Abstract
The beta-lactam-based antibiotics are among the most prescribed and effective antibacterial agents. Widespread use of these antibiotics, however, has created tremendous pressure for the emergence of resistance mechanisms in bacteria. The most common cause of antibiotic resistance is bacterial production of actamases that efficiently degrade antibiotics. The metallo-beta-lactamases are of particular clinical concern due to their transference between bacterial strains. We used molecular dynamics (MD) simulations to further study the conformational changes that occur due to binding of an inhibitor to the dicanzinc metallo-beta-lactamase from Bacteroides fragilis. Our studies confirm previous findings that the major flap is a major source of plasticity within the active site, therefore its dynamic response should be considered in drug development. However, our results also suggest the need for care in using MD simulations in evaluating loop mobility, both due to relaxation times and to the need to accurately model the zinc active site. Our study also reveals two new robust responses to ligand binding. First, there are specific localized changes in the zinc active site—a local loop flip—due to ligand intercalation that may be critical to the function of this enzyme. Second, inhibitor binding perturbs the dynamics throughout the protein, without otherwise perturbing the enzyme structure. These dynamic perturbations radiate outward from the active site and their existence suggests that long-range communication and dynamics may be important in the activity of this enzyme.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Among the most prescribed and effective antibacterial agents are beta-lactam based antibiotics such as the penicillins, cephalosporins, and carbapenems (Fig. 1) [1, 2]. These agents retard bacterial growth by inhibiting the transpeptidase activity necessary for bacterial cell wall synthesis [3, 4]. However, the widespread clinical and agriculture use of antibiotics has created tremendous selective pressure for the emergence of antibiotic-resistance mechanisms in pathogenic bacteria [5–9].
Bacteria develop resistance to antibiotics through a variety of mechanisms including production of antibiotic efflux pumps and modification of bacterial substrates such as cell wall components. However, the most common cause of antibiotic resistance is through the bacterial production of enzymes (lactamases) that intercept and degrade antibiotics with high efficiency.
Two well-established strategies to rescue the bactericidal activity of beta-lactam based antibiotics in infections with lactamase-producing bacteria have emerged. One is to develop new beta-lactam-based antibiotics through medicinal chemistry derivatization of existing agents in an effort to thwart lactamase recognition and subsequent hydrolysis. The other is to co-administer a lactamase inhibitor in conjunction with a traditional beta-lactam-based antibiotic. An example of this particular strategy is Augmentin, a widely prescribed combination of amoxicillin and clavulanic acid, a potent inhibitor of serine beta-lactamases.
To date, more than 500 distinct beta-lactamases have been described and grouped into four classes: A, B, C and D [10–12]. Class B (also known as group 3 [13]) includes the metal-containing (metallo) beta-lactamases (MBL). Although not common in the United States, MBL-harboring isolates represent a considerable public health concern as MBLs possess the widest substrate specificity of all beta-lactamases, and hydrolyze all current beta-lactam-based antibiotics. They tend to have high turnover rates (k cat) and broad specificity (high k cat/K m) [14–16], and exhibit little susceptibility to clinically approved beta-lactamase inhibitors [15].
Of great concern for clinicians, MBL genes are rapidly being transferred between bacterial strains by plasmid [17] and integron-borne [18–20] dissemination pathways. This has an impact both on the rate of mutation of MBL genes and on the range of affected bacterial strains. Furthermore, MBL and aminoglycoside resistance genes are frequently co-transferred [21–24]. Therefore outbreaks of MBL-associated infections are anticipated to continue to increase in severity, incidence, and in breadth of associated strains [25–28].
Metallo-beta-lactamases of subgroup B1 have two zinc sites: a “Zn1” site where the zinc is coordinated by three histidines and a bridging hydroxide; and a “Zn2” site, in which a second zinc is coordinated by an aspartic acid, a cysteine, a histidine, the bridging hydroxide, and a solvent molecule. Representative of subgroup B1 MBLs are CcrA from Bacteroides fragilis [29] and IMP-1 and VIM-2, both of which are secreted by several bacterial species [30]. B2 MBLs have the same zinc sites as B1, with the distinction that one Zn1-coordinating histidine is replaced by an asparagine. Representative of subgroup-B2 MBLs is ImiS from Aeromonas sobria [31]. B3 MBLs have the same two zinc sites as B1, with the distinction that the cysteine coordinating Zn2 is replaced by a histidine. Representative of B2 MBL is the L1 enzyme from Stenotrophomonas maltophilia [32]. The B1 and B3 MBL subgroups are most active when bound to two zinc ions [33–35], while the B2 enzymes are most active when bound to one zinc ion [36, 37].
Isolates from many bacterial strains in healthcare-associated infections have been reported to produce MBLs, including IMP-1- and VIM-1-positive clinical isolates of Pseudomonas (aeruginosa, putida, fluorscens, and stutzeri species), Serratia marcescens, Acinetobacter baumannii, Klebsiella (oxytoca and pneumoniae species), and Enterobacter (aerogenes and cloacae species) [38, 39] and L1-positive clinical isolates of Stenotrophomonas maltophilia [40–43]. The latter, for example, is a significant pathogen in healthcare-associated outbreaks of infection in immunosuppressed and/or immunocompromised patients [40–42]. In addition to possessing a subgroup B3 MBL, these organisms are typically resistant to most antibiotics due to low outer membrane permeability [43] and to the production of a non-metallo beta-lactamase [38, 39].
Since their initial description in Japan in 1988, the incidence of MBL-positive isolates has increased dramatically worldwide. MBL-producing isolates have been reported in 60.7% of Korean hospitals. In one series, 94% of randomly selected isolates of S. maltophilia were MBL-producing [44]. In Taiwan, 40 of 140 multiresistant K. pneumoniae isolates were IMP-positive by hybridization [45]. In Korea, 11.4% of imipenem-resistant P. aeruginosa and 14.2% of imipenem-resistant A. baumanii isolates produced MBLs [68]. The incidence of MBL-possessing P. aeruginosa in Brazilian hospitals is 20–30% [45, 46]. VIM-2-positive P. aeruginosa has been reported in France, Italy, Greece, Spain, Poland, Croatia, Germany, Belgium, Venezuela, Chile, Argentina, Korea, Japan, Taiwan, Saudi Arabia, and the United States. The first VIM-2-positive P. aeruginosa isolates were reported in Texas and New Mexico in 2004 and the first IMP-1-positive isolate was reported in New Mexico in 2006 [47, 48]. The first nosocomial outbreak of VIM-2-positive P. aeruginosa was reported in Chicago in 2005, affecting six patients in a trauma intensive care unit [49]. Upon implementation of MBL screening methods, a further 11 cases were identified over an 18-month period at this hospital alone. Clearly, the world-wide incidence of MBL production and positivity in clinical isolates is increasing dramatically.
