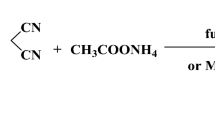Abstract
Dihydropyridines are the most extensively used drugs in the treatment of hypertension. Nifedipine is the prototype of calcium channel blocker. The dihydropyridine derivative compounds of diethyl 4-(4-bromophenyl)-2,6-dimethyl-1,4-dihydropyridine-3,5-dicarboxylate (DHPB), diethyl 4-(furan-2yl)-2,6-dimethyl-1,4-dihydropyridine-3,5-dicarboxylate (DHPF), and diethyl-4-phenyl-2,6-dimethyl-1,4-dihydropyridine-3,5-dicarboxylate (DHPP) were synthesized using the Hantzsch reaction. The DFT/B3LYP exchange–correlation function was employed to perform quantum chemical calculations such as molecular geometry optimization, vibrational analysis, frontier molecular orbital (FMO), molecular electrostatic potential (MEP), natural bond order (NBO), global reactive descriptors, and Fukui functions to determine the structural characteristics related to biological activity of the compounds. The molecular docking and molecular dynamics were employed to study the binding interaction and stability of protein–ligand complex in the docked site.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
1,4-Dihydropyridines are essential class of compound in the field of drugs and pharmaceuticals, which exhibit a wide range of biological activities such as antitumor [1], anti-inflammatory [2], anti-anginal [3,4,5], calcium channel antagonists [6], anti-tubercular [7], antithrombotic [8], antihypertensive [3,4,5], anticonvulsant [9], and cardio depressant [10]. Dihydropyridines are widely known drugs in the treatment of hypertension and cardiovascular diseases as effective calcium channel blockers (CCB) [11]. The ion channel which controls selective permeability of calcium ions enters the cell membrane and is also called the voltage-gated calcium channel. The voltage-gated calcium channels are different types such as L-type, P-type, N-type, R-type, and T-type; each type has different functions [12]. The L-type calcium channel is responsible for the excitation–contraction coupling of cardiac and skeletal smooth muscle. The L-type calcium channel has four subunits cav1.1, cav1.2. cav1.3. and cav1.4. The cav1.2 is a protein in humans; this protein binds and inhibited by dihydropyridine. The calcium channel blocker molecules interact with the calcium channels and reduce the influx of calcium ions entering the heart and the vessels, thereby lowering heart rate. The most commonly used drugs are amlodipine, nifedipine, isradipine, felodipine, nicardipine, nimodipine, nitrendipine, and lacidipine, which have been found as potent cardiovascular agents for the treatment of hypertension. Despite the fact that the Food and Drug Administration (FDA) approved numerous medications for calcium channel blockers, there is a necessity for the discovery of new drugs that should be safer for managing high blood pressure, chest pain, and heartbeat irregularities, it should be better than the existing drugs. Structural information of the compound is prerequisite in drug designing which reduce the cost and time. In this present study, the 1,4-dihydropyridine compounds DHPB, DHPF, and DHPP were synthesized by the classical Hantzsch route, and their spectral characteristics FT-IR, FT-Raman, UV–Vis, and NMR were explored through experimental and theoretical (DFT) methods. The biological activity of the compound was investigated using molecular docking and molecular dynamics simulations to predict the possible binding site of the compound in the targeted protein and stability of the compound in the active site.
Materials and methods
4-Dihydropyridine synthesize procedure
For the synthesis of compound DHPB from the reported Hantzsch reaction [13], 1.2 mmol of ethanolic solution of ammonium acetate, 1 mmol of 4-bromobenzaldehyde, 2 mmol of ethyl acetoacetate, and 100 mg of mesoporous acid catalyst AlFeTUD-1 (preheated at 80 ºC for 3 h) were taken in a 25-ml round bottom-flask fitted with reflux condenser. Then the flask was immersed in an oil bath, and the reaction was carried out under reflux condition. After completion of the reaction, the reaction mixture was quenched with crushed ice and stirred. The crude product was filtered and washed with distilled water followed by treatment with brine solution and mixed with ethyl acetate to separate the compound and dried over anhydrous Na2SO4.The crude mixture was dissolved in hot ethanol to separate the solid catalyst, and the crude product was further purified by recrystallization from ethanol. The scheme of the reaction is shown in Fig. 1. The rest of the compounds, DHPF and DHPP, were synthesized via the above route using 2-furaldehyde and benzaldehyde.
