Abstract
We have conducted a series of experiments to explore the surface of the polished pyrolytic graphite ‘edge’ electrode as routinely prepared for use in protein film voltammetry. Our investigations have included nitrogen porosimetry and scanning electron microscopy. The nitrogen adsorption revealed a Brunauer–Emmett–Teller surface area ∼104 times greater than the geometric surface area of the electrode. The pore-size distribution calculated by the Horváth–Kawazoe method showed that 10–18% of the pore volume arises from pores having widths >10 nm and, thus, should be accessible to enzymes, although much of the exposed ‘wall’ surface may be inactive for enzyme binding or electron transfer: for example, it may be mainly basal plane. Scanning electron micrographs of the abraded pyrolytic graphite edge showed differing scales of surface damage caused by the abrasion and the presence of many cracks in the surface where thin platelets had been removed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The ability of the polished edge ‘plane’ of pyrolytic graphite (PG) to adsorb a variety of proteins has made it a very useful electrode surface for electrochemical studies of redox-active proteins, including large enzymes [1]. With relatively easy pre-treatment, the pyrolytic graphite edge (PGE) provides a protein-compatible environment, rich in acidic C–O functionalities, at which many proteins adsorb and display fast electron transfer [2–4]. Adsorption is often dependent on the inclusion of polyions; in particular, the binding of proteins having negatively charged surfaces is greatly enhanced by the presence of polyamines such as neomycin or polymyxin, leading to ‘rational’ models invoking the importance of predictable electrostatic interactions between protein and electrode surface similar to the situation at biological membranes [5]. However, cartoons depicting protein molecules on the electrode surface (the ‘substrate’ in the language of surface science) present a misleading picture of uniformly oriented, featureless, spheroidal objects on a flat surface; the true picture is much more complex. Enzymes are large (typically with radii above 5 nm) and have extremely complex surfaces that are irregular both in terms of shape and distribution of hydrophobic, polar and charged amino-acid sidechains. In real experiments, ‘saturation coverage’ is usually much less than expected (from the crudest of calculations) for a structural monolayer, but provided the enzyme is highly active, this does not prevent useful information from being obtained because the electrochemical signatures of the active sites are amplified often in excess of 1,000-fold [6]. There are several observations common to protein film voltammetry studies on graphite: PGE is characterised by slow charging currents compared to metal electrodes [7], and cyclic voltammograms of adsorbed proteins show (a) non-zero peak separations even at scan rates of 1 mV s−1 [8, 9], (b) peak widths that are often considerably larger than the theoretical values and (c) reduction potentials that vary with scan rate [10]. Observations in this laboratory (unpublished) that some highly active enzymes (hydrogenases) remain adsorbed to the surface even after extensive polishing and rinsing suggest features that penetrate deep into the carbon surface. We have therefore explored the surface of the PGE electrode as it results from preparations routinely used in protein film voltammetry using nitrogen porosimetry and scanning electron microscopy.
Experimental
PG plates measuring 50.8×50.8×3.2 mm (2×2×1/8″, General Electric Quartz Europe) were cut into strips approximately 3 mm wide. The full length was used for analytical nitrogen porosimetry, whereas the strips were cut to shorter lengths for analysis by scanning electron microscopy. The geometric surface areas were calculated from multiple linear measurements using digital callipers with a precision of 0.01 mm. The geometric surface area of the strip that was sandpapered was 4.43 cm2, of which 3.10 cm2 was edge plane. The geometric surface area of the strip polished with alumina was 5.28 cm2, of which 2.88 cm2 was edge plane. Abrasives used for polishing the graphite surface were P800 Tufbak Durite sandpaper (Norton) and 1 μm α-alumina, 0.3 μm α-alumina, and 0.05 μm γ-alumina (Buehler). All water used for rinsing or sonicating the electrodes was purified by reverse osmosis and ion exchange to a resistivity of 18.2 MΩ cm.
A CE Instruments Sorptomatic 1990 was used to acquire nitrogen adsorption and desorption isotherms at 77 K. Rods of PG were abraded, then rinsed in a stream of water. The basal planes were repeatedly treated with Sellotape to remove any loosely bound basal layers. The rods were then sonicated in water for 30 min, changing the water twice during the treatment. The rods were outgassed in vacuum for at least 1 h at 110°C and <0.1 Pa. Isotherms were corrected for instrument hysteresis and dead volume in the burette based on the mass of the outgassed sample and assuming a density of 2.2 g cm−3 for PG. Surface areas were calculated using the method developed by Brunauer, Emmett and Teller [11], both in its linearised, two-parameter form (assuming N=\( \propto \)) and in its non-linear three-parameter form:
where V ads is the volume adsorbed, V m is the monolayer volume, p/p° is the nitrogen vapour pressure relative to its saturation pressure and C and N are the Brunauer–Emmett–Teller (BET) parameters related to adsorption strength and number of monolayers, respectively.