As noted above, beta-lactam antibiotic co-administration with a beta-lactamase inhibitor has proved to be an effective therapeutic strategy for antibiotic-resistant isolates that possess serine beta-lactamases (classes A and D). However, clinically approved combination beta-lactam antibiotics, including carbapenem combinations, are not effective against MBL-harboring strains. Development of combination therapeutics (such as Augmentin) to combat MBL-harboring strains has been complicated by a number of factors including subtle active-site heterogeneity among individual MBL sub-types [50] and intrinsic active-site plasticity, which greatly complicates derivatization of existing beta-lactam-based antibiotics [51–54].
The aggregate of broad specificity of MBLs, their ability to rapidly hydrolyze all clinically useful beta-lactam antibiotics, and apparent ease of dissemination between bacterial strains, when coupled with insufficient market to justify investment by the pharmaceutical sector and occurrence in a healthcare-associated setting could result in what has been characterized by Walsh and co-workers as a “clinical catastrophe” [45].
Given the relative dearth of commercial development of novel antibiotics, or of MBL inhibitors that can be used in combination therapeutics, it is important to explore efficient means by which potential therapeutics can be identified and commercially developed for clinical use. Computational approaches to model target protein structure, dynamics, and the virtual screening of compound libraries represents an effective strategy for identification of potential therapeutic scaffolds and lead compound generation to speed drug discovery while reducing development costs.
Computational efforts to develop novel antibiotics and lactamase inhibitors are highly dependent on reasonably accurate models of target structure, dynamics, and protein–drug interactions. For modeling of metal-containing enzymes, there exist a number of ways in which to treat metal–protein interactions relevant to the overall protein fold and catalysis. For zinc-containing proteins, the zinc can be treated purely as a charged van der Waals (vdW) sphere [55], or the atom can be directly bonded to its coordinating residues. For these methods, charges and other parameters are obtained from quantum mechanical calculations [53]; therefore, the force field model must be remade for every active site and every ligand. Other methods include a more general parameterization of the zinc interactions including nonadditivity and polarization [56]. A more recent model for zinc is the cationic dummy atom approach (CaDA) developed by Pang and co-workers [57, 58]. The approach, which is simple and general, has been reported to be more realistic than earlier models, which treated zinc atoms as charged vdW spheres [59, 60].
Of the MBLs studied thus far, the enzyme from Bacteriodes fragilis has been the subject of comprehensive computational and experimental characterization of the polypeptide-chain dynamics of both the free protein and in complex with the proprietary inhibitor 3-[2′-(S)-benzyl-3′-mercaptopropanoyl]-4-(S)-carboxy-5,5-dimethylthiazolidine (SB225666). Two essentially identical X-ray structures of the apo B. fragilis enzyme have been reported (PDB ID 1ZNB and 2BMI, [29, 61]. The overall structure consists of a four-layered αβ/βα motif [32]. The enzyme active site resides at one end of the beta-sandwich and contains the binuclear zinc site. A standard numbering scheme for MBLs was suggested initially by Gallini and co-workers and later expanded by Garau and coworkers [16, 62]. However, to simplify the comparison of the results reported here to previous computational investigations [53], we number the amino acids beginning with serine 1. Here, Zn1 is coordinated by three histidine residues (His79 Nε, His81 Nδ, and His142 Nε) and a single water molecule. Zn2 is coordinated by Asp83 Oδ2, Cys161 Sγ, and His203 Nε) and two water molecules. One water molecule (“Wat1”) is coordinated by both zinc ions. This bridging water is thought to exist in a hydroxide form and is believed to mount a nucleoophilic attack on the antibiotic substrate, ultimately leading to its hydrolysis [35, 63].
Flanking the enzyme active site is an extended beta-flap (also known as the “major flap”) consisting of residues Leu23 through Ser34. Another “minor loop” (residues Ala168 through Ser175) sits roughly opposite the major flap centered about the binuclear zinc site. Previous computational work, crystallographic temperature factors, as well as heteronuclear nuclear Overhauser effect (NOE) data derived from nuclear magnetic resonance (NMR) experiments suggest a high degree of conformational plasticity associated with the major flap and minor loop [29, 51–53]. This plasticity is clearly a deterrent to the development of novel beta-lactam based antibiotics. Simple derivatization of existing beta-lactam scaffolds may result in drugs that possess bactericidal activity, but the dynamics and active-site plasticity of this and similar MBLs allows for structural accommodation and hydrolysis of a wide range of structurally diverse antibiotic substrates [52].
In this study, we performed molecular dynamic (MD) simulations of the MBL from Bacteroides fragilis in the presence and absence of the inhibitor SB225666 using the CaDA approach as implemented in the AMBER MD suite [64]. We compare the results of these simulations to previously published investigations and extend our comparisons to experimentally derived dynamic data. Specifically, we report the results of three different nanosecond timescale simulations. One simulation is of the apo lactamase (apo refers to lack of inhibitor-binding, not absence of zinc). Two simulations are of the lactamase in complex with SB25666. The difference between the two bound simulations laid in different docking protocols leads to slightly different structures and dynamics.
Materials and methods
Initial model building and refinement, apo structure
The modeling of Zn-binding proteins presents several challenges, most notably how best to treat the Zn2+ ions computationally. For example, in recent efforts to model the IMP-1 MBL from B. fragilis, Irwin and co-workers [69] treated each Zn atom independently, placing the full +2 formal charge at each Zn center without spatially distributing the charge. However, Zn2+ can be modeled as being purely non-bonded, or treated as mixed bonded and non-bonded for two-zinc systems [53]. Zinc-bound atoms can also be modeled as cationic “dummy” atoms (CaDA) possessing a tetrahedral charge distribution [57, 58]. While each method has its advantages, the CaDA approach allows for modeling of the binuclear Zn active site without having to covalently attach the coordinating residues/side chains to the Zn atoms. Consequently, this may more accurately reflect the overall electrostatic landscape of the MBL binding pocket. In a study comparing non-bonded and CaDA approaches for modeling Zinc using the IMP-1 MBL from P. aeruginosa, Oelschlager and co-workers [59, 60] determined that a better representation of the IMP-1 MBL active site was obtained using the CaDA model, with either deprotonated or protonated inhibitor docked into the active site. Furthermore, they found that the overall architecture of the binuclear Zn site (internuclear distances and angles between Zn dummy atoms and the coordinating residues and bridging water molecule) was maintained over nanosecond-timescale simulations at a temperature of 300 K. For MD simulations of the MBL from B. fragilis, the coordinates from the 2BMI X-ray crystal structure (resolved to 2.0 Å, PDB entry 2BMI, chain “A” only [61]) were used to construct an initial model. Zn-coordinating histidines 79, 81, 142 and 203 were modeled as histidinates, while Asp83 and Cys161 sidechains were assumed to be deprotonated at the Oδ2 and Sγ positions, respectively. Asp176, which resides in the second coordination sphere for Zn1, was protonated to form a hydrogen bond to the side chain of histidinate 81. To simplify model construction, tetrahedral coordination geometry was assumed for both Zn1 and Zn2. Prior to modeling, all crystal waters except the bridging water oxygen were removed. The bridging water was assumed to exist in hydroxide form. Zinc dummy atoms were created as previously described [58, 59] using zinc, histidinate and hydroxide force field terms and electrostatic potential energy library files graciously provided by Y.P. Pang at the Mayo Clinic College of Medicine, Minneapolis.