Experimental characterization details
All the chemicals were purchased from Sigma-Aldrich and Merck products without any further purification except 2-furaldehyde.The FT-IR spectra of the synthesized compounds were recorded within the mid-IR region 4000–400 cm−1 using the KBr pellet technique on a Perkin Elmer FT-IR spectrometer. The FT-Raman spectra of the compounds were recorded using Bruker RFS27 in the region of 4000–50 cm−1 using an Nd:YAG laser source of 1064 nm. The UV–Visible absorption spectra of the synthesized compounds were recorded in the range of 190–900 nm with ethanol as the solvent using a Perkin Elmer Lambda 35 UV–Vis spectrometer.
Computational details
All the DFT calculations were performed using the Gaussian09 software program suit and Gauss view as a visualization program [14, 15]. The optimization and vibrational wavenumbers were predicted using the B3LYP/6–311 + G(d,p) basis set. The potential energy distribution (PED) was employed to identify the vibrational assignments quantitatively using the Veda4X program [16]. The time-dependent density functional theory (TD-DFT) was adopted to compute the electronic properties such as the UV absorption, the highest occupied molecular orbital (HOMO), and the lowest unoccupied molecular orbital (LUMO) energy gap. The natural bond analysis (NBO) was carried out with the Gaussian NBO version 3.1 program [17] which is implemented in the Gaussian09 software program suit. For the compounds DHPB, DHPF, and DHPP, the molecular docking calculations were executed with the Autodock4 Tools version 1.5.6 program [18], and the docking results were analyzed using Pymol [19]. Molecular dynamics simulations were done with the DESMOND program suit [20].
Results and discussion
Molecular geometry
The optimized molecular structures of DHPB, DHPF, and DHPP are shown in Fig. 2 with their atom numbering schemes. The title compound belongs to the C1 point group symmetry, and their minimized energy is listed in Table 1. The optimized parameters like bond length, bond angle, and torsional angles of DHPB, DHPF, and DHPP are listed in supplementary Table S1. In the present study, the molecular structure of ethyl, methyl, carboxylate groups, and pyridine ring are common to DHPB, DHPF, and DHPP compounds. Only the bromophenyl, furan, and phenyl substituted to the fourth position of the 1,4-dihydropyridine ring vary. The single-crystal XRD analysis of DHPB has been reported by Boulcina R et al. [21]. From this report, the bond length, bond angle, and dihedral angle are compared with theoretically calculated values, which are in good agreement with each other. The dihedral angle of 4-bromophenyl C21-C22-C23-Br1 and C25-C24-C23-Br1 are 180° conformal to the planar structure of 4-bromophenyl, which is consistent with the experimental data. The calculated bond length of DHPF are C–C, C = C, C-O, C = O, C-H, C-N, and N–H and are almost similar to the experimental XRD value [21, 22]. The bond angles of DHPF, C13-O17-C16, C14-C15-C16, and C15-C16-O17, are 107.28, 106.15, and 110.30 Å, respectively, which is similar to the literature value. In DHPP, the bond length of C–C, C = C, C-O, C = O, C-H, C-N, and N–H are compared with the experimental XRD value reported by M.S Bai et al. [23]. The bond angles of the phenyl ring C13-C14-C16, C14-C16-C18, and C20-C22-C13 of DHPP are almost 120° and the dihedral angles of C13-C14-C16-C18 and C13-C22-C20-C18 of DHPP are at zero degree. Thus the experimental and theoretical values are in good concurrence with one another.
