Abstract
Copahue is a natural geothermal field (Neuquén province, Argentina) dominated by the Copahue volcano. As a consequence of the sustained volcanic activity, Copahue presents many acidic pools, hot springs and solfataras with different temperature and pH conditions that influence their microbial diversity. The occurrence of microbial biofilms was observed on the surrounding rocks and the borders of the ponds, where water movements and thermal activity are less intense. Microbial biofilms are particular ecological niches within geothermal environments; they present different geochemical conditions from that found in the water of the ponds and hot springs which is reflected in different microbial community structure. The aim of this study is to compare microbial community diversity in the water of ponds and hot springs and in microbial biofilms in the Copahue geothermal field, with particular emphasis on Cyanobacteria and other photosynthetic species that have not been detected before in Copahue. In this study, we report the presence of Cyanobacteria, Chloroflexi and chloroplasts of eukaryotes in the microbial biofilms not detected in the water of the ponds. On the other hand, acidophilic bacteria, the predominant species in the water of moderate temperature ponds, are almost absent in the microbial biofilms in spite of having in some cases similar temperature conditions. Species affiliated with Sulfolobales in the Archaea domain are the predominant microorganism in high temperature ponds and were also detected in the microbial biofilms.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
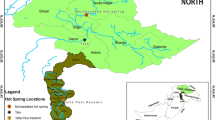
The Copahue geothermal area is located in the Cordillera de Los Andes in the North West of Neuquén province (Patagonia) Argentina. The map in Fig. 1 shows the location of the study area in Argentina and the sampling sites. Copahue is crowned by the still active Copahue volcano, located 2965 m above sea level. Approximately 100 m below the crater there are two hydrothermal springs that are the source of Río Agrio, an acidic river that flows down the Copahue–Caviahue valley. Associated with the sustained volcanic activity, the area presents geothermal activity, which is reflected in ponds, pools, hot springs and solfataras with different pH, and temperature conditions. The Copahue geothermal area has very interesting geochemical features; especially attractive for studying prokaryotic biodiversity are the elevated acidity of the aquatic environments (Río Agrio and the springs) and the accumulation of different sulphur and iron compounds and minerals (Mas et al. 1996; Parker et al. 2008; Varekamp et al. 2009). These extreme conditions impact on the microbial community that colonise the different habitats found in Copahue (Urbieta et al. 2014). In the past few years our research group has been studying the prokaryotic biodiversity in Copahue by multiple approaches, either by using molecular ecology techniques, such as FISH and 16S rRNA gene sequencing, on environmental samples (Urbieta et al. 2014, 2012), or by enrichment cultures searching for acidophilic bacteria (Lavalle et al. 2005; Chiacchiarini et al. 2010) or novel thermoacidophiles, like the recently presented archaea Acidianus copahuensis (Giaveno et al. 2013). Our results in previous studies on water samples from Río Agrio and the geothermal ponds showed the presence of different acidophilic bacteria and archaea, most of them related to sulphur and iron cycles. So far, the microbial community that inhabits acidic microbial biofilms in Copahue has not been studied.
Microbial biofilms are common in diverse habitats, from soil, terrestrial and coastal habitats (Cohen and Gurevitz 1992), to others with extreme conditions, such as hypersaline areas, alkaline lakes or hot springs (Ward et al. 1989), although there are not many reports of microbial mats or biofilms from acidic environments. Microbial biofilms and mats in hot springs commonly occur as gelatinous or calcareous mats of several centimetres in thickness and often with various colours. The uppermost layer is generally composed of photosynthetic Cyanobacteria and diatoms and the orange, yellow, red or flesh colour layer usually of filamentous bacteria, primarily Chloroflexi (Ward et al. 1998). Cyanobacteria from geothermal springs can produce extracellular pigments capable to prevent the cells from UV radiation damages and generally are the primary producers in these kind of habitats where grazing is limited (Roeselers et al. 2007). In this way bacteria inhabiting in the deeper layers benefit from this shield and also from the carbon compounds synthesised by Cyanobacteria (Stal 2000). Environmental factors such as temperature or water chemistry may affect the species composition and community structure of hot spring microbial biofilms and mats (Skirnisdottir et al. 2000). For instance, Cyanobacteria are not usually observed in hot springs with pH below 4.0, and their ability to develop in acidic environments has been questioned by several authors (Brock 1973; Steinberg et al. 1998; González-Toril et al. 2003).