The apo structure was further refined in two steps using the AMBER8 suite of MD simulation algorithms [64], an NPT ensemble and the ff03 force field [70]. The structure was initially energy minimized by performing a steep-decent in vacuo minimization (1,000 steps, conjugate gradient) to relieve bad steric interactions. The structure was then charge neutralized and solvated with the LEAP module of AMBER8, which required adding ten sodium counter ions and a TIP3P water box with 9,007 water molecules in an isometric truncated octahedral box of 76.937 Å. A solvated MD simulation was then performed in four steps. A minimization with a tightly restrained protein (1,000 steps, conjugate gradient, 500 Kcal/mol Å2 force constant) was first performed to relieve bad contacts in the surrounding solvent, followed by an unrestrained minimization (1,000 steps, conjugate gradient) to relieve bad contacts in the entire system. This was followed by a 20 ps simulation, with weak restraints (10 Kcal/mol Å2 force constant) on the protein, raising the temperature from 0 to 300 K to relax the position of the solvent molecules. This was then followed by a 5 ns constant-temperature (Langevin dynamics 2 ps time-constant 1/ps collision frequence) production simulation . The simulations all shared the following common parameters as implemented in the SANDER module of AMBER842: constant pressure (1 atm) with isotropic position scaling using a Langevin barostat with a collision frequency of 1.0/picosecond and 2 ps time-constant; the SHAKE algorithm to constrain hydrogen bond lengths; a 2 fs timestep, 10 Å nonbonded cutoff, coordinate saving every 1 ps, and Particle Mesh Ewald with default parameters (81 grid points in each dimension, cubic spline interpolation). Any parameters not specified were unchanged from the defaults.
Initial model building and refinement, inhibitor-bound structure
Initial structural models for the inhibitor-bound form of the enzyme were created as follows. A 3D structure of SB225666 was generated by first using the Java Molecular Editor (JME) to produce a molecule SMILES representation of SB225666 [73]. The SMILES representation of the inhibitor was then input into the online SMILES translator and 3D structure coordinate file generation resource available through the Computer-Aided Drug Design group of the Laboratory of Medicinal Chemistry at the National Cancer Institute, National Institutes of Health [77]. The 3D structure coordinate file of SB225666 was then manually edited to match atom number and naming conventions consistent with pdb format for input into the Antechamber module of AMBER 8. The inhibitor was modeled to carry a net −2 charge, by deprotonating the carboxylic acid moiety and by removing the proton associate with the thiol portion of the inhibitor. The carboxylic acid moiety would be deprotonated at physiologic pH, and the thiol group would be deprotonated, poised to displace the negatively charged shared hydroxide, consistent with structures of other thiol centric inhibitors [74]. Force field terms and preparatory files necessary to model the inhibitor were obtained using the Antechamber module of AMBER, the general AMBER force field [75, 76] and employing a AM1-BCC charge method.
The 3D structure coordinate file for SB225666 was docked manually into the active site of the lactamase (PDB ID 2BMI) by placing the negatively charged sulfur at the position of the shared hydroxide at the binuclear zinc site. The inhibitor was oriented such that the phenyl group was directed toward the putative hydrophobic patch thought to accommodate the beta substituent of beta-lactam-based antibiotics [29]. For the inhibitor-bound model, the binuclear zinc active site was treated identically as described for the apo form.
Following in vacuo minimization, the complex was then charge neutralized (11 sodium counterions), solvated using a TIP3P water box (8,975 water molecules in an isometric truncated octahedral box ), and the system was minimized in steps identical to those procedures described for the apo structure. The choice to first charge-neutralize, solvate and system-minimize versus first solvate, then charge-neutralize should in principle generate comparable starting structures. However, we noted that the sequence of neutralizing, then solvating resulted in a sodium counterion being placed well within the active site of the enzyme, poised to counter the negative charge of the carboxylic acid moiety of the inhibitor (approximately 2.2 Å distant). Interestingly, the process of solvating, and then charge-neutralizing resulted in a counterion being placed in the proximity of the carboxylic acid moiety, approximately 1.1 Å more distant, and displaced out of the enzyme active site. The two procedures (neutralize-solvate versus solvate-neutralize) each followed by system minimization also resulted in two different inhibitor orientations within the active site. For the first pose (neutralize-solvate), the two methyl groups at the 5 position of the thiazolidine ring were opposite Trp29, while the phenyl moiety of the inhibitor oriented out from the beta sandwich. The second pose (solvate-neutralize) resulted in the phenyl group of the inhibitor interacting closely with a hydrophobic patch located at the base of the major flap formed by residues Ala24, Ile26, Val32 and Ile52. This hydrophobic patch is thought to accommodate the beta substituent of many beta-lactam antibiotics [29], and may be a more realistic representation of the actual orientation of SB252666 within the enzyme active site. This particular observation is noteworthy in itself; however, the choice to charge-neutralize then solvate or vice versa should be considered when employing such procedures. Nonetheless, we pursued subsequent nanosecond timescale MD simulations of each pose so as not to bias the outcomes of our subsequent analyses. Due to the time-scale of apparent equilibration of pose two, a total of 10 ns simulation was conducted.
Analysis of simulations
In order to ensure that the analysis of the different simulations compared equivalent equilibrated trajectories, where averaging was required, i.e., Figs. 5 and 7, only the initial portions of the simulations were not considered, i.e., 2.5 ns–5 ns for the apo and the first docked pose, and 5–10 ns for the second pose. For the remaining measures, the entire production simulation was analyzed.