Vibrational assignments
The molecules DHPB, DHPF, and DHPP exhibit a total number of 47, 44, and 47 atoms and have 135, 126, and 135 normal modes of vibration. The experimental FT-IR and FT-Raman spectra vibrational frequencies along with their potential energy distribution (PED) for each normal modes of vibrations of DHPB, DHPF, and DHPP compounds are summarized in supplementary Table S2.1–2.3. The experimental FT-IR and FT-Raman spectra of DHPB, DHPF, and DHPP compounds were contrasted with the stimulated spectra as shown in Fig. 3a–c. The C-H stretching vibrations of saturated hydrocarbons are mostly observed in the range 3000–2840 cm−1 [24]. In FT-IR spectra, the compounds DHPB, DHPF, and DHPP were found at 2979, 2978, and 2978 cm−1, respectively, which is nearly identical to the computed values. The strong N–H stretching vibrations of DHPB, DHPF, and DHPP were found at 3352, 3340, and 3333 cm−1 in FT-IR spectra [25]. The aromatic C-H stretching vibration of FT-IR and FT-Raman bands is usually observed in the region of 3100–3000 cm−1 [26]. The vibrational band observed at 3085, 3092, and 3084 cm−1 in FT-IR and 3058, 3123, and 3059 cm−1 in FT-Raman was attributed to the aromatic C-H stretching vibrations accordingly. The compound DHPF is slightly higher than the usual range, which is due to the highly electronegative oxygen atom present in the furan ring. The DHPB, DHPF, and DHPP compounds have carboxylic group C = O stretching vibrations in FT-IR spectra at 1691, 1697, and 1685 cm−1, respectively [27]. The calculated frequencies are slightly higher than the observed values due to the anharmonicity and overestimation of force constants, and these values can be reduced by the scaling factor of 0.966, since theoretical calculations were done in the isolated system [28, 29].
UV–visible analysis
UV–Visible spectral analysis has been investigated by experimental as well as theoretical techniques. The electronic transition properties such as absorption wavelength (λ), excitation energy (E), and oscillator strength (f) were theoretically calculated using TD-DFT/B3LYP/6–311 + G(d,p) basis level with ethanol as a solvent using the IEFPCM method for the compounds DHPB, DHPF, and DHPP which were reported in Table 2. From Table 2, the experimental UV–Vis spectra absorption maximum (λmax) values for the compounds DHPB, DHPF, and DHPP are observed at 365, 342, and 350 nm, and their corresponding theoretical values are 350, 345, and 345 nm, respectively. The compounds show n-π* transitions, which reveals that lone pair electrons have a tendency to donate electrons and also interacts with the protein molecules and enhance the biological activity [30, 31]. All other possible transition details are explained in NBO analysis.
NMR spectral studies
NMR analysis is usually used in chemistry and materials to enhance the information regarding the structure. Utilizing the chemical shift values, it is easier to find the corresponding functional group. The gauge independent atomic orbital (GIAO) has been implemented for 1H NMR chemical shift calculations using the B3LYP/6–311 + + G(2d,p) basis set with TMS as the reference material. The GIAO procedure reveals a quick convergence of the theoretical properties upon extension [32]. The experiment and theoretical proton chemical shifts of compounds DHPB, DHPF, and DHPP are listed in supplementary Table S3.
Global reactive descriptors
The global reactivity descriptors such as ionization potential (I), electron affinity (A), electrophilicity (ω), chemical potential (µ), electronegativity (χ), hardness (Ƞ), and softness (S) of the molecules are calculated from their HOMO and LUMO energy values using the basis set DFT/B3LYP/6-311G + (d,p) and are listed in Table 3. According to Koopmans’ theory of closed shells [33], the negative of HOMO is the ionization potential, and the negative of LUMO is the electron affinity. High chemical reactivity and low kinetic stability have a smaller energy gap (ΔE), and it is a soft molecule, while low chemical reactivity and high kinetic stability have a larger energy gap and vice versa [34]. From Fig. 4, the HOMO orbitals of the compounds DHPB, DHPF, and DHPP are mainly localized on the pyridine ring and carbonyl group, while the LUMO orbitals cover the entire region of the molecules. From Table 3, the compounds have higher ionization energy than the electron affinity. It is clear that compounds have a tendency to accept electrons and exhibit nucleophile behavior; hence, these compounds may be expected to interact with the targeted protein molecules [35]. While comparing the compounds, DHPF has higher ionization potential, electron affinity, electrophilicity, electronegativity, hardness, and softness than DHPB and DHPP.