The non-linear form was fit by a least-squares minimisation using the Solver Visual Basic add-in for Microsoft Excel; the values of C and V m from the two-parameter fit were used as seed values for the minimisation algorithm. Relative pressures of p/p°=0.05–0.35 were used for the two-parameter fit and p/p°=0–0.8 were used for the three-parameter fit. The cross-sectional area of a N2 molecule was taken to be 0.162 nm2. The method introduced by Horváth and Kawazoe was used to calculate the pore-size distribution (PSD) [12]. Because this method was originally applied to nitrogen adsorption isotherms determined on molecular sieve carbons which were assumed to have slit-shaped graphitic pores, it was deemed most suitable for analysing the isotherms in this work.
Scanning electron micrographs were recorded on a JEOL JSM 6300 with a LaB6 filament operating at 7 kV. Samples were mounted on 12-mm Al stubs with adhesive conductive carbon discs. The samples were sufficiently conductive so that no metal coating was required to prevent sample charging. The elemental composition was determined using an integrated X-ray energy dispersive spectrometer running Inca software (Oxford Instruments), with a super-atmospheric thin window and a quoted resolution of 138 eV at the Mn K edge.
Results and discussion
Nitrogen adsorption
Typical adsorption and desorption isotherms for PG strips with abraded edge planes are shown in Fig. 1. The sample that was polished with sandpaper (Fig. 1a) shows a type IIa isotherm, according to the International Union of Pure & Applied Chemistry 1985 convention, with no hysteresis [13]. Such isotherms are normally associated with monolayer–multilayer adsorption on an open and stable external surface of a powder, which may be non-porous or macroporous [14]. The adsorption and desorption isotherm of the PG strip polished with 1 μm α-alumina (Fig. 1b) is type IIb with a type-H3 hysteresis loop, and shows an approximately fourfold lower uptake of gas with respect to the sandpapered strip. This hysteresis can be observed in aggregated powders such as clays, but is more generally seen with materials that contain non-rigid, slit-shaped pores. Because of delayed capillary condensation, multilayer adsorption cannot proceed until high relative pressure is achieved. However, when condensation has occurred, the state of the adsorbate has changed, and the desorption follows a different path until the critical relative pressure is reached and the condensate becomes unstable again [14]. Neither adsorption isotherm shows a well-defined ‘point B’, when there is a clear transition from monolayer to multilayer adsorption. The absence of this point is indicative of a low degree of adsorbate–adsorbant interactions and a significant overlap between monolayer and multilayer adsorption.
Both adsorption isotherms fit well to the BET equation, in both its three- and two-parameter forms. The values for the surface area, number of statistical monolayers (N) and interaction parameter (C) are listed in Table 1. Most notable is the huge excess in the BET surface area relative to the geometric surface area. For both the sandpapered and alumina-polished samples, the ratio between the two is on the order of 104. There was approximately 20% variability in the BET surface areas from different experiments, presumably due to subtle differences in polishing.
The BET surface area is in agreement with that reported by Bañares-Muñoz et al., who reported surface areas of ca 4.2 m2 g−1 on PG ground into ca 34-μm particles [15]. In their work, however, they reported a much higher C value, a very clear B point in the adsorption isotherm and a much clearer type-H3 hysteresis. They suggested that their hysteresis may be due to capillary spaces within packets of layers or interparticular spaces.
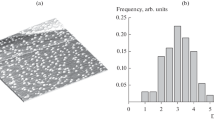
Not all the pores in the material are likely to be accessible to enzyme-size macromolecules. The relative and cumulative PSDs for both samples are shown in Fig. 2. The modal pore size in the case of the sandpapered PG strip is ca 2.7 nm, with about 10% of the pores having widths greater than 10 nm. For alumina-polished PG, the PSD has peaks around 2 nm, with about 18% of the pores having diameters greater than 10 nm. This is large enough to accommodate enzymes such as [NiFe]-hydrogenase, laccase and many others exhibiting strong adsorption and fast electron transfer. However, much of the surface area that is exposed, if it has a primarily basal character, will be unproductive for enzyme catalysis because the electron transfer rate is very low, perhaps zero, and certainly orders of magnitude lower than that of edge plane [16].