Root mean square deviation calculations
Each trajectory (5 ns for the apo and pose 1, 10 ns for pose 2), was aligned to the initial structure of the corresponding production simulation to minimize the alpha-carbon root mean square deviation (RMSD) from the initial structure. The RMSD for each frame, saved every 1 ps, was then recorded.
Clustering analysis
K-means clustering was performed in order to confirm the apparent equilibration found from the RMSD calculations and to identify representative structures for visualization. First, the centers of geometry for each residue in each aligned frame from each simulation were calculated. This resulted in 229 3D time series for each simulations. These time-series were then clustered using k-means clustering on the Euclidean distance between each time-series to obtain three sets of clusters, one for each simulation. Cutoffs of 1.0, 1.1, and 1.5 Å were used to construct different sets; the results were robust with regards to finding a single dominate cluster after an initial time period, which varied among the simulations, as discussed in the Results. The structures closest to the center of the equilibrated clusters were used for visualization.
Covariance analysis
In order to determine the presence of correlated fluctuations, equal-time correlations of position fluctuations were calculated for the alpha-carbons in each simulation. To prevent the equilibration period from adding rare fluctuations to the correlations and thereby biasing them, the initial half of each simulation was discarded during this analysis. The remaining portion of each aligned simulation was realigned to the average structure obtained from the remaining portion of the aligned simulation in order to minimize artifacts from rotation or translation. Then, from each simulation, a new average structure was calculated, alpha-carbon fluctuations calculated and the correlations calculated and normalized to obtain three covariance matrices.
Although covariance matrices are often compared visually to ensure an unbiased comparison, the ligand covariance matrices were subtracted from the apo covariance matrix and those covariances whose differences were 3 standard deviations above the mean were identified as the most likely to be significant.
Root mean square fluctuations
As with the covariance analysis, the last half of each simulation was used to calculated atom root mean square fluctuations, which were then averaged over each residue. Note that these are directly proportional to B-factors.
Hydrogen bond analysis
There are 722 hydrogen bond donors and acceptors within the protein. In each frame, a value of 0 or 1 was assigned to each hydrogen bonding partner depending on whether it was in a hydrogen bond, as defined using standard CHARMM cutoffs of 2.4 Å for the heavy atom-hydrogen distance and 180° for the angle cutoff, in that particular frame. This was performed across all three simulations and the correlation coefficient between each of the 722 hydrogen bond time-series and a time-series of the Zn–Zn distances were calculated. Those hydrogen bond partners with Pearson correlation coefficients greater than 0.4 were selected as highly correlated and the identity of the hydrogen bonding pairs confirmed by recalculating the hydrogen bonding partners for an identified hydrogen bond acceptor/donor over for the course of the simulations.
Results and discussion
Simulations equilibrate within at most 4.5 ns, but with different convergence behaviors
A comparison of the three simulations by either RMSDs (Fig. 2), or a clustering analysis (Fig. 3) demonstrates that the three simulations are all equilibrated within, at most, 4.5 ns. In order to consider the simulations as properly equilibrated, we would expect to see a leveling off of the RMSD (although possibly with fluctuations representing conformational changes) and either a single cluster in clustering analysis, or several clusters through which the protein fluctuates. The clustering analysis should be a more precise measure of equilibration as it is an all-on-all analysis as opposed to a comparison to a single reference point. The length of the apparent equilibration period does suggest a problem with earlier simulations, conducted by the present authors and others, which were on shorter timescales.
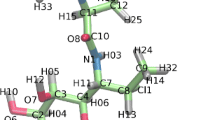
Structural comparisons of apo protein and bound complex. The centroids of the most populated cluster from each of the three simulations are depicted: white apo, blue pose 1, red pose 2. The clusters were obtained from a 1.1 Å radius k-means clustering of snapshots saved every 1 ps in the simulation. The protein is depicted in the new cartoon representation, the inhibitor in a bonded representation, and the zinc atoms in van der Waals (vdW) representation. Note, the blue and red zinc atoms overlap almost completely. This figure was prepared using VMD (http://www.ks.uiuc.edu/Research/vmd/)
The apo protein equilibrates within 2–3 ns, as suggested by the RMSD, and a clustering analysis (not shown) indicates that the protein is in a single cluster (Fig. 3), with only rare excursion to other clusters, after ∼3 ns. This relaxation to a single cluster of conformations, along with the reasonable overall RMSD of ∼2 Å suggests that the MD simulation accurately represents the overall conformational flexibility of the apo protein, after an initial equilibration period, and that the protein does not undergo significant overall conformational changes on the nanosecond time-scale; this does not rule out local conformational changes, as we will discuss later.
The two docked structures exhibit very different behaviors. The first docked pose appears to equilibrate quickly; within 1 ns according to either RMSD or clustering, although there are greater fluctuations afterwards than in the apo simulation (Fig. 2). However, the second docked structure suggests a different picture, in which the conformational relaxation requires ∼4.5 ns. This suggests that the ligand induces conformational changes in the protein that require a more extensive equilibration period, but which result in a single global protein conformation (Fig. 3), as confirmed by clustering analysis. These two apparently different effects of ligand-binding will be reconciled in the remainder of the article.
The protein conformational response is localized predominately in the active site and zinc-binding loops
A comparison of the structures of the centroids of the equilibrated clusters from the three simulations clearly shows the localization of the protein response in two different loops in the active site. First, the major loop (residues 22–35) samples different conformations depending on the presence or absence of the inhibitor (Fig. 3). Even depending on how the inhibitor is docked (pose 1 versus pose 2), the precise conformations sampled by the active site loop vary. Second, in the inhibitor-bound structures there is a localized flip away from the zinc atoms in a loop that appears to be indirectly involved in complex formation. The flip is localized to residues 80, and 82–84; beyond this region the differences between the alpha-carbon positions of the different amino acid cluster centroids are less than the sum of the alpha-carbon root-mean fluctuations. The flip is largest at residue 82, 4.5 Å difference between the apo and pose 2 cluster, and decreases to between 1.3 and 1.9 Å for residues 80, 83, and 84. The flip occurs in the residues surrounding two zinc-binding residues, 79 and 81. This change is independent of the inhibitor docking pose. The conformational changes in the major loop are not unexpected; they have been reported before and are expected to be involved as part of catalysis. However, our simulations suggest that the ensemble of conformations populated depends critically on the nature of the ligand binding pose, and are different from those adopted in the apo protein (Fig. 3). This may explain the differences observed among the different bound simulations; the loop response depends critically upon the ligand orientation, which does not interconvert on a 5–10 ns timescale.