MEP
Molecular electrostatic potential (MEP) is used to study the net electrostatic effect produced at a point in space by the total charge distribution over the molecule [36]. It helps to predict the reactivity of molecules towards electrophilic and nucleophilic reactions and also to study drug–receptor interactions, while the protein and ligand interactions are mainly formed through their potentials. The different colors on the surface of the molecule indicate their electrostatic potential [37, 38]. The potential is in increasing order of red < orange < green < blue, the higher potential red color indicates the abundance of electrons, and the blue color indicates the absence of electrons in that region, while the green color indicates zero potential or neutral. The MEP map of DHPB, DHPF, and DHPP is generated using the basis set B3LYP/6–311 + G(d,p) and were presented in Fig. 5a–c. From Fig. 5, it is obvious that the deep red color around the oxygen atoms represents an abundance of electrons (nucleophilic) and the blue color around the hydrogen atoms represents electron deficiency (electrophilic), which is similar in all the three compounds. From these results, the compounds are more prone to nucleophilic attack. The detailed reactivity sites of electrophilic and nucleophilic sites are discussed in the Fukui function.
NBO analysis
NBO is a useful tool to study the factors such as inter- and intra-molecular hydrogen bonding, intermolecular charge transfer (ICT), electron density transfer, and hyper-conjugative interactions in the molecular system. The NBO calculations were performed with B3LYP/6-311G + (d,p) basis level for DHPB, DHPF, and DHPP. The second-order perturbation theory of the Fock matrix was carried out to evaluate the donor–acceptor interactions present in the compounds along with hyper-conjugative interactions E(2), which is given by Eq. (1) as follows:
where qi is the donor orbital occupancy, εj and εi are diagonal elements, and Fij is the off-diagonal NBO Fock matrix element. The intra-molecular interaction occurred by orbital overlap which results in intra-molecular charge transfer causing the stabilization of the system; the higher E(2) value indicates more stable interaction between the promoter and acceptor and hence the greater the extent of conjugation of the entire system [39]. From Table 4 part DHPB, the sigma bond σ(C25-H47) to anti-sigma bond σ*(C25-H47) shows the strongest stabilization energy of 39.72 kJmol−1, and other interactions such as π(C20-C21) to π*(C22-C23), π (C11-C12) to π*(O4-C14), and π(C8-C9) to π*(O2-C17) and their corresponding stabilization energies are 22.99 kJmol−1, 22.34 kJmol−1, and 21.94 kJmol−1, respectively. For DHPF, the strongest stabilization energy of 31.63 kJmol−1 was observed for σ(C23-H42) to σ*(C19-H37), where as in DHPP, the strongest stabilization energy shows for π(C11-C12) to π*(C11-C12) is 22.1 kJmol−1 and where summarized in Table 4 parts DHPF and DHPP, respectively. From the NBO analysis, it revealed that there is charge transfer occurring within the molecule. It is evident that the compounds have bioactivity in nature and are suitable for pharmaceutical applications [40].
Fukui function
Fukui function (or) local reactive descriptors provides information about the reactivity site within a molecule by adding or removing a certain amount of charge to the molecular system. It helps to predict where the maximum electrophilic and nucleophilic sites on the molecule and also used to analyze chemical reactivity and site selectivity in organic reactions [41]. Here, we calculated the local reactive descriptors such as Fukui function, local softness, and local electrophilicity using DFT/B3LYP/6–311 + G(d,p) basis set from their Mulliken atomic charges, and their corresponding values are listed in supplementary Tables S4.1, S4.2, and S4.3. Nucleophilic attacks are measured by adding an electron to the electron density, while electrophilic attacks are measured by removing an electron from the electron density. The Fukui function on the kth atomic site is described by Eqs. (2), (3), and (4), where q is the Mulliken atomic charge at the kth atomic site and where N, N – 1,and N + 1 denote the total number of electrons existing in the neutral, anion, and cation states of the molecules, respectively. Local softness Sk+, Sk−, and Sk0 as well as local electrophilicity indices ωk+, ωk−, and ωk0 are also used to define the local reactivity of atoms in molecules, where + , − , and 0 signs represent the nucleophilic, electrophilic, and radial attack, respectively [42, 43]:
The predicted reactivity upon nucleophilic attack f(k+) for the compounds DHPB, DHPF, and DHPP is in increasing order of Br > C11 > C9 > O2, C1 > C5 > N3 > O20, and C8 > C11 > C22 > N1, respectively. It shows that there is a stronger possibility of electron transfer from the electrophilicity reactivate sites to these atoms. On the other hand, for electrophilic attack f(k−), it was observed that C11 > C9 > O4 > O2, C5 > C1 > O20 > 021, and C8 > C11 > O6 > O4 sites are suitable for nucleophilic reactivate sites to these atoms. From the Fukui function calculation, the DHPB, DHPF, and DHPP show higher value for f(k+) nucleophilic attack, so all these molecules are more prone to electrophilic substitution reactive sites.