The aim of this study is to compare the prokaryotic biodiversity found in the water of the ponds and hot springs and in microbial biofilms, highlighting the presence of photosynthetic species as important members of the prokaryotic community in microbial biofilms in the Copahue geothermal springs.
Materials and methods
Sample collection and physicochemical determinations
Samples of water and microbial biofilms were collected in December 2009 from four different ponds in the Copahue geothermal area. The four ponds were selected to represent the different physicochemical conditions, mainly temperature and pH, found in the Copahue geothermal area. Temperature and pH were measured in situ with a Hanna HI 8424 NEW portable instrument properly calibrated against calibration standards.
Samples for chemical analysis
The samples were analysed by inductively coupled plasma mass spectrometry (ICP-MS). None of the most relevant elements related with volcanic origin or geothermal activity (Mg, Ca, Mn, Ni Cu, Pb, Cr, Zn, Cd, B, As) were detected in concentrations over 1.0 mg/L. The concentration of sulphate was determined by a turbidimetric method using an excess of barium chloride.
Water samples for DNA extraction
Approximately 1 litre of water was collected in sterile plastic jars and kept on ice. As soon as possible samples were filtered through 0.22 µm Millipore membranes. Filtrates were used for chemical analysis and material retained on membranes for DNA extraction. Filters were washed with pH 2 sterile water and TE buffer (10 mM Tris–HCl pH 8.0, 1 mM EDTA) to remove any acidic water containing heavy metals that may cause DNA hydrolysis (Herrera and Cockell 2007). Filters were stored at −20 °C until further processing.
Water samples for fluorescence in situ hybridisation
Samples were fixed on the field. Between 200 and 500 µL of water sample were incubated with the corresponding volume of paraformaldehyde (PFA) to reach a 4 % final concentration. Samples were incubated for 4–12 h, then diluted in approximately 15 mL of pH 2 sterile water and finally filtered through a GTTP 0.25 Millipore filter (0.22 µm) using a filtration column. Filters were washed and neutralised with 20 mL of PBS buffer (130 mM NaCl, 7 mM Na2HPO4, 3 mM NaH2PO4, pH 7.2) and air dried. Fixed samples were stored at −20 °C until hybridisation reaction.
Microbial biofilms samples for DNA extraction
Portions of the microbial biofilms were collected in separate 50 mL sterile polypropylene tubes and kept at 4 °C until further processing.
DNA extraction, amplifications and 16S ribosomal RNA clone library construction
Total genomic DNA was extracted from the water and microbial biofilms samples using the Fast DNA Spin kit for soil (Bio 101, Carlsbad, CA, USA) according to the manufacturer’s instructions. Cells were disrupted using the mixture of ceramic and silica beads provided in the kit and a laboratory vortex at maximum speed for 10 min. Clone libraries of complete 16S rRNA genes for Bacteria and Archaea domains were generated from DNA templates. 16S rRNA genes were amplified by PCR using forward primers 8F: 5′-AGAGTTTGATC(A/C)TGGC-3′ for Bacteria and 25F: 5′-TCYGGTTGATCCYGCCRG-3′ for Archaea; reverse primer for both was 1492R: 5′-TACCTTGTTACGACTT-3′ (Lane 1991; Achenbach and Woese 1999). Primer numbers correspond to Escherichia coli positions. PCR conditions were as follows: initial denaturation at 95 °C for 5 min, followed by 38 cycles of denaturation at 95 °C for 1 min, annealing temperature was 46 °C for Bacteria domain primers and 50 °C for Archaea domain primers and maintained for 1 min, finally extension at 72 °C for 1 min. Amplification reactions contained 20–30 ng of DNA per 50 µL reaction volume, 1× PCR buffer (Promega Biotech), 2.5 mM of each of the deoxynucleotides, 2.5 mM MgCl2, 500 mM of the forward and reverse primers and 0.025 U/mL of Taq DNA polymerase (Promega Biotech). PCR amplification was checked by 1.2 % agarose gel electrophoresis stained with ethidium bromide. Amplified 16S rRNA gene products (>1400 bp) were cloned using the Topo Ta Cloning Kit (Invitrogen CA, USA) and sequenced using a Big-Dye sequencing kit (Applied Biosystem) following the manufacturer’s instructions.