The observation of a conformation change in the loop lateral to the zinc center is new, and consistent with NMR chemical shifts of residues within the vicinity of the enzyme active site that are not directly involved in substrate binding, in particular for the Trp80 indole [51, 52]. Again, the conformational change of this loop appears independent of the ligand docking pose (Fig. 3), suggesting that the change is a response to the intercalation of the sulfur from the inhibitor between the zinc atoms. We believe that this flip is likely due to intercalation of the negatively charged sulfur moiety of the inhibitor, which is supported by the observed increase in zinc–zinc distance (Fig. 4). As this flip appears to be the local response to the increased zinc–zinc distance, it suggests that this region may play a role in controlling the zinc coordination. Otherwise we would have expected a more delocalized response to sulfur binding in the zinc coordination shell. If the change in the zinc–zinc distance is a general phenomena in ligand binding, which would be expected if ligands interacts with both zincs, the loop would allow for ligand binding without disruption of zinc coordination. If this region were to become more rigid, then the increase in zinc–zinc distance induced by binding should either disrupt other portions of the active site, or the zinc–zinc distance change would be prevented due to the overall rigidity of the active site. A disruption of the active site could occur through a different loop flip; however, the loop affected is a unique loop in the active site. We suspect that the more likely outcome would be a more general disruption of the active site geometry. As a result, we hypothesize that making this loop more rigid should disrupt the function of the enzyme, as articulation of the enzyme within the catalytic transition states and/or distance response in the zinc coordination shell would be less-than-optimal for efficient lactam hydrolysis.
In pose 2, there is a brief excursion to exceptionally large zinc–zinc distances of up to 5.2 Å. This excursion is short and occurs only once, and so its significance is unknown. However, it is interesting to see if this excursion is due to another specific conformational change in the zinc active site or in the protein as a whole. This does not appear to be the case; the greatest change in conformation is in the same zinc binding loop that flips in response to ligand binding, although there are other slight rearrangements throughout the active site (Figures S1, S2). This further suggests that the lateral zinc-binding loop may be especially important in controlling the binding of zinc in the binding site, but that the remainder of the zinc active site is sufficiently flexible to allow for changes in the zinc–zinc distances once the lateral zinc binding loop has changed conformation.
Changes in correlated motions involve residues radiating out from the zinc ions
Another measure of the response to inhibitor binding and the dependence of the response on orientation of the inhibitor is the residue–residue covariance matrix (Fig. 5), which measures correlations between the positional fluctuations of the different residues. The covariance matrices show limited variability among the different simulations (Fig. 5a–c), especially when the two different ligand simulations are compared (Fig. 5a,b). A calculation of the covariances that change most significantly between the apo and bound simulations (Fig. 5d,e) shows that the protein dynamic response to ligand binding is limited, and largely independent of the binding pose (Fig. 5d–f). A detailed analysis of the differences in correlated motions due to ligand binding shows that there are several small regions of difference, many of which show changes in correlated motions with each other. These regions are, approximately, residues 28–34, 49–58, 80–86, 104–110, 115–125, 130–133, 146–148, and 180–186.
a–f Correlated fluctuations. The covariance matrix of atomic fluctuations for the alpha-carbons calculated across the three simulations (apo, pose 1 and pose 2, respectively) are displayed in panels a–c, with intense red indicating perfect correlation, intense blue indicating perfect anticorrelation, and yellow no correlation. d, e Covariance matrices for the bound simulations (pose 1 and pose 2, respectively) are subtracted from the apo covariance matrix, and those covariances that change by more than 3 STDs are then depicted in blue if the apo simulation showed higher correlation, and in red if the bound exhibited higher correlations. f The matrices in panels d and e are added and divided by two to demonstrate the consistency of the dynamic response of the protein to binding
The location of these residues (Fig. 6) in the protein shows that they form chains that connect to the binding site either directly or through residues in areas with perturbed conformations, but not perturbed dynamics. This suggests a generic delocalized dynamic response to binding that alters the coupling of motions across the protein. The functional significance of these changes is currently unknown. They could be involved in mediating catalysis [65, 66], or could possibly be a side effect of ligand binding. If these are indeed involved in mediating catalysis, they could do so by directly driving the catalysis, or by controlling the conformational freedom of the complex. Direct driving would require that the motions involving these residues directly “push” the ligand–protein complex over an activation barrier, whereas conformational control would require these residues to be involved in a network of interactions that preferentially stabilize specific protein–ligand conformations that are optimal for catalysis. At this stage, neither role can be ruled out.
Structural mapping of the most dynamically perturbed residues. Regions that exhibit the greatest perturbation in correlated fluctuations are placed into their structural context. The three conformations from Fig. 1 are depicted in blue (first pose), gray with red inhibitor (second pose) and white (apo) . Residues exhibiting the greatest perturbation in their correlated motions (Fig. 4f) are depicted in orange, and those residues that connect the latter to the binding site are depicted in yellow. For clarity, the orange and yellow residues are shown in the conformations adopted in the centroid for pose 2
Hydrogen bonding analysis indicate correlation between zinc–zinc distances and specific hydrogen bonds
In addition to the localized changes in the active site, hydrogen bonding analysis indicates that there are three hydrogen bonds whose formation or rupture are correlated with the changes in the zinc–zinc distance. The loss of a hydrogen bond between the backbone oxygen of His 142, one of the zinc-coordinating residues, and Asp 176 is correlated with increasing zinc–zinc distance (correlation coefficient −0.59). The gain of a hydrogen bond between the backbones of Ala 177 and Val 179 is correlated with changing zinc–zinc distance (correlation coefficient 0.43). The loss of a hydrogen bond between the side chain of Trp 182 with either the backbone of Met 162 or with side chain of Leu 186 is also correlated with increasing zinc–zinc distance (correlation coefficient −0.41). This suggests that there are specific hydrogen bonds between involving residues in a secondary shell about the di-zinc active site connecting the active site, His 142 specifically, to solvent exposed residues Val 179 and Leu 186. Most of the residues involved in these changing hydrogen bonds are in or near a specific region (residue 180–186) implicated in changes in correlation motions upon ligand binding, further suggesting that this region may have functional importance and so should be the subject of additional experimental studies.