Molecular docking
Molecular docking is a key tool to identify the protein–ligand interaction theoretically [44]. Based on the structure of compounds, their biological activities can be predicted. Before docking, the evaluation of physicochemical properties and bioactivity score of the synthesized compound is of utmost importance, which reduces the time and cost by using pre-optimized fragments. The bioactivity score and physicochemical properties were predicted with the online tool Molinspiration given in supplementary Table S5 and S6 [45]. The mechanism of the calcium channel membrane is to control the passage of solutes such as small molecules and ions through biological cell membranes. In the present study, the flow of calcium ions through the cell membrane was inhibited by the calcium channel antagonist. These compounds reduce the influx of calcium ions entering the cell membrane. All three compounds are suitable for L-type calcium channel blocker which was taken as target protein [46]. The target protein structure in complex with amlodipine (PDB:5KMD) was obtained from X-ray crystallography data reported by Tang Lin et al. [47]. The preparation of the protein was followed by removing water molecules, co-factors, and co-crystallized ligands in the protein, later polar hydrogen, and kollman charges were added to the prepared protein using the Autodock4 program suite; the Lamarckian genetic algorithm (LGA) was used to evaluate the binding sites. The energy minimized compounds of DHPB, DHPF, and DHPP obtained from the Gaussian B3LYP/6-311G + (d,p) level were designated as ligand. The complete ligand profile of the compounds such as rotational bonds, torsion angles, and RMSF within the docked site is shown in supplementary Fig. S1a–c. After completing the docking procedures, the docking parameters’ binding energy and inhibitory constants of DHPB, DHPF, and DHPP from the 100 conformers the best conformer depend upon the energy and interaction with the protein and were taken for further analysis which is listed in Table 5. The protein ligand interactions were visualized via PyMOL software is presented in Figs. 6a–c. Figure 7 shows the ligands DHPB, DHPF, and DHPP bind to the active site of the targeted protein (5KMD). In order to compare the docked results, the similar 1,4-dihydropyridine derivative compounds such as amlodipine, nifedipine, and felodipine were taken as ligands and docked with the same protein (5KMD), and their binding energies are presented in Table 6. The result shows that the binding energy − 4.71, − 5.26, and − 4.75 kcal/mole for the compounds DHPB, DHPF, and DHPP has higher binging energy than felodipine.
Molecular dynamics
From the results of molecular docking, the best docked protein–ligand complex has been taken for molecular dynamics simulation studies. Molecular dynamics (MD) was performed using DESMOD software to analyze the effect of macromolecular motion on protein–ligand interactions [48]. The solvated system was neutralized by adding counter ions, and the system was relaxed through an energy minimization process. The system was considered as an isothermal isobaric NPT ensemble. A simple point charge was used for the solvent system, and other parameters such as temperature 300 K, pressure 1.0322 bar, and cut-off radius of 10 Å are included in the simulation calculation. From the results, RMSD and RMSF are evaluated, and also ligand–protein interactions with amino acid residues throughout simulations are shown in supplementary Fig. S2a–c.
Root mean square deviation (RMSD)
RMSD is most commonly used to analyze the equilibration period of MD trajectories. It provides insight into the conformational stability of the protein–ligand complex and their motions during simulation within the hydrated lipid atmosphere. Figure 8a–c shows the RMSD analysis of the protein–ligand complex of the DHPB, DHPF, and DHPP compounds for the period of 40 ns. From the MD trajectory figure, the complexes DHPB, DHPF, and DHPP reach equilibration at around 10 ns, 15 ns, and 9 ns, respectively. After the 15 ns from their initial fluctuations, all the compounds became relatively stable and have less deviation throughout the simulation of 40 ns. While comparing the RMSD simulation results, the compound DHPP is more stable than DHPB and DHPF.
Root mean square fluctuation
The RMSF was analyzed to retrieve the flexibility of the protein backbone structure and motion of the individual amino acid residues. The high RMSF value indicates more flexibility, whereas the lower value represents more stability with limited movements under appropriate conditions [49]. The RMSF value of the protein–ligand complex of DHPB, DHPF, and DHPP evaluated from MD simulation result is shown in Fig. 9a–c. The residues in the position 80–100 fluctuate high, while the other residues in protein remain stable with limited fluctuation. The green color vertical lines indicate the ligand interaction with residues. It is clear from the figure that the compound DHPP has less fluctuation and more stable than DHPB and DHPF.