Scanning electron microscopy
Scanning electron microscopy provides much insight into the reasons for the large difference between geometric and actual surface area for an ostensibly non-porous material. Figure 3 shows images of sandpapered PGE. At lower magnification (Fig. 3a) the thin, roughly parallel but undulating graphite layers that have been exposed by sandpaper abrasion are visible. The scale of the roughness on the surface is comparable to the ca 25-μm-grit particles that make up the sandpaper. The image at higher magnification shows many places where the layers of graphite have been split or where individual plates of graphite have been removed. The edges of stacks of graphene sheets, typically 100–200 nm thick and around 5–10 μm wide, can be seen pushing out of the surface. Under the best imaging conditions (e.g. Au on C at 30 kV) the lateral resolution of the microscope approaches 6 nm; however, using lower-atomic-number material and lower operating voltages means that any of the smaller mesopores indicated by the porosimetry would be obscured.
A different perspective on the effect of abrading the electrode with sandpaper comes from Fig. 4. The PGE surface shown in these images has only been rinsed with water rather than sonicated and is thus covered with much of the smaller debris from polishing. Many small graphite plates, about 100–300 nm thick and 1–3 μm across, still litter the surface. Larger particles may have been washed away in the water rinse before analysis. This image suggests that sandpaper abrasion dislodges some of the plates, leaving a large number of high-aspect-ratio trenches that are both deep and wide on the scale of the enzymes. In addition, judging solely from the scale of the surface roughness, the comparison between the images in Figs. 3 and 4 suggests that different polishing techniques can give rise to very different surface structures and, presumably, different amounts of enzyme adsorption.
Scanning electron micrographs at a lower and b higher magnification of the edge plane of PG abraded with P800 sandpaper and only rinsed with water. The scale on these images is the same as those in Fig. 3
The PGE surface polished with 1 μm α-alumina (Fig. 5) is relatively smooth in comparison to the sandpapered samples. The magnifications of the images in Fig. 5a, b match those in Figs. 3 and 4. At the lowest magnification (Fig. 5a), the sample is fairly planar, with some trenches from the cutting process remaining on the surface and not polished out. The polishing appears to have splayed apart the layers on the surface, in contrast with the sandpaper abrasion, which left more of the graphite platelets in apparent physical contact. The stacks of graphene sheets on the surface that was polished by 1 μm α-alumina have about the same thickness as those that were abraded with sandpaper, but the average width was smaller, about 2 μm. The highest magnification image (Fig. 5c) shows some of the damage done to the graphite platelets, as evidenced by the number of holes in the surface layer and their ‘fuzzier’ edges. The smaller scale tears in the graphite stacks are 50–100 nm long; the number of edges lacking sharp contrast suggests that there is an even finer-scale roughness that is beyond the resolution of the instrument.
Although the surface area of the alumina-polished sample is lower than that of the sandpapered sample, this additional roughness may contribute additional surface oxo species (such as ketones, carboxylic acids and phenolic groups) that facilitate enzyme binding. The roughness of the surfaces also helps to explain why the densities of the abraded PG (Table 1) are much lower than the theoretical density of PG (2.2 g cm−3): the sample polished to a mirror finish with 1 μm α-alumina has a density about 8% lower and the sandpapered sample is about 30% lower.
Also evident in Fig. 5b are the large number of alumina particles remaining on the surface despite sonicating the sample in water. Metal oxide particles, such as the alumina abrasive, are not only known to adsorb proteins (which could influence the experiments by removing protein from very dilute film-forming solutions) but could also physically block sites on the electrode that are suited for binding more hydrophilic proteins [17–19].
Polishing with progressively finer alumina particles can produce a surface, such as the one shown in Fig. 6, in which the edges of the platelets of graphite are nearly invisible. Some boundaries between groups of sheets are faintly visible in Fig. 6a, b, running from the top left to the bottom right of the image; however, the contrast was too low to get a measurement of their thickness. The size of the particles (50 nm) and the presence of aluminium in the X-ray spectrum suggests that these are residual γ-alumina particles from the last stage of polishing. Despite sonicating in water for 90 s, and changing the water twice during that time, the surface still has about a 40% coverage of alumina (as measured by image analysis).
Scanning electron micrographs of the edge plane of PG, polished with decreasingly smaller diameters of alumina particles down to 0.05 μm γ-alumina, then rinsed in water and sonicated for 90 s. The scale of each image matches those in Fig. 5
The X-ray energy spectrum shown in Fig. 7 reveals a few more characteristics of the edge plane surface. In addition to carbon and oxygen, all of the samples examined show the presence of a similar amount of iron or cobalt (the L lines cannot be discriminated in the spectrum given the detector resolution). Both alumina-polished samples show a significant amount of aluminium in their spectra, which was expected because of the spherical particles visible in the scanning electron micrographs. The sample polished with the 1-μm particles also shows the presence of sodium, magnesium, sodium and chlorine, which may be left from the abrasive particle synthesis or may have been added to adjust its colloidal interactions. In contrast, the sample abraded with sandpaper showed no silicon in the X-ray spectrum, and no residual particles were visible in the scanning electron micrographs.