Differences in fluctuations further suggest specific locales dynamically affected by ligand-binding
The final measure of the structural response to inhibitor binding is determined from the root-mean-square (RMS) fluctuations (Fig. 7). RMS fluctuations show changes that are independent of binding-mode in two regions. First, there is rigidification of the helix encompassed by residues 109 though 115. Interestingly, this region overlaps and connects with two other regions (residues 104–110, and 115–125) that exhibit perturbed correlated motions. This region also interacts with the zinc-binding loop (residues 80–85). These two observations suggests that changes in the zinc-binding loop lead to the rigidification of this region, which in turn leads to changes in correlated motions. Second, the major flap (residues 22–35) shows increased flexibility upon binding of SB225666. Here, the two ligand-bound simulations show surprisingly similar changes despite the differences in major loop conformations. These changes are especially significant in residues 25–31, including the apex of the major flap, which strongly interacts with the inhibitor in the complex. The magnitude of the changes, and the similarity between the two bound simulations further suggest that ligand binding changes and broadens the ensemble of loop conformations adopted away from the apo structure. These results further indicate, at least the in two orientations sampled here, that the change in conformational flexibility is similar regardless of inhibitor pose. There is one region (for residues ∼50–55) that shows significant differences depending on ligand orientation. There is a general increase in flexibility due to binding, but it is significantly more in the first binding pose. This predominately helical region also interacts with the zinc-binding loop (residues 80–85), and so the change in flexibility is likely due to the perturbation of the zinc-binding loop, which arises from the intercalation of the inhibitor between the zinc atoms. This region can also interact with the major loop in some conformations, especially those adopted when the inhibitor is in pose 2. This likely explains the variability in flexibility between the two inhibitor-bound simulations.
Conclusions
The results of this study have implications for the molecular function of the MBLs, for the use of molecular simulations in probing the biological function of key residues in the action of MBL hydrolysis of antibiotic substrates, and in the design of novel MBL inhibitors.
Our findings show that the timescales needed for system relaxation suggest that previous computational efforts may have focused on timescales too short to capture key aspects of MBL dynamics and structural response to substrate/inhibitor binding. Around 5–10 ns appears to be the minimum time necessary to relax an inhibitor-bound MBL, even around a single binding pose. As we have seen, two different binding poses exhibit different loop conformation indicating that longer simulation times are necessary to explore conformational interchange among different binding poses. Furthermore, different initial conditions are required to adequately probe the possible effects of substrate/inhibitor binding.
Our findings also show that an accurate representation of the electrostatic landscape of the binuclear zinc site is required to reasonably perform MD simulations of MBL structure in order to obtain information on the conformational changes and dynamics of the entire protein complex. These simulations clearly show that there is a change in the zinc–zinc distance, which in turn causes a specific response in the zinc-binding loop. This then propagates throughout the enzyme structure to cause subtle changes in dynamics and flexibility. These delocalized changes, discussed in detail below, are generally in agreement with experimental studies that have shown the importance of interactions within the secondary sphere surrounding the metal center in metallo-beta-lactamases [67]. For example, mutations in secondary shell residues not directly involved in zinc binding can affect the catalytic efficiency and/or substrate specificity of an MBL while retaining a di-zinc active site [68, 71, 72]. Our results suggest that some of the changes in dynamics occur beyond the secondary shell around the di-zinc active site. In order to observe these aspects of MBL dynamics, a flexible zinc representation is clearly required as previously used mixed bonded/nonbonded models of zinc binding were apparently insufficiently flexible [53]. Our observations suggest caution in the use of MD simulations to study binuclear zinc active sites. However, careful analysis suggests that several conclusions regarding the molecular function of these enzymes and for future drug discovery efforts can be drawn.
There are at least two implications for future drug discovery efforts. First, despite the lack of conformational exchange with regards to the major flap and the inhibitor binding pose, the dynamics of the protein are rather robust. The perturbations due to ligand binding can be traced back primarily to the increased zinc–zinc distance. This strongly suggests that the response to binding by most of the protein is rather generic. Furthermore, our results also suggest that the bridging hydroxide becomes bound to only one metal during binding. The implication of this is that the resulting hydroxide should be nucleophilic enough for attack on the beta-lactam carbonyl, whereas a bridging hydroxide may not be. This is encouraging for future drug design efforts as efforts to include conformational sampling in drug discovery can focus on including variable major flap conformations. Second, we show that the conformation of the tip of the major flap is highly dependent on the precise binding pose adopted by the inhibitor. This confirms that the major flap is indeed the primary source of plasticity within the active site, and so should be considered in drug development. In the future, these conformations will be further used in drug screening.
In addition, our results have two notable implications for possible molecular function, which may or may not be relevant to drug discovery efforts. First, the molecular conformational response to the increased zinc–zinc distance from inhibitor binding is concentrated in the flip of a loop indirectly involved in substrate/inhibitor binding, rather then being delocalized across the entire active site. This suggests that this loop may be critical in the function of the enzyme. However, the zinc active site is then capable of accommodating larger fluctuations in the zinc–zinc distance, at least as rare fluctuations, although the biological relevance of such fluctuations is unknown. We hope that interested experimentalists will be able to rigidify this loop either by site-directed mutagenesis or chemical modification, and determine if the function of the enzyme is indeed disrupted. Such a disruption could occur through alteration of enzyme–substrate interactions within the catalytic transition state(s) or by perturbing a required response in the zinc coordination shell. In either case, the expectation is that the active site would be less-than-optimal for efficient lactam hydrolysis.
Second, we observe networks of residues whose dynamics are perturbed, without corresponding perturbations in the enzyme structure. This is the first prediction for perturbed dynamics in the MBLs. Previous MD studies [53] did not uncover such perturbations, presumably for a number of reasons. The simulations were insufficiently long to have confidence in any differences between the apo and inhibitor-bound dynamics. Also, prior simulations did not have sufficient flexibility in the zinc representations to allow for perturbations in the zinc-binding loop. The latter is particularly important with respect to changes in dynamics for residues that interact with this loop. The functional significance of these changes is currently unknown. They do appear to comprise several potential “molecular wires” that could be involved in mediating catalysis [65, 66] either directly or through molecular interaction networks that may stabilize catalytically important conformations. Or these could merely be a side effect of ligand binding without functional significance. The characterization of alterations in MBL dynamics that occur upon ligand binding cannot distinguish between these hypotheses as the changes that appear to trigger these dynamic changes are also robust with respect to binding pose. Mutations in these regions may disrupt the connections and change the dynamics away from those found in ligand-bound simulations. This could determine if these changes are indeed functionally significant, which would add the MBLs to the growing list of proteins for which dynamics are functionally important. If functional significance is established for these dynamical perturbations, then further theoretical investigations should be able to provide insight into the precise role of dynamics in the catalysis of the metallo-beta-lactamase.