Conclusion
The synthesized compounds DHPB, DHPF, and DHPP were studied in detail with FT-IR, FT-Raman, NMR, and UV–Visible spectra experimentally, and the results were compared with theoretically simulated FT-IR, FT-Raman, NMR, and UV–Visible spectral data which are in good agreement with each other. The quantum chemical studies such as electron affinity, ionization potential, chemical potential, hardness, and softness of the material results prefer high ionization potential. The MEP map predicts reactive sites for nucleophilic and electrophilic attacks, and the Fukui function results infer that the compounds were prone to nucleophilic reactivity sites. The frontier molecular orbital HOMO–LUMO energy gap confirms electronic property and charge transfer within the molecules and also enhances the biological activity of the compounds. The NBO investigation ensures the intra-molecular charge transfer (ICT) appears within the molecules. The results obtained from molecular docking and molecular dynamics show that the compound has good binding affinity and low inhibition value towards the calcium channel activity receptor. Therefore, these compounds may be treated as calcium channel antagonist drugs. However, necessary biological tests and clinical trials should be conducted.
Data availability
The data used in this study are available from the corresponding author upon interest.
References
Mohamed MF, Ibrahim NS, Elwahy AHM, Abdelhamid IA (2018) Molecular studies on novel antitumor bis 1,4-dihydropyridine derivatives against lung carcinoma and their limited side effects on normal melanocytes. Anti-Cancer Agents Med Chem (Formerly Curr Med Chem Agents) 18:2156–2168
Briukhanov VM, Zverev I, Elkin VI (1994) The effect of calcium antagonists on the development of inflammatory edema in rats. Eksp Klin Farmakol 57:47–49
Loev B, Goodman MM, Snader KM et al (1974) Hantzsch-type dihydropyridine hypotensive agents. J Med Chem 17:956–965
Bossert F, Meyer H, Wehinger E (1981) 4-Aryldihydropyridines, a new class of highly active calcium antagonists. Angew Chemie Int Ed English 20:762–769
Breitenbucher JG, Figliozzi G (2000) Solid-phase synthesis of 4-aryl-1, 4-dihydropyridines via the Hantzsch three component condensation. Tetrahedron Lett 41:4311–4315
Triggle DJ (2007) Calcium channel antagonists: clinical uses—past, present and future. Biochem Pharmacol 74:1–9
Wächter GA, Davis MC, Martin AR, Franzblau SG (1998) Antimycobacterial activity of substituted isosteres of pyridine-and pyrazinecarboxylic acids. J Med Chem 41:2436–2438
Sunkel CE, Fau de Casa-Juana M, Santos L et al (1990) 4-Alkyl-1, 4-dihydropyridine derivatives as specific PAF-acether antagonists. J Med Chem 33:3205–3210
Tusell JM, Barro S, Serratosa J (1993) Anticonvulsant activity of δ-HCH, calcium channel blockers and calmodulin antagonists in seizures induced by lindane and other convulsant drugs. Brain Res 622:99–104
Budriesi R, Ioan P, Locatelli A et al (2008) Imidazo[2,1-b]thiazole system: a scaffold endowing dihydropyridines with selective cardiodepressant activity. J Med Chem 51:1592–1600
Xu L, Li D, Tao L et al (2016) Binding mechanisms of 1,4-dihydropyridine derivatives to L-type calcium channel Ca v 1.2: a molecular modeling study. Mol Biosyst 12:379–390
Birnbaumer L, Campbell KP, Catterall WA et al (1994) The naming of voltage-gated calcium channels. Neuron 13:505–506
Hantzsch A (1882) Ueber die synthese pyridinartiger verbindungen aus acetessigäther und aldehydammoniak. Justus Liebigs Ann Chem 215:1–82
Gaussian09 RA (2009) 1, mj frisch, gw trucks, hb schlegel, ge scuseria, ma robb, jr cheeseman, g. Scalmani, v. Barone, b. Mennucci, ga petersson et al., gaussian. Inc, Wallingford CT 121:150–166
