive (a) and sec-positive (b) Staphylococcus aureus clinical isolates. The 85% cut-off value of the Dice coefficient is shown by a vertical dotted line. Date of isolation, hospital ward, and antimicrobial resistance and toxin gene profiles are also indicated. OXA, oxacillin; KM, kanamycin; TM, tobramycin; LVX, levofloxacin; ERY, erythromycin; LIN, lincomycin; PT, pristinamycin; FA, fusidic acid; FOS, fosfomycin; RA, rifampicin
Clinical significance of toxin-producing strains
The overall prevalence of toxin-producing strains among UI patients (27.8%) and SI patients (37.1%) was not statistically different (p = 0.3044) (Table 3). In addition, none of the detected genes was more prevalent in one of the two groups. Nonetheless, SI tended to be associated with a higher prevalence of the sec gene (25.7% vs. 12.5%; p = 0.0655).
Discussion
To the best of our knowledge, this is the first study that has evaluated the prevalence of several major toxins on a collection of consecutive (methicillin-resistant or -susceptible) S. aureus clinical isolates. To do that, we optimized a multiplex real-time PCR assay that appeared to be a useful tool for the detection of S. aureus toxin genes during epidemiological surveys, as previously reported [17, 18]. Overall, 29.6% of all tested isolates were positive for one or more toxin genes, this prevalence being similar to those reported in previous studies [10, 11, 13, 19]. More specifically, only a small proportion of strains were positive for pvl (0.5%) and eta (0.5%) genes and none for the etb gene. Such low prevalence has also been reported in other studies [11, 13, 20]. The single pvl-positive strain harbored a typical antimicrobial resistance pattern (resistance to methicillin, kanamycin, and fusidic acid, and susceptibility to tobramycin, gentamicin, and fluoroquinolones), suggesting that it could belong to the MRSA sequence type 80 (ST80) European clone that has emerged and spread in Europe since the late 1990s [21]. We found a lower proportion of tst gene (2.8%) in comparison with other studies (8.7 to 30% of infections) [10, 11, 13, 19], but in Peacock et al.’s study, this prevalence decreased after adjustment for effects of clonality [13]. Among the five tst-positive strains (also sec-positive), three isolates had a particular antimicrobial resistance phenotype (phenotype 1: resistance to methicillin, kanamycin, tobramycin, and fusidic acid, and susceptibility to gentamicin and fluoroquinolones; phenotype 2: resistance to methicillin and fusidic acid, and susceptibility to kanamycin, tobramycin, gentamicin, and fluoroquinolones) assumed to belong to the MRSA ST5 Geraldine clone, which was recently reported in France [22]. The analysis of PFGE patterns showed the polyclonality of circulating strains in our hospital during the period of the study. Only one major cluster of sec-positive isolates (46.4%) was identified, suggesting that these strains were epidemiologically related. However, no outbreak was identified during the two-month study period.
In this study, we demonstrated no statistically significant association between the severity of infection and the presence of the six detected toxin genes, even if the sec gene tended to be more common in SI patients than in UI patients (25.7% vs. 12.5%; p = 0.065). This tendency has not yet been reported [10, 11, 13, 19] and remains to be confirmed.
Our study presented several limitations: (i) this study was a single-center investigation, (ii) clinical data have been retrospectively collected, and (iii) the SI patients group (n = 35) was relatively small compared to the UI patients group. Furthermore, the molecular analysis of strains does not ascertain the in vivo production of toxins.
Other toxins have been shown to play a role in severity. For instance, Ferry et al. demonstrated in S. aureus bacteremia a link between the presence of the sea gene and septic shock, whereas the presence of the egc genes was negatively correlated with the severity of infection [10]. In addition, several virulence factors could act in combination for enhancing the clinical severity of infections [13]. Even if only one pvl-positive strain was identified in this study, the role of PVL as a major virulence determinant in severe infections remains controversial [9, 23–28]. Finally, the interaction between the host immune system (including genetic predispositions) and the pathogen may play a major part in S. aureus toxin-mediated infections’ pathogenesis. Not all colonized or infected patients with a toxin-producing strain of S. aureus go on to develop STSS. A lack of detectable antibodies to STSS-associated superantigens seems to be a major risk for the development of STSS [2, 6]. These individuals may be predisposed to repeated episodes of STSS. In addition, toll-like receptor type 2 (TLR-2) polymorphism may place individuals at a higher risk of Gram-positive septic shock, as well as certain HLA haplotypes can also impact on the clinical susceptibility to toxic effects of superantigens [29].
In conclusion, we did not observe a statistically significant relation between the severity of the infection and the presence of toxin gene(s). However, it would be interesting to complete these first results by a multicenter investigation with a larger population. So far, the detection of toxin genes in S. aureus has been considered to be of interest for the diagnosis of toxin-mediated diseases leading to an early antitoxinic therapy, such as clindamycin, linezolid, and/or intravenous immunoglobulins. Although these molecules have been shown to be effective for reducing exotoxin production in vitro [30–33], the clinical benefits should be more thoroughly evaluated.