References
Bush K (1998) Clin Infect Dis 27[Suppl 1]:S48–S53
Bush K, Mobashery S (1998) In: Rosen PB, Mobashery S (eds) Resolving the antibiotic paradox: progress in understanding drug resistance and development of new antibiotics. Kluwer/Plenum, New York, pp 71–98
Knowles JR (1985) Acc Chem Res 18:97–104. doi:10.1021/ar00112a001
Knowles JR (1980) In: Brodbeck U (ed) Enzyme inhibitors. Verlag Chemie, Weinheim, pp 163–167
Neu HC (1992) Science 257(5073):1064–1073. doi:10.1126/science.257.5073.1064
Ash C (1996) Trends Microbiol 4(10):371–372. doi:10.1016/0966-842X(96)30028-0
Wilkins AS (1996) Bioessays 18(10):847–848. doi:10.1002/bies.950181012
Levy SB (1998) Sci Am 3:46–53
Barbosa TM, Levy SB (2000) Drug Resist Updat 3:303–311. doi:10.1054/drup.2000.0167
Ambler RP (1980) Philos Trans R Soc Lond B Biol Sci 289:321–331. doi:10.1098/rstb.1980.0049
Jaurin B, Grundstrom T (1981) Proc Natl Acad Sci USA 78:4897–4901. doi:10.1073/pnas.78.8.4897
Mederios A (1984) Br Med Bull 40:18–27
Bush K (1989) Antimicrob Agents Chemother 33(3):259–263
Carfi A, Duee E, Galleni M, Frère JM, Dideberg O (1998) Acta Crystallogr D Biol Crystallogr 54(Pt 3):313–323. doi:10.1107/S0907444997010627
Toney JH, Moloughney JG (2004) Curr Opin Investig Drugs 5(8):823–826
Garau G, Garcia-Saez I, Bebrone C, Anne C, Mercuri P, Galleni M et al (2004) Antimicrob Agents Chemother 48(7):2347–2349. doi:10.1128/AAC.48.7.2347-2349.2004
Ito H, Arakawa Y, Ohsuka S, Wacharotayankun R, Kato N, Ohta M (1995) Antimicrob Agents Chemother 39(4):824–829
Levy SB, Miller A (1989) Gene transfer in the enviroment. McGraw Hill, New York
Senda K, Arakawa Y, Ichiyama S, Nakashima K, Ito H, Ohsuka S, Shimokata K, Kato N, Ohta MJ (1996) Clin Microbiol 34:2909–2913
Levy SB, Marshall B (2004) Nat Med 10(12):S122–S129. doi:10.1038/nm1145
Bush K, Miller GH (1998) Curr Opin Microbiol 1:509–515. doi:10.1016/S1369-5274(98)80082-9
Tsakris A, Ikonomidis A, Spanakis N, Poulou A, Pournaras S, Antimicrob J (2007) Chemother 59(4):739–741
Mendes RE, Toleman MA, Ribeiro J, Sader HS, Jones RN, Walsh TR (2004) Antimicrob Agents Chemother 48(12):4693–4702. doi:10.1128/AAC.48.12.4693-4702.2004
Riccio ML, Pallechi L, Fontana R, Rossolini GM (2001) Antimicrob Agents Chemother 45(4):1249–1253. doi:10.1128/AAC.45.4.1249-1253.2001
Gales AC, Menezes LC, Silbert S, Sader HS, Antimicrob J (2003) Chemother 52(4):699–702
Giakkoupi P, Petrikkos G, Tzouvelekis LS, Tsonas S, Legakis NJ, Vatopoulos AC (2003) J Clin Microbiol 41(2):822–825. doi:10.1128/JCM.41.2.822-825.2003
Giakkoupi P, Xanthaki A, Kanelopoulou M, Vlahaki A, Miriagou V, Kontou S et al (2003) Clin Microbiol 41(8):3893–3896. doi:10.1128/JCM.41.8.3893-3896.2003
Lee K, Lee WG, Uh Y, Ha GY, Cho J, Chong Y (2003) Emerg Infect Dis 9(7):868–871
Concha NO, Rasmussen BA, Bush K, Herzberg O (1996) Structure 4:823–836. doi:10.1016/S0969-2126(96)00089-5
Walsh TR (2005) Clin Microbiol Infect 11(Suppl 6):2–9. doi:10.1111/j.1469-0691.2005.01264.x
Walsh TR, Neville WA, Haran MH, Tolson D, Payne DJ, Bateson JH et al (1998) Antimicrob Agents Chemother 42(2):436–439
Ullah JH, Walsh TR, Taylor IA, Emery DC, Verma CS, Gamblin SJ et al (1998) J Mol Biol 284:125–136. doi:10.1006/jmbi.1998.2148
Crowder MW, Wang Z, Franklin SL, Zovinka EP, Benkovic SJ (1996) Biochemistry 35:12126–12132. doi:10.1021/bi960976h
Crowder MW, Walsh TR, Banovic L, Pettit M, Spencer J (1998) Antimicrob Agents Chemother 42(4):921–926
Wang Z, Benkovic SJ (1998) J Biol Chem 273(35):22402–22408. doi:10.1074/jbc.273.35.22402
Valladares MH, Felici A, Weber G, Adolph HW, Zeppezauer M, Rossolini GM et al (1997) Biochemistry 36:11534–11541. doi:10.1021/bi971056h
Crawford PA, Sharma N, Chandrasekar S, Sigdel T, Walsh TR, Spencer J et al (2004) Protein Expr Purif 36:272–279. doi:10.1016/j.pep.2004.04.017
Walsh TR, Hall L, Assinder SJ, Nichols WW, Cartwright SJ, MacGowan AP et al (1994) Biochim Biophys Acta 1218:199–201
Walsh TR, Payne DJ, Neville T, Tolson D, MacGowan AP, Bennett PM (1997) Antimicrob Agents Chemother 41:1460–1462
Muder RR, Yu VL, Dummer JS, Vinson C, Lumish RM (1987) Arch Intern Med 147:1672–1674. doi:10.1001/archinte.147.9.1672
Khardori N, Elting L, Wong E, Schable B, Bodey GP (1990) Rev Infect Dis 12(6):997–1003
Villarino ME, Stevens LE, Schable B, Mayers G, Miller JM, Burke JP et al (1992) Infect Control Hosp Epidemiol 13(4):201–206
Mett H, Rosta S, Schacher B, Frei R (1988) Rev Infect Dis 10(4):765–769
Miller LA, Ratnam K, Payne DJ (2001) Curr Opin Pharmacol 1(5):451–458. doi:10.1016/S1471-4892(01)00079-0
Walsh TR, Toleman MA, Poirel L, Nordmann P (2005) Clin Microbiol Rev 18(2):306–325. doi:10.1128/CMR.18.2.306-325.2005
Marra AR, Pereira CA, Gales AC, Menezes LC, Cal RG, de Souza JM et al (2006) Antimicrob Agents Chemother 50(1):388–390. doi:10.1128/AAC.50.1.388-390.2006
Toleman MA, Rolston K, Jones RN, Walsh TR (2004) Antimicrob Agents Chemother 48(1):329–332. doi:10.1128/AAC.48.1.329-332.2004
