Abstract
Migraine is a prevalent neurovascular disease with a significant genetic component. Linkage studies have so far identified migraine susceptibility loci on chromosomes 1, 4, 6, 11, 14, 19 and X. We performed a genome-wide scan of 92 Australian pedigrees phenotyped for migraine with and without aura and for a more heritable form of “severe” migraine. Multipoint non-parametric linkage analysis revealed suggestive linkage on chromosome 18p11 for the severe migraine phenotype (LOD*=2.32, P=0.0006) and chromosome 3q (LOD*=2.28, P=0.0006). Excess allele sharing was also observed at multiple different chromosomal regions, some of which overlap with, or are directly adjacent to, previously implicated migraine susceptibility regions. We have provided evidence for two loci involved in severe migraine susceptibility and conclude that dissection of the “migraine” phenotype may be helpful for identifying susceptibility genes that influence the more heritable clinical (symptom) profiles in affected pedigrees. Also, we concluded that the genetic aetiology of the common (International Headache Society) forms of the disease is probably comprised of a number of low to moderate effect susceptibility genes, perhaps acting synergistically, and this effect is not easily detected by traditional single-locus linkage analyses of large samples of affected pedigrees.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Migraine is a debilitating neurovascular disorder, characterised by recurrent headache that is associated with nausea and/or vomiting, photophobia and phonophobia. The prevalence of migraine varies between different racial groups and tends to be highest in Caucasian populations (12–14%). The International Headache Society (IHS) has formally classified migraine into two main subtypes: migraine with aura (MA) and migraine without aura (MO). These two subtypes have substantial symptomatic overlap, but MA sufferers experience distinguishing neurological disturbances (the aura) that usually precede the headache phase of an attack [1]. Strong familial aggregation of migraine and an increased concordance rate in MZ twins over DZ twins suggest a significant genetic component for the disease. An epidemiologic study by Russell et al. [2] showed that the risk of migraine for first-degree relatives of MO probands is ∼1.9 and ∼3.8 for MA probands. Heritability is estimated to be between 40 and 65%, with MA tending to be the more heritable IHS subtype [3–5]. Segregation analyses indicate that the mode of genetic transmission of migraine is likely to be multifactorial, although autosomal dominant inheritance with reduced penetrance is evident in some affected pedigrees [6, 7]. The MA and MO subtypes exhibit symptomatic heterogeneity implying that different modifying factors may contribute to the variable expression of these clinical end points. However, both MA and MO often occur within the same individual and within the same families. Therefore, it is plausible that the two IHS subtypes have some genetic determinants in common with possibly a major gene(s) initiating general disease pathogenesis [5].
In an attempt to dissect the genetics of the headache continuum, Nyholt et al. [5] analysed headache symptom data collected from a large survey of 6,265 Australian twin individuals. They employed latent class analysis (LCA) to identify three major classes of headache (mild, moderate and severe) in this large population-based sample. Interestingly, the LCA “migrainous” phenotype (i.e. the moderate and severe classes) was shown to have an increased heritability over the IHS-affected phenotype in this twin sample (0.41 vs 0.33). Moreover, the relative risk for the “severe” class of migraineurs in DZ twins (RR=2.16) was larger than the risk for MA (RR=1.77) suggesting that this new LCA phenotyping scheme may be more useful in studies specifically aimed at uncovering disease susceptibility genes [5].
Genome mapping studies for migraine have so far indicated the existence of susceptibility regions for MA and/or MO on chromosomes 1q, 4q, 6p, 11q, 14q, 19p and Xq [7–15]. However, with the exception of the INSR gene on chromosome 19p13 [16], the predisposing gene(s) within these implicated regions has not yet been identified. Several putative modifying genes including dopamine 2 receptor (DRD2), dopamine beta-hydroxylase (DBH) and, more recently, the methylenetetrahydrofolate reductase (MTHFR) have also been implicated through case-control association studies with some independent confirmation [17–19]. In an effort to identify major susceptibility loci for migraine, we performed a full genome scan in Australian pedigrees phenotyped for migraine using standard IHS criteria as well as the new LCA severe criteria of Nyholt et al. [5].