References
Lowy FD (1998) Staphylococcus aureus infections. N Engl J Med 339:520–532
Dinges MM, Orwin PM, Schlievert PM (2000) Exotoxins of Staphylococcus aureus. Clin Microbiol Rev 13:16–34
Lina G, Piémont Y, Godail-Gamot F, Bes M, Peter MO, Gauduchon V, Vandenesch F, Etienne J (1999) Involvement of Panton–Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis 29:1128–1132
Gillet Y, Issartel B, Vanhems P, Fournet JC, Lina G, Bes M, Vandenesch F, Piémont Y, Brousse N, Floret D, Etienne J (2002) Association between Staphylococcus aureus strains carrying gene for Panton–Valentine leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients. Lancet 359:753–759
Bocchini CE, Hulten KG, Mason EO Jr, Gonzalez BE, Hammerman WA, Kaplan SL (2006) Panton–Valentine leukocidin genes are associated with enhanced inflammatory response and local disease in acute hematogenous Staphylococcus aureus osteomyelitis in children. Pediatrics 117:433–440
McCormick JK, Yarwood JM, Schlievert PM (2001) Toxic shock syndrome and bacterial superantigens: an update. Annu Rev Microbiol 55:77–104
Descloux E, Perpoint T, Ferry T, Lina G, Bes M, Vandenesch F, Mohammedi I, Etienne J (2008) One in five mortality in non-menstrual toxic shock syndrome versus no mortality in menstrual cases in a balanced French series of 55 cases. Eur J Clin Microbiol Infect Dis 27:37–43
Ladhani S (2003) Understanding the mechanism of action of the exfoliative toxins of Staphylococcus aureus. FEMS Immunol Med Microbiol 39:181–189
Bae IG, Tonthat GT, Stryjewski ME, Rude TH, Reilly LF, Barriere SL, Genter FC, Corey GR, Fowler VG Jr (2009) Presence of genes encoding the Panton–Valentine leukocidin exotoxin is not the primary determinant of outcome in patients with complicated skin and skin structure infections due to methicillin-resistant Staphylococcus aureus: results of a multinational trial. J Clin Microbiol 47:3952–3957
Ferry T, Thomas D, Genestier AL, Bes M, Lina G, Vandenesch F, Etienne J (2005) Comparative prevalence of superantigen genes in Staphylococcus aureus isolates causing sepsis with and without septic shock. Clin Infect Dis 41:771–777
Heymans F, Fischer A, Stow NW, Girard M, Vourexakis Z, Des Courtis A, Renzi G, Huggler E, Vlaminck S, Bonfils P, Mladina R, Lund V, Schrenzel J, François P, Lacroix JS (2010) Screening for staphylococcal superantigen genes shows no correlation with the presence or the severity of chronic rhinosinusitis and nasal polyposis. PLoS ONE 5:e9525
Lalani T, Federspiel JJ, Boucher HW, Rude TH, Bae IG, Rybak MJ, Tonthat GT, Corey GR, Stryjewski ME, Sakoulas G, Chu VH, Alder J, Steenbergen JN, Luperchio SA, Campion M, Woods CW, Fowler VG (2008) Associations between the genotypes of Staphylococcus aureus bloodstream isolates and clinical characteristics and outcomes of bacteremic patients. J Clin Microbiol 46:2890–2896
Peacock SJ, Moore CE, Justice A, Kantzanou M, Story L, Mackie K, O’Neill G, Day NP (2002) Virulent combinations of adhesin and toxin genes in natural populations of Staphylococcus aureus. Infect Immun 70:4987–4996
Bone RC, Balk RA, Cerra FB, Dellinger RP, Fein AM, Knaus WA, Schein RM, Sibbald WJ (1992) Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM Consensus Conference Committee. American College of Chest Physicians/Society of Critical Care Medicine. Chest 101:1644–1655
Horan TC, Andrus M, Dudeck MA (2008) CDC/NHSN surveillance definition of health care-associated infection and criteria for specific types of infections in the acute care setting. Am J Infect Control 36:309–332
Maslow JN, Mulligan ME, Arbeit RD (1993) Molecular epidemiology: application of contemporary techniques to the typing of microorganisms. Clin Infect Dis 17:153–162; quiz 163–4
McDonald RR, Antonishyn NA, Hansen T, Snook LA, Nagle E, Mulvey MR, Levett PN, Horsman GB (2005) Development of a triplex real-time PCR assay for detection of Panton–Valentine leukocidin toxin genes in clinical isolates of methicillin-resistant Staphylococcus aureus. J Clin Microbiol 43:6147–6149
Letertre C, Perelle S, Dilasser F, Fach P (2003) Detection and genotyping by real-time PCR of the staphylococcal enterotoxin genes sea to sej. Mol Cell Probes 17:139–147