Hanson ND, Hossain A, Buck L, Moland ES, Thomson KS (2006) Antimicrob Agents Chemother 50(6):2272–2273. doi:10.1128/AAC.01440-05
Lolans K, Queenan AM, Bush K, Sahud A, Quinn JP (2005) Antimicrob Agents Chemother 49(8):3538–3540. doi:10.1128/AAC.49.8.3538-3540.2005
Daiyasu H, Osaka K, Ishino Y, Toh H (2001) FEBS Lett 503(1):1–6. doi:10.1016/S0014-5793(01)02686-2
Scrofani SDB, Chung J, Huntley JJA, Benkovic SJ, Wright PE, Dyson HJ (1999) Biochemistry 38:14507–14514. doi:10.1021/bi990986t
Huntley JJA, Scrofani SDB, Osborne MJ, Wright PE, Dyson HJ (2000) Biochemistry 39:13356–13364. doi:10.1021/bi001210r
Salsbury FR, Crowley MF, Brooks CL (2001) Protein Struct Funct Genet 44(4):448–459. doi:10.1002/prot.1110
Huntley JJA, Fast W, Benkovic SJ, Wright PE, Dyson HJ (2003) Protein Sci 12:1368–1375. doi:10.1110/ps.0305303
Stote RH, Karplus M (1995) Proteins 23(1):12–31. doi:10.1002/prot.340230104
Gresh N, Sponer J (1999) J Phys Chem B 103(51):11415–11427. doi:10.1021/jp9921351
Pang YP, Xu K, Yazal JE, Prendergas FG (2000) Protein Sci 9(10):1857–1865
Pang YP (2001) Proteins 45(3):183–189. doi:10.1002/prot.1138
Oelschlaeger P, Schmid RD, Pleiss J (2003) Protein Eng 16(5):341–350. doi:10.1093/protein/gzg049
Oelschlaeger P, Schmid RD, Pleiss J (2003) Biochemistry 42(30):8945–8956. doi:10.1021/bi0300332
Carfi A, Duee E, Paul-Soto R, Galleni M, Frere JM, Dideberg O (1998) Acta Crystallogr D Biol Crystallogr 54:45–57. doi:10.1107/S090744499700927X
Galleni M, Lamotte-Brasseur J, Rossolini GM, Spencer J, Dideberg O, Frere JM (2001) Antimicrob Agents Chemother 45(3):660–663. doi:10.1128/AAC.45.3.660-663.2001
Wang Z, Fast W, Benkovic SJ (1998) J Am Chem Soc 120(41):10788–10789. doi:10.1021/ja982621m
Case DA, Cheatham TE III, Darden T, Gohlke H, Luo R, Merz KM et al (2005) J Comput Chem 26(16):1668–1688. doi:10.1002/jcc.20290
Agarwal PK, Billeter SR, Rajagopalan PT, Benkovic SJ, Hammes-Schiffer S (2002) Proc Natl Acad Sci USA 99:2794–2799. doi:10.1073/pnas.052005999
Pineda JR, Schwartz SD (2006) Philos Trans R Soc Lond B Biol Sci 361:1433–1438. doi:10.1098/rstb.2006.1877
Campos-Bermudez VA, Leite NR, Krog R, Costa-Filho AJ, Soncini FC, Oliva G et al (2007) Biochemistry 46(39):11069–11079. doi:10.1021/bi7007245
Tomatis PE, Rasia RM, Sergobia L, Vila AJ (2005) Proc Natl Acad Sci USA 102(29):13761–13766. doi:10.1073/pnas.0503495102
Oelschlaeger P, Schmid P, Pleiss J (2003) Biochemistry 42(30):8945–8956
Oelschlaeger P, Mayo S, Pleiss J (2005) Protein Sci 14(3):765–774
Irwin JJ, Raushel FM, Shoichet BK (2005) Biochemistry 44(37):12316–12328. doi:10.1021/bi050801k
Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W et al (2003) J Comput Chem 24:1999–2012. doi:10.1002/jcc.10349
Ertl P, Muhlbacher J, Rohde B, Selzer P (2003) SAR QSAR Environ Res 14(5–6):321–328 http://www.molinspiration.com/jme/ doi:10.1080/10629360310001673917
Concha NO, Janson CA, Rowling P, Pearson S, Cheever CA, Clarke BP et al (2000) Biochemistry 39:4288–4298. doi:10.1021/bi992569m
Kurosaki H, Yamaguchi Y, Higashi T, Soga K, Matsueda S, Yumoto H et al (2005) Angew Chem Int Ed Engl 44:3861–3864. doi:10.1002/anie.200500835
Wang J, Wolf RM, Caldwell JW, Kollamn PA, Case DA (2004) J Comput Chem 25(9):1157–1174. doi:10.1002/jcc.20035
Acknowledgments
Some of these calculations were performed on the DEAC cluster at Wake Forest University (http://www.deac.wfu.edu). We thank the WFU Information Systems for their support of the cluster, WFU’s Department of Physics and Associate Provost for Research for their funding of the cluster, and IBM for their generous support through a SUR grant to provide disk space. J.H. acknowledges partial support of this publication by NIH Grant Number RR-16480 from the NM-INBRE Program of the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH). We thank an anonymous reviewer for suggesting the hydrogen bond analysis.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1
Structural comparison of Apo, pose 2 and the structure with the longest zinc–zinc distance. The centroids of the most populated clusters in the apo (gray), and pose 2 (red) simulations are depicted along with the structure with the largest zinc–zinc distance (orange), which is found in pose 2. The protein is depicted in the new cartoon representation, and the zinc atoms in vdW representation (GIF 819 KB).
Figure S2
Detailed structural comparison of Apo, pose 2 and the structure with the longest zinc–zinc distance. As Fig. S1, but zoomed in with all atoms within 5 Å of the zinc atoms depicted in a bonded representation to highlight the loop flip and the slight structural changes at the zinc site (GIF 109 MB).
Rights and permissions
About this article
Cite this article
Salsbury, F.R., Crowder, M.W., Kingsmore, S.F. et al. Molecular dynamic simulations of the metallo-beta-lactamase from Bacteroides fragilis in the presence and absence of a tight-binding inhibitor. J Mol Model 15, 133–145 (2009). https://doi.org/10.1007/s00894-008-0410-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-008-0410-0