Abstract
We report three cases of psittacosis in staff working in a veterinary surgery, which was related to exposure to a sick, wild psittacine bird. Chlamydial genus- and chlamydial species-specific DNA was detected in clinical specimens, including throat swabs, whole blood and urine. The organism load was quantified by real-time PCR (RT-PCR), which revealed 105-fold more organisms in conjunctival swabs from the source bird than in the human samples. One clinic attendant was infected despite using personal protective equipment when handling the bird. This is the first report of PCR analyses of blood and urine samples being used to diagnose human psittacosis, and the first time that the organism load in humans has been compared to that of the infecting bird.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chlamydophila psittaci is the cause of psittacosis, a multi-system disease in humans. It is common in birds of many types and can be identified in avian specimens using antigen detection and molecular methods. In contrast, the organism is seldom identified by culture in humans, and molecular diagnostic techniques are not readily available. Most human cases are diagnosed by detecting Chlamydophila-specific antibodies using complement fixation or species-specific microimmunofluorescence (MIF) in acute and convalescent sera; consequently, laboratory diagnosis lags behind the clinical presentation by several weeks. Moreover, serology can be difficult, delayed and relatively non-specific [1]. A rapid, sensitive and specific diagnostic nucleic acid test (NAT) that is applicable to different specimen types (e.g. blood, urine and upper airway samples) is a clinical necessity.
Chlamydiaceae is a family of intracellular organisms found in both humans and animals. It has recently been separated into two genera, the Chlamydiae, including Chlamydia trachomatis, and the Chlamydophilae, including C. pneumoniae and C. psittaci [2]. Chlamydophilae psittaci is a zoonotic infection that is usually acquired through close contact with captive or commercial birds, but it has also been reported as a cause of community-acquired pneumonia following exposure to wild birds [3]. There are eight known serovars of C. psittaci, and the corresponding genotypes have been identified [4]. Human disease has been described with serovars A, C, D and E. Serovar A is endemic among psitticine birds, and serovar E is associated with pneumonitis in humans [2, 4, 5].
Human-to-human spread of C. psittaci is rare [6–8], but contact with birds can result in a high infectivity rate. Transmission occurs by inhaling dried bird secretions and excreta or following close contact with an infected bird [9]. The likelihood of infection is presumably related to exposure to a high organism load, but the C. psittaci serotype may also be a factor. The main clinical presentation of psittacosis is a generalised febrile illness with pneumonia. Neurological disease, hepatitis, endocarditis and renal disease have also been reported.
We describe the natural history of C. psittaci infection in three patients who acquired the infection following contact with a sick wild bird. Molecular techniques were used to examine the tissue distribution and organism load during the acute infection phase. High infectivity was demonstrated despite the use of personal protective equipment.
Case reports
Case 1
A 57-year-old male veterinary surgeon examined a sick Crimson Rosella (Platycercus elegans – a psitticine parrot) from the Blue Mountains region, west of Sydney, Australia. The bird had been found by the roadside and brought into the clinic by a member of the public. The bird was sick, thin and had moderately weepy eyes, all symptoms suggestive of psittacosis. The bird was examined without the use of protective clothing or a facemask and placed in a covered cage; it subsequently died overnight.
Eight days later the veterinary surgeon became unwell with headaches, myalgia, fever, sweats, cough and dyspnoea. These symptoms worsened for 3 days. A regimen of oral roxithromycin was commenced, and the patient progressively improved over the next 5 days. A chest X-ray was not performed. Blood, throat and urine samples were collected 4 days after the roxithromycin treatment regimen was initiated.
Case 2
Case 2 was a 55-year-old female assistant in the veterinary practice of case 1, and the relationship partner of case 1. She only handled the bird after death and wore protective clothing, namely gloves and a surgical mask. She removed the bird from the cage, wrapped it for autopsy and washed her hands afterwards. She became unwell 11 days later with severe headaches, myalgia, fatigue, cough and dyspnoea. The results of a chest X-ray performed on the fourth day of her illness were normal. Oral doxycycline was commenced the same day, and throat, blood and urine samples were obtained. She began improving after another 48 h and recovered completely within 3 weeks.
Case 3
Case 3 was a 35-year-old female veterinary nurse who was employed at the practice of case 1. She initially received the bird and handled it briefly before it was examined by case 1. She also hosed out the cage after the bird had died. She used no personal protective equipment at either time. She became unwell 10 days after contact, with signs of weakness, myalgia and headache. No chest X-ray was performed. She recovered completely within 6 days without treatment and has remained well.