Dennington RD, Keith TA, Millam JM (2008) GaussView 5.0. Gaussian, Inc, Wallingford
Jamroz MH (2004) Vibrational energy distribution analysis VEDA 4
Weinhold F, Landis CR (2001) Natural bond orbitals and extensions of localized bonding concepts. Chem Educ Res Pract 2:91–104
Morris GM, Huey R, Lindstrom W et al (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791
Schrodinger LLC (2010) The PyMOL molecular graphics system. Version 1:0
Schrodinger LLC (2019) Schrodinger Release 2019–3: Desmond Molecular Dynamics System. DE Shaw Research. Maestro-Desmond Interoperability Tools, Schrodinger, LLC, New York
Boulcina R, Bouacida S, Roisnel T, Debache A (2007) Diethyl 4-(4-bromophenyl)-2, 6-dimethyl-1, 4-dihydropyridine-3, 5-dicarboxylate. Acta Crystallogr Sect E Struct Reports Online 63:o3635–o3636
Sagar MB, Ravikumar K, Sadanandam YS (1999) Crystal and molecular structure of 2, 6-dimethyl-3, 5-di [N-methyl]-carbamoyl-4-[3, 4-methoxy] phenyl-1, 4-dihydropyridine hemihydrate and 2, 6-dimethyl-3, 5-di [N-methyl] carbamoyl-4-[furan]-1, 4-dihydropyridine
Bai M-S, Chen Y-Y, Niu D-L, Peng L (2009) Diethyl 2, 6-dimethyl-4-phenyl-1, 4-dihydropyridine-3, 5-dicarboxylate. Acta Crystallogr Sect E Struct Reports Online 65:o799–o799
Kavitha T, Velraj G (2016) Structural, spectroscopic (FT-IR, FT-Raman, NMR) and computational analysis (DOS, NBO, Fukui) of 3, 5-dimethylisoxazole and 4-(chloromethyl)-3, 5-dimethylisoxazole: a DFT study. J Theor Comput Chem 15:1650039
Chung TW, Tureček F (2008) Electronic properties of charge-tagged peptides upon electron capture. Eur J Mass Spectrom 14:367–378
Scott AP, Radom L (1996) Harmonic vibrational frequencies: an evaluation of Hartree−Fock, Møller−Plesset, quadratic configuration interaction, density functional theory, and semiempirical scale factors. J Phys Chem 100:16502–16513
Kalsi PS (2007) Spectroscopy of organic compounds. New Age International
Govindasamy P, Gunasekaran S (2015) Quantum mechanical calculations and spectroscopic (FT-IR, FT-Raman and UV) investigations, molecular orbital, NLO, NBO, NLMO and MESP analysis of 4-[5-(4-methylphenyl)-3-(trifluoromethyl)-1H-pyrazol-1-yl] benzene-1-sulfonamide. J Mol Struct 1081:96–109
Johnson RD (2006) NIST computational chemistry comparison and benchmark database. http://srdata.nist.gov/cccbdb. Accessed 15 Mar 2021
Turan N, Buldurun K, Alan Y et al (2019) Synthesis, characterization, antioxidant, antimicrobial and DNA binding properties of ruthenium(II), cobalt(II) and nickel(II) complexes of Schiff base containing o-vanillin. Res Chem Intermed 45:3525–3540. https://doi.org/10.1007/s11164-019-03806-3
Chaitanya K (2012) Molecular structure, vibrational spectroscopic (FT-IR, FT-Raman), UV–vis spectra, first order hyperpolarizability, NBO analysis, HOMO and LUMO analysis, thermodynamic properties of benzophenone 2, 4-dicarboxylic acid by ab initio HF and density functional. Spectrochim Acta Part A Mol Biomol Spectrosc 86:159–173
Uthayakumar M, Jeyakumari AP, Dhandapani A, et al (2019) Synthesis, experimental and computational spectroscopic investigations of third-order nonlinear optical material (E)-N′-(benzo[d][1,3]dioxol-5-ylmethylene)benzohydrazide. J Phys D Appl Phys 52: https://doi.org/10.1088/1361-6463/ab284b
Koopmans T (1934) Über die Zuordnung von Wellenfunktionen und Eigenwerten zu den einzelnen Elektronen eines Atoms. Physica 1:104–113
Sridevi C, Shanthi G, Velraj G (2012) Structural, vibrational, electronic, NMR and reactivity analyses of 2-amino-4H-chromene-3-carbonitrile (ACC) by ab initio HF and DFT calculations. Spectrochim Acta Part A Mol Biomol Spectrosc 89:46–54