Materials and methods
Subject phenotyping and pedigrees
The protocol for this research was approved by the Griffith University Ethics Committee. All subjects included in the study were Caucasian and of UK descent and gave informed consent prior to participation. Affected probands were ascertained by the Genomics Research Centre Recruitment Clinic through extensive media advertising and resided mostly on the East Coast of Australia. Affected family members were identified, and pedigrees were constructed following detailed proband interview. Clinical evaluation of all subjects was conducted carefully and thoroughly by qualified clinical neurologist using a validated headache questionnaire. All affected individuals were formally diagnosed as having either MA or MO using the IHS criteria [1]. Assuming a major underlying locus (loci) to be operating and considering MA to be the more heritable of the IHS subtypes, we phenotyped those patients experiencing both MA and MO intermittently (<5%) as being affected with MA.
Latent class analysis (LCA) is a statistical method for identifying subtypes of patients (latent classes) from multivariate categorical data. Applying LCA to the headache symptom data, the twin study of Nyholt et al. [5] identified three major classes of headache (mild, moderate and severe). According to this analysis, a headache sufferer was categorised as having severe migraine if they experienced most of the IHS symptoms (nausea/vomiting, photophobia, etc.) and usually experienced the aura. Membership in this headache class was calculated probabilistically by LCA (please refer to Nyholt et al. [5] for details of the LCA models used here and the definition of latent classes relative to IHS symptoms). Incorporating the symptom data from our clinic-based genome scan sample into the population-based twin sample of Nyholt et al., we re-phenotyped our subjects under the new LCA scheme. Reassuringly, this analysis revealed that nearly all (98.5%) of the IHS-diagnosed migraineurs were also affected with migrainous headache (including moderate and severe) under the LCA scheme.
In total, 433 subjects were included in our genome scan study, 380 of these being originally diagnosed with migraine based on IHS criteria. Of this migraine-affected group, 289 (76%) were phenotyped as having MA, whilst 247 (65%) were classified as having severe migraine under the LCA scheme (LCA-severe). This LCA-severe group was mostly comprised of MA sufferers (83%), but interestingly, 17% of the severe group were also phenotyped as IHS-based MO. The subjects formed a total of 92 pedigrees. The pedigree structures ranged in complexity from just two affected sibpairs (with no parents) to multigenerational pedigrees with many affected members. The total pedigree sample consisted of 598 affected relative pairs (Table 1). In an effort to manually (rather than statistically) control for heterogeneity, we also examined a subsample of larger multigenerational pedigrees (n=21) arbitrarily defined as having at least five affected non-founders (genetically related family members). This subsample consisted of 242 individuals with 155 subjects affected with migraine (∼80% with MA, ∼70% LCA-severe). The mode of disease transmission for these larger pedigrees was apparently a non-Mendelian (complex) mode of inheritance.
Markers and genotyping
Genomic DNA was extracted from blood samples collected from the 433 subjects enrolled in the study [20]. We utilised 400 DNA markers from the ABI PRISM Human Linkage Mapping Sets (version 2). This set of highly polymorphic dinucleotide microsatellites had an average heterozygosity of 0.80 and a resolution of ∼10 cM. Fluorescently labelled primers were used to amplify the DNA using standard PCR protocols. All PCR products were multiplexed where possible, and fragments were separated and genotyped using ABI PRISM Genetic Analyzers and GeneScan Software (Applied Biosystems, Foster City, CA). Centre d’Etude du Polymorphisms Humain (CEPH) DNA samples were used as known positive controls for allele sizing and were run with each batch of samples (in a 96-well microtitre plate) for each tested marker. We also genotyped a subset of DNA samples from each batch in duplicate to ensure consistent allele calling. Negative (water) controls were also incorporated to guard against DNA sample contamination.