Material and methods
Clinical samples
Throat swabs, blood (5 mL in EDTA) and 10 mL urine were collected 15 days after exposure and 4–7 days after the onset of symptoms. The samples were stored at 4°C and processed within 2 weeks. Patient samples were collected 4 (case 1) and 1 (case 2) days after the antibiotic regimen had been initiated and in the absence of antibiotic therapy (case 3). Blood was taken for chlamydia serology at 2 and 6 weeks post onset of illness.
A limited autopsy of the bird was performed with appropriate safety precautions by staff other than those from the veterinary practice. Conjunctival swabs and a sample of liver and spleen were collected from the bird cadaver.
Chlamydial culture
Specimens were cultured in a BSL3 facility with African Buffalo Green Monkey Kidney cells (BGM). To optimise growth, the cultures were subcultured after 72 h, then fixed and stained using the Pathfinder Chlamydia genus immunofluorescent stain (Bio-Rad, Hercules, CA) after a further 72 h. Isolate identity was confirmed by species-specific PCR [10, 11].
PCR amplification
Human throat swabs and bird swabs were soaked in 200 μL of 0.9% NaCl and vortexed periodically for several hours at room temperature before DNA extraction. Urine was centrifuged and the pellet resuspended in 200 μL 0.9% NaCl before extraction. The buffy coat from EDTA blood specimens was treated with Bio-Rad Instagene (Bio-Rad) in order to lyse cells and inactivate the DNAse and RNAse enzymes, then resuspended in 200 μL 0.9% NaCl. DNA was extracted from the specimens using the QiaAmp DNA Mini-kit (Qiagen Germantown, MA), according to the manufacturer’s instructions. Prior to all PCRs, bird samples were diluted 100-fold as the target organism was too concentrated in neat specimens (data not shown).
PCR was performed on a Corbett 3000 real-time (RT)-PCR machine (Corbett Research, Sydney, Australia) using three primer sets targeting either the 16 s rRNA gene or the 16–23 S rRNA interspacer gene. One set was genus-specific (5′-GGGGTTGTAGGGTYGAGRAIAWRRCATC-3′ and 5′-GAGAGTGGTCTCCCCAGATTCARACTA-3′) [10], and two sets were specific for C. psittaci [11, 12]. Primers for C. psittaci were 5′-ATAATGACTTCGGTTGTTATT-3′, 5′-TGTTTTAGATGCCTAAACAT3′ [12] and 5′-CCCAAGGTGAGGCTGATGAC-3′, 5′-CAAACCGTCCTAAGACAGTTA-3′ [11]. An additional C. trachomatis-specific set of primers (5′-GGCGTATTTGGGCATCCGAGTAACG-3′, 5′-TCAAATCCAGCGGGTATTAACCGCCT-3′) was used on urine specimens.
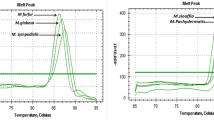
Samples for PCR were prepared in a class 2 laminar flow hood. The reaction mixture contained 5 μL Qiagen SYBRgreen master mix, including hot start polymerase (http://www.Qiagen.com), a dNTP mix and SYBRgreen, 0.1 μL of primers (3 pmol/mL) and 5 μL of extracted DNA. The cycling protocol consisted of an initial denaturation for 15 min at 95°C, followed by 50 cycles of 10 s at 95 °C, 15 s at 55 °C and 20 s at 72°C. Amplicons were detected using SYBRgreen and melt curve analysis and confirmed with gel electrophoresis . Amplicons of bird origin were sequenced by the Sydney University Prince Alfred Macromolecular Analysis Centre (SUPAMAC; http://www.supamac.com), and a BLAST search (http://130.14.29.110/BLAST/) confirmed the identity as C. psittaci (Gen Bank accession numbers: EF612704, EF612705). An avian specimen, AP1, was used as a positive control [13]. Genotyping was performed according to a published PCR method [4] in which variation in the OMP gene was targeted. We did not genotype beyond the A-B_E group.
DNA concentrations were determined by spectrophotometry, and copy numbers were calculated by reference to a standard curve that had been constructed using serial dilutions of control DNA in an approach similar to a published method [14].
Serology
Sera were tested for chlamydial species-specific IgG using a microimmunofluorescence (MIF) assay (Chlamydia IgG SeroFIA; Savyon Diagnostics, Ashdod, Israel). Microimmunofluorescence titres >1:64 for C. psittaci and C. trachomatis, and >1:512 for C. pneumoniae were regarded as indicative of current or recent infection. All paired sera were tested in parallel in a single laboratory.
Results
The results of testing the human and avian samples are expressed in Table 1.