Becker K, Friedrich AW, Lubritz G, Weilert M, Peters G, Von Eiff C (2003) Prevalence of genes encoding pyrogenic toxin superantigens and exfoliative toxins among strains of Staphylococcus aureus isolated from blood and nasal specimens. J Clin Microbiol 41:1434–1439
Prevost G, Couppie P, Prevost P, Gayet S, Petiau P, Cribier B, Monteil H, Piémont Y (1995) Epidemiological data on Staphylococcus aureus strains producing synergohymenotropic toxins. J Med Microbiol 42:237–245
Vandenesch F, Naimi T, Enright MC, Lina G, Nimmo GR, Heffernan H, Liassine N, Bes M, Greenland T, Reverdy ME, Etienne J (2003) Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton–Valentine leukocidin genes: worldwide emergence. Emerg Infect Dis 9:978–984
Durand G, Bes M, Meugnier H, Enright MC, Forey F, Liassine N, Wenger A, Kikuchi K, Lina G, Vandenesch F, Etienne J (2006) Detection of new methicillin-resistant Staphylococcus aureus clones containing the toxic shock syndrome toxin 1 gene responsible for hospital- and community-acquired infections in France. J Clin Microbiol 44:847–853
Bubeck Wardenburg J, Palazzolo-Ballance AM, Otto M, Schneewind O, DeLeo FR (2008) Panton–Valentine leukocidin is not a virulence determinant in murine models of community-associated methicillin-resistant Staphylococcus aureus disease. J Infect Dis 198:1166–1170
Crémieux AC, Dumitrescu O, Lina G, Vallee C, Côté JF, Muffat-Joly M, Lilin T, Etienne J, Vandenesch F, Saleh-Mghir A (2009) Panton–Valentine leukocidin enhances the severity of community-associated methicillin-resistant Staphylococcus aureus rabbit osteomyelitis. PLoS ONE 4:e7204
Diep BA, Palazzolo-Ballance AM, Tattevin P, Basuino L, Braughton KR, Whitney AR, Chen L, Kreiswirth BN, Otto M, DeLeo FR, Chambers HF (2008) Contribution of Panton–Valentine leukocidin in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. PLoS ONE 3:e3198
Labandeira-Rey M, Couzon F, Boisset S, Brown EL, Bes M, Benito Y, Barbu EM, Vazquez V, Höök M, Etienne J, Vandenesch F, Bowden MG (2007) Staphylococcus aureus Panton–Valentine leukocidin causes necrotizing pneumonia. Science 315:1130–1133
Löffler B, Hussain M, Grundmeier M, Brück M, Holzinger D, Varga G, Roth J, Kahl BC, Proctor RA, Peters G (2010) Staphylococcus aureus Panton–Valentine leukocidin is a very potent cytotoxic factor for human neutrophils. PLoS Pathog 6:e1000715
Voyich JM, Otto M, Mathema B, Braughton KR, Whitney AR, Welty D, Long RD, Dorward DW, Gardner DJ, Lina G, Kreiswirth BN, DeLeo FR (2006) Is Panton–Valentine leukocidin the major virulence determinant in community-associated methicillin-resistant Staphylococcus aureus disease? J Infect Dis 194:1761–1770
Silversides JA, Lappin E, Fergusson AJ (2010) Staphylococcal toxic shock syndrome: mechanisms and management. Curr Infect Dis Rep 12:392–400
Darenberg J, Söderquist B, Normark BH, Norrby-Teglund A (2004) Differences in potency of intravenous polyspecific immunoglobulin G against streptococcal and staphylococcal superantigens: implications for therapy of toxic shock syndrome. Clin Infect Dis 38:836–842
Dumitrescu O, Badiou C, Bes M, Reverdy ME, Vandenesch F, Etienne J, Lina G (2008) Effect of antibiotics, alone and in combination, on Panton–Valentine leukocidin production by a Staphylococcus aureus reference strain. Clin Microbiol Infect 14:384–388
Gauduchon V, Cozon G, Vandenesch F, Genestier AL, Eyssade N, Peyrol S, Etienne J, Lina G (2004) Neutralization of Staphylococcus aureus Panton Valentine leukocidin by intravenous immunoglobulin in vitro. J Infect Dis 189:346–353
Stevens DL, Ma Y, Salmi DB, McIndoo E, Wallace RJ, Bryant AE (2007) Impact of antibiotics on expression of virulence-associated exotoxin genes in methicillin-sensitive and methicillin-resistant Staphylococcus aureus. J Infect Dis 195:202–211
Acknowledgments
We thank Nancy Bourdon for her excellent assistance in the pulsed-field gel electrophoresis (PFGE) bioinformatic analysis.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nhan, TX., Leclercq, R. & Cattoir, V. Prevalence of toxin genes in consecutive clinical isolates of Staphylococcus aureus and clinical impact. Eur J Clin Microbiol Infect Dis 30, 719–725 (2011). https://doi.org/10.1007/s10096-010-1143-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-010-1143-4