Statistical analysis
The PEDMANAGER program was used to test for Mendelian errors and relationship inconsistencies in the pedigrees checked using the PEDCHECK program. Since there was no clear or consistent mode of disease inheritance within or between the migraine pedigrees, model-free linkage analysis of affected relatives was performed using the MERLIN program [21]. This program computes multipoint, allele sharing LOD* scores and associated P values according to the methods of Kong and Cox [22]. These LOD* scores are interpreted in the same way as traditional LOD scores. Linkage peaks having LOD* scores ≥0.59 (corresponding to a P value of 0.05) indicate excess allele sharing. Peaks yielding LOD* scores ≥2.2 (P=7.4×10−4) are suggestive of linkage, whilst LOD* scores ≥3.3 (P=2.2×10−5) indicate significant linkage [23]. For the linkage analysis, MERLIN calculates the allele frequencies automatically from unaffected founders in the pedigrees.
We analysed the genotype data under three phenotyping schemes. Firstly, we analysed all individuals with MA and/or MO as being affected (referred to as the “MA/MO” phenotype). Secondly, since the majority (76%) of our case group was affected with MA, we analysed the data considering only those with MA as being affected (referred to as the “MA-only” phenotype). Those individuals with MO only were phenotyped as unknown under this IHS classification scheme. Thirdly, we analysed the severe (and most heritable) headache phenotype identified by LCA (referred to as LCA-severe, 65% of affected). In addition to analysing the entire pedigree sample (n=92), we also analysed the subset of the 21 larger pedigrees.
Results
Analysis of all migraine-affected pedigrees
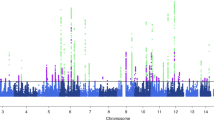
First, we conducted multipoint linkage analyses on the entire pedigree set considering only those phenotyped “LCA-severe” as being affected. Suggestive linkage was revealed on chromosome 18p11 for the LCA-severe phenotype with a maximum LOD* score of 2.32 for marker D18S53 (P=0.0006) (Fig. 1). This analysis also showed excess allele sharing on chromosomes 1q, 3q, 6p, 7p, 9q, 14q, 16p and q and 19p (P<0.05). An additional analysis of the entire migraine pedigree sample under the two IHS phenotyping schemes was conducted. Analysis of the MA/MO phenotype revealed excess allele sharing at eight chromosomal regions—1q, 3q, 6p, 7q, 9p, 14q, 16p and 18q, whilst analysis of the MA-only phenotype showed excess allele sharing at three regions—9p, 16p and 18p (P<0.05). There were no LOD* scores greater than 1 for the entire pedigree sample for either of the MA/MO and MA-only IHS phenotypes (LODs not shown).
Multipoint non-parametric linkage analysis of chromosome 18 in 92 migraine families. The Y-axis shows the allele sharing LOD* scores resulting from analyses of the migraine phenotypes (MA/MO, dotted line) and MA (dashed line) and LCA-severe (unbroken line). The X-axis shows the genetic distance in Kosambi cM across these markers
Analysis of the large pedigrees
We also conducted genome-wide linkage analyses on the subset of larger multigenerational pedigrees (n=21). For the LCA-severe phenotype, suggestive linkage for this phenotype was again observed for chromosomes 18p (LOD*=2.30, P=0.0006) and also for chromosome 3q (LOD*=2.28, P=0.0006) (Table 2). The excess allele sharing peaks localised to chromosomal regions 1q, 2q, 4q, 10p, 13p, 14q, 15q, 16q, 17p and q (P<0.05). For analysis of the MA/MO phenotype, the linkage peaks localised to chromosomal regions 1q, 2p, 4q, 10p, 13p, 14q and 17q and, for the MA-only phenotype, to chromosomal regions 1q, 2q, 4q, 10p, 14p and 18p. The LOD* scores for our linkage peaks on chromosomes 1q, 4q and 14q, as well as for the peaks on chromosomes 2p and 17q, were all greatest for the MA/MO phenotype, although the MA-only phenotype also displayed excess allele sharing for markers in the same region. Alternatively, the LOD* scores for linkage peaks identified on chromosome 10p(tel) and 18p11 were greatest for the MA-only phenotype. It is important to emphasise that the linkage peaks identified for each migraine phenotype overlapped in most cases, yet the LCA-severe phenotype typically yielded the largest LOD scores.