Conclusions
These measurements have interesting implications for both general studies involving protein film voltammetry and technological applications. The results show that PGE, the polished edge plane of PG (the ‘substrate’ as understood by surface scientists), actually has a larger and more easily accessible surface than many porous materials with a largely internal surface area. Thus, a N2 molecule ‘sees’, on PGE, an area that is about 104 times the area given by the ‘ruler’. Much of the additional surface that is revealed in these studies will be inaccessible to typical enzyme molecules with minimum diameters >5 nm because it forms the walls of deep narrow trenches (<10 nm wide). Taking this into account, the results suggest that the maximum coverage that one can achieve on PGE electrodes is still potentially one to two orders of magnitude higher than the normal expectation, i.e. a maximum of approximately 10 pmol cm−2 for a typical electrode surface that is assumed to be planar. The physically accessible fraction of the electrode surface will most likely have a primarily unreactive basal character and be unproductive for protein film voltammetry, either because enzyme molecules do not bind to areas of such extensive hydrophobicity without unfolding or because electron transfer is too slow. However, with appropriate surface modifications, much more of the electrode surface area may be made accessible for productive enzyme adsorption and binding, greatly improving the signal in electrochemical studies and power output in technological applications.
References
Léger C, Elliott SJ, Hoke KR, Jeuken LJC, Jones AK, Armstrong FA (2003) Biochemistry 42:8653–8662
Boehm HP (2001) In: Delhaès P (ed) World of carbon. Gordon and Breach Science, Amsterdam, pp 141–178
Schloegl R, Boehm HP (1983) Carbon 21:345–358
Takahagi T, Ishitani A (1984) Carbon 22:43–46
Barber J (1982) Annu Rev Plant Physiol 33:261–295
Armstrong FA, Bond AM, Buchi FN, Hamnett A, Hill HAO, Lannon AM, Lettington OC, Zoski CG (1993) Analyst 118:973–978
Jeuken LJC, Armstrong FA (2001) J Phys Chem B 105:5271–5282
Armstrong FA, Camba R, Heering HA, Hirst J, Jeuken LJC, Jones AK, Léger C, McEvoy JP (2000) Faraday Discuss 116:191–203
Hirst J, Armstrong FA (1998) Anal Chem 70:5062–5071
Léger C, Jones AK, Albracht SPJ, Armstrong FA (2002) J Phys Chem B 106:13058–13063
Brunauer S, Emmett PH, Teller E (1938) J Am Chem Soc 60:309–319
Horváth G, Kawazoe K (1983) J Chem Eng Jpn 16:470–475
Sing KSW, Everett DH, Haul RAW, Moscou L, Pierotti RA, Rouquérol J, Siemieniewska T (1985) Pure Appl Chem 57:603–619
Rouquérol F, Rouquérol J, Sing KSW (1999) Adsorption by powders and porous solids: principles, methodology and applications. Academic, San Diego
Bañares-Muñoz MA, Llorente JMM, Gonzalez LVF (1988) Carbon 26:681–685
Davies TJ, Hyde ME, Compton RG (2005) Angew Chem Int Ed Engl 44:5121–5126
Kondo A, Mihara J (1996) J Colloid Interface Sci 177:214–221
Norde W (1986) Adv Colloid Interface Sci 25:267–340
Rezwan K, Studart AR, Voros J, Gauckler LJ (2005) J Phys Chem B 109:14469–14474
Acknowledgements
The authors wish to acknowledge Mike Jenkins for the use of the scanning electron microscope. This research was supported by grants from the Biotechnology and Biological Sciences Research Council (43/B19096) and the Leverhulme Trust (F/08 699/C).
Author information
Authors and Affiliations
Corresponding author
Additional information
This work is dedicated to Prof. Alan Bond on the occasion of his 60th birthday. Alan’s enthusiasm for the complexities of diffusion control persuaded one of us (F.A. Armstrong) to try and avoid it altogether in protein electrochemical studies.
Rights and permissions
About this article
Cite this article
Blanford, C.F., Armstrong, F.A. The pyrolytic graphite surface as an enzyme substrate: microscopic and spectroscopic studies. J Solid State Electrochem 10, 826–832 (2006). https://doi.org/10.1007/s10008-006-0183-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10008-006-0183-2