Chlamydial genus-specific DNA was detected in the throat swab from case 1, in the blood and urine specimens from case 2 and in the urine specimen from case 3. The C. psittaci-specific RT-PCR was positive for the throat swab of case 1 and the blood specimen from case 2.
All three patients had increased C. psittaci MIF titres – up to 256 in cases 1 and 2 and 512 in case 3.
Chlamydia trachomatis-specific antibodies were not detected in any of the three cases.
Repeat throat swabs and blood samples were collected on all three patients approximately 10 days after the initial specimens. Chlamydial genus- and C. psittaci-specific RT-PCR analyses were negative on all samples. Chlamydia was not cultured from human specimens, and the C. trachomatis PCR was negative in all urine samples.
Chlamydia psittaci was cultured from the liver, spleen and conjunctiva of the source bird. The isolate was typed as genotype E, but due to lack of control strains for each genotype we were unable to separate Type A, B and E. The C. psittaci-specific PCR analysis was positive for a diluted specimen of liver/spleen homogenate as well as for both conjunctival specimens. All three primer sets rapidly became positive (within about 20 cycles) for the bird samples, in contrast to human specimens, which tested PCR positive between 35 and 40 cycles. Quantification against the standard curve revealed that bird liver and conjunctival specimens contained at least a 105-fold greater load of organisms than clinical specimens. The DNA in clinical specimens was close to the limit of detection as determined from serial dilutions of controls. Organisms cultured from the bird were amplified using the three primer sets and confirmed as C. psittaci by melt curve analysis and sequencing.
Discussion
This case cluster is the first time that C. psittaci has been quantified using RT-PCR in both the source bird and human patients with psittacosis. The results illustrate the high level of infectious units that can occur in avian secretions, which was 105-fold greater than that in human samples, and the value of DNA-based methods for rapid diagnosis. All specimens from the bird were PCR positive for C. psittaci DNA using all three primer sets. Results in human specimens were inconsistent, as one would expect when a PCR analysis is operating close to the limit of detection. Increasing the specimen numbers, sites and volume may improve the diagnostic yield. In this way urine may add to the value of blood and upper respiratory specimens.
New molecular technology is expanding our understanding of human psittacosis
Chlamydophila DNA was detected by RT-PCR in all three patients exposed to a heavily infected wild bird in the Blue Mountains of New South Wales, an area from which outbreaks of psittacosis have previously been reported [3]. The diagnosis was confirmed by chlamydia-specific serology.
Chlamydia psittaci is not often isolated or detected in human cases of psittacosis. Due to the possibility of laboratory acquisition, cultures are not usually performed. Chlamydia psittaci has only rarely been isolated from blood [15, 16] and never from urine. This case cluster indicates that low concentrations of organisms from human cases can be quantitated rapidly and directly from respiratory, urine and blood samples. If the low concentrations found are representative of human infection, this and the fact of inactivation during initial DNA extraction mean that diagnosis by NAT should not pose a risk to laboratory staff in the event of a lapse in protective procedures.
High loads of C. psittaci in birds result in high infectivity
The 100% case rate in staff at the veterinary practice in this cluster reveals the highly infective nature of this episode, which correlated with a high load of P. psittaci in the source bird. One patient was infected despite using gloves and a standard surgical mask. Handling a bird in this situation is clearly dangerous, and all steps to prevent infection must be taken. Eye protection and the fastidious use of correctly fitting N95 particulate masks (rather than surgical masks) may prevent infection. By contrast, the very low levels of Chlamyophila DNA in human samples provides an explanation for the difficulty in achieving human-to-human transmission outside of intimate contact. The high infectivity but mild illness in this cluster may relate to organism phenotype as well as load. Genotype E, also known as Cal-10, MP or MN, was isolated from human cases in the early 1900s and has subsequently been isolated from a range of birds, but its relative virulence is unknown. Serovar A is endemic in psittacine birds, and serovar B has been described in pigeons and turkeys [17].
Clinical features associated with low organism loads
These cases were all mild, with symptom onset 8–11 days post-exposure. None of the patients required hospitalisation, and one recovered spontaneously. Cases 1 and 2 had specimens collected after the commencement of the antibiotic regimen, perhaps attenuating illness and antibody response. It remains to be established whether severe cases of psittacosis are associated with a high organism load and an increased risk of human to human transmission.
In summary, the spread of infection in these three veterinary workers exposed to the same sick bird highlights the occupational risk of psittacosis for people in the veterinary industry or those handling sick birds. Timely clinical diagnosis of psittacosis may be assisted using NAT of urine, blood or other samples.