Overall, our genome scan analyses revealed several promising linkage peaks yielding LOD* scores >1. Table 2 illustrates the markers that produced the maximum LOD* scores at these regions, the phenotype for which these LOD* scores were obtained, as well as the corresponding list of migraine susceptibility regions implicated in previous studies. For our genome scan, suggestive linkage was detected on chromosome 18p11 for the LCA-severe phenotype when all pedigrees were analysed. Interestingly, this peak remained positive in the analysis of the larger pedigree subsample for both the LCA-severe and MA-only phenotype. Suggestive linkage was also observed at chromosome 3q-tel in the large pedigree sample. This signal was not detected in the entire pedigree set and may be the result of reducing the genetic heterogeneity.
Discussion
Migraine is a prevalent neurovascular disease with a complex genetic aetiology. Genetic studies into FHM, a rare autosomal dominant variant of MA, have successfully identified causative mutations in the CACNA1A and ATP1A2 genes located on chromosomes 19p13 and 1q23, respectively [24, 25]. Although these ion channel genes have not yet been shown to influence the more common types of migraine, several other modifying genes have been implicated through case-control association studies [16–19]. However, all the genes implicated thus far only seem to confer a modest independent effect on disease susceptibility in the general population and therefore are not likely to represent major risk factors for migraine.
Linkage (or allele sharing) studies utilising samples of affected pedigrees remain a popular approach for detecting genomic regions likely to harbour genes conferring a more substantial effect on disease susceptibility. To date, there have been five genome scans published for migraine (with and without aura). These describe the existence of migraine susceptibility loci on chromosomes 4, 6, 11 and 14, respectively [7, 9–12]. Here we present a full genome scan study using 92 independent Australian pedigrees. We did not perform linkage analyses based on particular disease model parameters given the apparent complexity of migraine transmission within and between the different pedigrees we examined. Instead, and like Bjornsson et al. [9], we chose to perform the so-called “model-free” analyses, which test for excess allele sharing between pairs of affected relatives. At present, there is conflicting evidence regarding the underlying aetiology of MA and MO and the degree of genetic commonality for these subtypes [5]. Therefore, we also opted to stratify our analyses according to clinical phenotype and examine MA and MO as a single entity as well as testing MA as a distinct phenotype. A novel feature of our study is the inclusion of a more heritable form of severe migraine not previously investigated in a genome scan [5].
Multipoint analyses of our entire pedigree sample for the LCA-severe phenotype revealed suggestive evidence for chromosome 18p11. This peak may be important since the marker yielding the greatest LOD* score (D18S53) is only about 2 cM away from a positive marker for MO (D18S453) reported by Bjornsson et al. (LOD*=1.5). Although the migraine phenotypes tested for the two studies are not identical, the substantial symptomatic overlap suggests that this region on chromosome 18p11 may harbour a fundamental susceptibility locus for migraine and certainly warrants further investigation. We speculate that variants in the myo-inositol monophosphatase gene on 18p11, which have previously been associated with bipolar disorder and febrile seizures, may also influence the migrainous phenotypes in these pedigrees. The IHS phenotypes revealed excess allele sharing at several regions in the entire pedigree set, although there were no LOD scores above 1.