Lakshmi A, Balachandran V, Janaki A (2011) Comparative vibrational spectroscopic studies, HOMO–LUMO and NBO analysis of 5,7-dibromo-8-hydroxyquinoline and 5,7-dichloro-8-hydroxyquinoline based on Density Functional Theory. J Mol Struct 1004:51–66
Thul P, Gupta VP, Ram VJ, Tandon P (2010) Structural and spectroscopic studies on 2-pyranones. Spectrochim Acta Part A Mol Biomol Spectrosc 75:251–260
Scrocco E, Tomasi J (1978) Electronic molecular structure, reactivity and intermolecular forces: an euristic interpretation by means of electrostatic molecular potentials. In: Advances in quantum chemistry. Elsevier, 115–193
Luque FJ, López JM, Orozco M (2000) Perspective on “Electrostatic interactions of a solute with a continuum. A direct utilization of ab initio molecular potentials for the prevision of solvent effects.” Theor Chem Acc 103:343–345
James C, Ravikumar C, Sundius T et al (2008) FT-Raman and FTIR spectra, normal coordinate analysis and ab initio computations of (2-methylphenoxy) acetic acid dimer. Vib Spectrosc 47:10–20
Kavitha T, Velraj G (2017) Density functional theory analysis and molecular docking evaluation of 1-(2, 5-dichloro-4-sulfophenyl)-3-methyl-5-pyrazolone as COX2 inhibitor against inflammatory diseases. J Mol Struct. https://doi.org/10.1016/j.molstruc.2017.03.061
Fleming I (1977) Frontier orbitals and organic chemical reactions. Wiley
Parr RG (1980) Density functional theory of atoms and molecules. In: Horizons of quantum chemistry. Springer, pp 5–15
Lehr GF, Lawler RG (1984) Quantitative CIDNP evidence for the SH2 reaction of alkyl radicals with Grignard reagents. Implication to the iron catalyzed Kharasch reaction. J Am Chem Soc 106:4048–4049
Jorgensen WL (2004) The many roles of computation in drug discovery. Science (80-) 303:1813–1818
Molinspiration C (2011) Calculation of molecular properties and bioactivity score. http://www.molinspiration.com/cgi-bin/properties. Accessed 18 Mar 2021
Daina A, Michielin O, Zoete V (2019) SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res 47:W357–W364. https://doi.org/10.1093/nar/gkz382
Tang L, El-Din TMG, Swanson TM et al (2016) Structural basis for inhibition of a voltage-gated Ca 2+ channel by Ca 2+ antagonist drugs. Nature 537:117–121
Sebhaoui J, El Bakri Y, Lai CH, et al (2021) Unexpected synthesis of novel 2-pyrone derivatives: crystal structures, Hirshfeld surface analysis and computational studies. J Biomol Struct Dyn 39:4859–4877. https://doi.org/10.1080/07391102.2020.1780943
El Bakri Y, Anouar EH, Subramani K et al (2020) Synthesis, spectroscopic characterizations, DFT, molecular docking and molecular dynamics simulations of a novel 2-methyl-3H-benzimidazolo[1,2-b][1,2,4]triazepin-4(5H)-one. J Mol Struct 1202:127317. https://doi.org/10.1016/j.molstruc.2019.127317
Acknowledgements
The authors gratefully acknowledge the Anna University, Chennai, and IIT Madras for providing facilities FT-IR, FT-Raman, and UV-Vis spectrometer to record the spectra of the samples.
Author information
Authors and Affiliations
Contributions
R. Karthick and S. Karthikeyan have performed simulation studies, and M. P. Pachamuthu has carried out synthesis of compounds. All these work are done under the guidance of Dr. G. Velraj.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Karthick, R., Velraj, G., Pachamuthu, M.P. et al. Synthesis, spectroscopic, DFT, and molecular docking studies on 1,4-dihydropyridine derivative compounds: a combined experimental and theoretical study. J Mol Model 28, 5 (2022). https://doi.org/10.1007/s00894-021-04939-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-021-04939-2