References
Centers for Disease Control and Prevention (2000) Compendium of measures to control Chlamydia psittaci infection among humans (psittacosis) and pet birds (avian chlamydiosis), 2000. MMWR Recomm Rep 49(RR-8):3–17
Everett KD, Bush RM, Andersen AA (1999) Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int J Syst Bacteriol 49(Pt 2):415–440
Telfer BL, Moberley SA, Hort KP, Branley JM, Dwyer DE, Muscatello DJ, Correll PK, England J, McAnulty JM (2005) Probable psittacosis outbreak linked to wild birds. Emerg Infect Dis 11(3):391–397
Geens T, Dewitte A, Boon N, Vanrompay D (2005) Development of a Chlamydophila psittaci species-specific and genotype-specific real-time PCR. Vet Res 36(5–6):787–797
Andersen AA, Grimes JE, Shivaprasad HL (1998) Serotyping of Chlamydia psittaci isolates from ratites. J Vet Diagn Invest 10(2):186–188
Saito T, Ohnishi J, Mori Y, Iinuma Y, Ichiyama S, Kohi F (2005) Infection by Chlamydophilia avium in an elderly couple working in a pet shop. J Clin Microbiol 43(6):3011–3013
Borel N, Doherr MG, Vretou E, Psarrou E, Thoma R, Pospischil A (2002) Ovine enzootic abortion: seroprevalence in Switzerland using a competitive enzyme-linked immunosorbent assay (cELISA). Schweiz Archiv Tierheilkunde 144(9):474–482
Hughes C, Maharg P, Rosario P, Herrell M, Bratt D, Salgado J, Howard D (1997) Possible nosocomial transmission of psittacosis. Infect Control Hosp Epidemiol 18(3):165–168
Smith KA, Bradley KK, Stobierski MG, Tengelsen LA (2005) Compendium of measures to control Chlamydophila psittaci (formerly Chlamydia psittaci) infection among humans (psittacosis) and pet birds, 2005. J Am Vet Med Assoc 226(4):532–539
DeGraves FJ, Gao D, Hehnen HR, Schlapp T, Kaltenboeck B (2003) Quantitative detection of Chlamydia psittaci and C. pecorum by high-sensitivity real-time PCR reveals high prevalence of vaginal infection in cattle. J Clin Microbiol 41(4):1726–1729
Madico G, Quinn TC, Boman J, Gaydos CA (2000) Touchdown enzyme time release-PCR for detection and identification of Chlamydia trachomatis, C. pneumoniae, and C. psittaci using the 16S and 16S-23S spacer rRNA genes. J Clin Microbiol 38(3):1085–1093
Messmer TO, Skelton SK, Moroney JF, Daugharty H, Fields BS (1997) Application of a nested, multiplex PCR to psittacosis outbreaks. J Clin Microbiol 35(8):2043–2046
Timms P, Eaves FW, Girjes AA, Lavin MF (1988) Comparison of Chlamydia psittaci isolates by restriction endonuclease and DNA probe analyses. Infect Immun 56(1):287–290
Menard A, Clerc M, Subtil A, Megraud F, Bebear C, de Barbeyrac B (2006) Development of a real-time PCR for the detection of Chlamydia psittaci. J Med Microbiol 55(Pt 4):471–473
Shapiro DS, Kenney SC, Johnson M, Davis CH, Knight ST, Wyrick PB (1992) Brief report: Chlamydia psittaci endocarditis diagnosed by blood culture. New Engl J Med 326(18):1192–1195
Harris AA, Pottage JC, Jr., Kessler HA, Zeihen M, Levin S (1984) Psittacosis bacteremia in a patient with sarcoidosis. Ann Intern Med 101(4):502–503
Geens T, Desplanques A, Van Loock M, Bonner BM, Kaleta EF, Magnino S, Andersen AA, Everett KD, Vanrompay D (2005) Sequencing of the 1Chlamydophila psittaci ompA gene reveals a new genotype, E/B, and the need for a rapid discriminatory genotyping method. J Clin Microbiol 43(5):2456–2461
Acknowledgements
We would like to acknowledge the kind assistance of the Staff at the Veterinary Surgery involved, Peter Timms from Queensland University of Technology for providing avian control strain, Sue Alderson from ICPMR for Chlamydial culture, Sau Wan Chan from ICPMR for performing serology and Kathryn Weston for manuscript review.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Branley, J.M., Roy, B., Dwyer, D.E. et al. Real-time PCR detection and quantitation of Chlamydophila psittaci in human and avian specimens from a veterinary clinic cluster. Eur J Clin Microbiol Infect Dis 27, 269–273 (2008). https://doi.org/10.1007/s10096-007-0431-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-007-0431-0