As is probably the case for many complex disorders of a neuro/psychiatric nature, migraine is likely to be highly heterogenous at the genetic level with sequence variation in multiple different genes influencing the disease within and between families. In an attempt to reduce the possible “diluting” effect that genetic heterogeneity between many small nuclear pedigrees may have on the global LOD* scores, we repeated our analyses on a subset of larger multigenerational migraine pedigrees. These analyses highlighted multiple separate regions showing evidence for linkage to all three phenotypes. In addition to the locus on 18p11, which remained suggestive of linkage, markers at chromosome 3q also yielded LODs suggestive of linkage in the large pedigree sample. This genomic region has not been previously implicated in migraine and may possibly represent a novel susceptibility region. Several of the other peaks are also of potential importance considering they overlap with regions previously implicated in independent genome scans (Table 2). In particular, we revealed a linkage peak on chromosome 1q23 for this subset of migraine pedigrees. This region harbours the recently discovered FHM gene (ATP1A2) [25]. It is also relatively close to a migraine susceptibility region we have previously identified at 1q31 in a single large pedigree [8]. Therefore, we are currently extending our linkage analyses in this general region and testing the ATP1A2 gene for mutations in migraine probands from our implicated pedigrees. Also warranting further investigation is the linkage peak we identified on chromosome 14q22. This peak happens to be in close proximity (<5 cM) to a locus linked to MO in a large Italian pedigree [12]. In addition, we also detected a linkage peak on chromosome 4q32. This peak is on the same arm as, yet somewhat distinct from (at least 65 cM telomeric), an MA locus at 4q24 and an MO locus on 4q21 [7, 9]. Thus, the chromosome 4q32 region should be considered as independent from the 4q21–24 region previously identified. Another genome scan of 43 Canadian pedigrees segregating MA revealed a significant susceptibility locus for this phenotype on chromosome 11q24 [11]. However, our results do not offer support for these findings. It is intriguing to note that the genome scans of Wessman et al. [7] and Cader et al. [11] both examined a large number of multigenerational pedigrees segregating IHS-defined MA. Both studies also performed two-point parametric linkage analyses based on the apparent autosomal dominant inheritance of the disease in the respective pedigree sets. Despite the design similarities, these two studies revealed different MA loci (both significant at the genome wide level) on chromosomes 4q and 11q, respectively. This discordance in results is perhaps indicative of the genetic heterogeneity between different affected population groups and may partially explain the different findings we present for our Australian migraine genome scan.
Overall, our genome scan provides evidence for severe migraine susceptibility loci on chromosomes 18p11 and 3q-tel. We also identified several other linkage peaks that might be important because they support the findings of previous migraine linkage studies. We conclude that the genetic aetiology of the common forms of migraine is probably comprised of a number of low to moderate effect susceptibility genes, perhaps acting synergistically, and this effect is not easily detected by single-locus linkage analyses of large samples of affected pedigrees. However, further dissection of the migraine phenotype in patients, such as we have done using LCA, may be helpful for identifying susceptibility genes that influence the more heritable clinical (symptom) profiles in affected pedigrees.
References
Headache Classification Committee of the International Headache Society (1988) Classification and diagnostic criteria for headache disorders, cranial neuralgias and facial pain. Cephalalgia 8(Suppl 7):19–28
Russell MB, Iselius L, Olesen J (1996) Migraine without aura and migraine with aura are inherited disorders. Cephalalgia 16(5):305–309
Honkasalo ML, Kaprio J, Winter T, Heikkila K, Sillanpaa M, Koskenvuo M (1995) Migraine and concomitant symptoms among 8167 adult twin pairs. Headache 35:70–78
Larsson B, Bille B, Pedersen NL (1995) Genetic influence in headaches: a Swedish twin study. Headache 35(9):513–519
Nyholt DR, Gillespie NA, Heath AC, Merikangas KR, Duffy DL, Martin NG (2004) Latent class analysis does not support migraine with aura and migraine without aura as separate entities. Genet Epidemiol 26:231–244
Kalfakis N, Panas M, Vassilopoulos D, Malliara-Loulakaki S (1996) Migraine with aura: segregation analysis and heritability estimation. Headache 36(5):320–322
Wessman M, Kallela M, Kaunisto MA, Marttila P, Sobel E, Hartiala J, Oswell G, Leal SM, Papp JC, Hamalainen E, Broas P, Joslyn G, Hovatta I, Hiekkalinna T, Kaprio J, Ott J, Cantor RM, Zwart JA, Ilmavirta M, Havanka H, Farkkila M, Peltonen L, Palotie A (2002) A susceptibility locus for migraine with aura, on chromosome 4q24. Am J Hum Genet 70(3):652–662
Lea RA, Shepherd AG, Curtain RP, Nyholt DR, Quinlan S, Brimage PJ, Griffiths LR (2002) A typical migraine susceptibility region localizes to chromosome 1q31. Neurogenetics 4(1):17–22
Bjornsson A, Gudmundsson G, Gudfinnsson E, Hrafnsdottir M, Benedikz J, Skuladottir S, Kristjansson K, Frigge ML, Kong A, Stefansson K, Gulcher JR (2003) Localization of a gene for migraine without aura to chromosome 4q21. Am J Hum Genet. Sep 25 (epublication)
Carlsson A, Forsgren L, Nylander PO, Hellman U, Forsman-Semb K, Holmgren G, Holmberg D, Holmberg M (2002) Identification of a susceptibility locus for migraine with and without aura on 6p12.2–p21.1. Neurology 59(11):1804–1807
Cader ZM, Noble-Topham S, Dyment DA, Cherny SS, Brown JD, Rice GP, Ebers GC (2003) Significant linkage to migraine with aura on chromosome 11q24. Hum Mol Genet 12(19):2511–2517
Soragna D, Vettori A, Carraro G, Marchioni E, Vazza G, Bellini S, Tupler R, Savoldi F, Mostacciuolo ML (2003) A locus for migraine without aura maps on chromosome 14q21.2–q22.3. Am J Hum Genet 72(1):161–167
Nyholt DR, Lea RA, Goadsby PJ, Brimage PJ, Griffiths LR (1998) Familial typical migraine-linkage to chromosome 19p13 and evidence for genetic heterogeneity. Neurology 50:1428–1432
Jones KW, Ehm MG, Pericak-Vance MA, Haines JL, Boyd PR, Peroutka SJ (2001) Migraine with aura susceptibility locus on chromosome 19p13 is distinct from the familial hemiplegic migraine locus. Genomics 78(3):150–154
Nyholt DR, Curtain RP, Griffiths LR (2000) Familial typical migraine: significant linkage and localization of a gene to Xq24–28. Hum Genet 107(1):18–23
McCarthy LC, Hosford DA, Riley JH, Bird M, White NJ, Hewett DR, Peroutka SJ et al (2001) Single nucleotide polymorphism (SNP) alleles in the insulin receptor (INSR) gene are associated with migraine. Genomics 78(3):135–149
Peroutka SJ, Wilhoit T, Jones K (1997) Clinical susceptibility to migraine with aura is modified by dopamine D2 receptor (DRD2) NcoI alleles. Neurology 49:201–206
Lea R, Dohy A, Jordan K et al (2000) Evidence for allelic association of the dopamine beta-hydroxylase gene (DBH) with susceptibility to typical migraine. Neurogenetics 3(1):35–40
Lea RA, Ovcaric M, Sundholm J, Solyom L, Griffiths LR (2004) The methylenetetrahydrofolate reductase gene variant C677T influences susceptibility to migraine with aura. BMC Med 2:3
Miller SA, Dykes DD, Plensky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16(3):1215
Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2002) Merlin-rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30(1):97–101
Kong A, Cox NJ (1997) Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 61:1179–1188
Nyholt DR (2000) All LODs are not created equal. Am J Hum Genet 67(2):282–288
Ophoff RA, Terwindt GM, Vergouwe MN, Van Eijk R, Oefner PJ, Hoffman SM et al (1996) Familial hemiplegic migraine and episodic ataxia type-2 are caused by mutations in the Ca2+ channel gene CACNL1A4. Cell 87:543–552
De Fusco M, Marconi R, Silvestri L, Atorino L, Rampoldi L, Morgante L, Ballabio A, Aridon P, Casari G (2003) Haploinsufficiency of ATP1A2 encoding the Na+/K+ pump alpha2 subunit associated with familial hemiplegic migraine type 2. Nat Genet 33(2):192–196
Acknowledgements
This research was supported by funding by GlaxoSmithKline and the National Health and Medical Research Council (NHMRC) of Australia. Dr. Rod Lea is supported by an NHMRC research fellowship (CJ Martin). The authors wish to thank Ralph McGinnis and Pam St Jean for statistical computing assistance and the GRC clinical staff and volunteers for aiding with collection and management of blood samples and pedigree data.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lea, R.A., Nyholt, D.R., Curtain, R.P. et al. A genome-wide scan provides evidence for loci influencing a severe heritable form of common migraine. Neurogenetics 6, 67–72 (2005). https://doi.org/10.1007/s10048-005-0215-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10048-005-0215-6