Sequences were checked for potential chimeras using Bellerophon Chimera Check program (http://greengenes.lbl.gov/cgi-bin/nph-bel3_interface.cgi) and Mallard software (Ashelford et al. 2006). Sequences detected as chimeras were retrieved from further analysis. A distance matrix generated using Greengenes online tool (http://greengenes.lbl.gov/cgi-bin/nph-distance_matrix.cgi) was used as the input file to distance-based OTU and richness software (Schloss 2005) which assigns sequences to operational taxonomic units (OTUs) for every possible distance (rarefaction curves are presented in Figures S1A and S1B of Supplementary Information). Phylogenetic classification of representative OTUs was done using the Classifier and Taxomatic online tools of the ribosomal database project (RDP) (http://rdp.cme.msu.edu). Nearly full-length (>1400 bp) sequence were imported to a data base of over 50000 prokaryotic 16S rRNA primary structures using the ARB software package aligning tool (http://www.arb-home.de) (Ludwig et al. 2004). OTUs were defined at 97 % sequence similarity using ARB software package. The rRNA alignments were corrected manually and alignment uncertainties were omitted in the phylogenetic analysis. Phylogenetic trees were constructed using ARB tools with Maximum likelihood and PhyML correlation. Filters, which excluded highly variable positions, were used. The retained positions were more than 1000 bp in all the cases. The 16S rRNA sequences representing the OTUs selected were deposited on NCBI data base under the accession numbers JX989227 to JX989264 and KP204487 to KP204546.
FISH and CARD-FISH
Fluorescent in situ hybridisation (FISH) was performed on water samples. Hybridisations were done using fluorescent-labelled probes as described by (Amann 1995). The probes used in this study are listed in Table S1 in the Supplementary Information section. Due to the presence of large amount of autoflorescence material, hybridisations on Las Maquinitas (LMi) water sample were performed using catalysed reported deposition fluorescent in situ hybridisation (CARD-FISH). The protocol reported by Pernthaler et al. (2002) was used except that no overnight treatment with active diethyl pyrocarbonate was done, as the samples did not show high endogenous peroxidase activity. For further cell permeabilisation filters were treated with achromopeptidase (0.6 U/mL final concentration; buffer contained 0.01 M NaCl, 0.01 M Tris–HCl pH 8.0; incubation at 37 °C for 30 min) and then washed with ultra pure water for 1 min. Peroxidases were inhibited by treating the filters with 20 % methanol and 0.015 % H2O2 solution for 30 min at room temperature. 4′,6′-Diamidino-2-phenylindole (DAPI ) stain was used in all hybridisations to evaluate total cells number. Vectashield Mounting Medium (Vector Laboratories Inc. CA, USA) was added to preparations to avoid fluorescence fading. A Leica DM 2500 epifluorescence microscope was used to visualise hybridisation. Images were taken using a Leica DFC 300 FX camera and its corresponding software (Leica Microscopy Systems Ltd, Heerburgg, Switzerland). Total cell density was calculated as the average of at least 50 DAPI stained fields. Hybridisation percentages for universal probes were calculated as the quotient of the average count of 20 hybridised fields over the average count of those same fields stained with DAPI. Non-specific hybridisation was discarded in all the samples by the use of NON338 probe.
Results and discussion
Sampling sites
Biodiversity and community structure were analysed in microbial biofilms and water samples from Copahue geothermal area. Figure 1 shows photographs of the sampling sites: Las Máquinas (LMa), Baño 9 (B9), Laguna Verde Este (LVE) and LMi. In a geothermal environment like Copahue, microbial biofilms tend to develop in the vicinity of ponds, hot springs and solfataras, where thermal activity is less intense. The four microbial biofilms sampled presented moderate temperature and pH values between 2.7 and 4.8. The water of the ponds were more acidic (pH values between 2.0 and 3.2) and their temperature greatly differed, from moderate to very high values, close to water boiling point at Copahue’s altitude. Table 1 presents temperature and pH values, as well as sulphate and iron concentrations measured in the samples analysed in this study.
Biodiversity in microbial biofilms
The OTUs found in microbial biofilm samples, together with the number of clones of each OTU, their closest BLAST hit and their accession number, are listed in Table S2 in the Supplementary Information section. Figure 2 shows the relative abundances of the OTUs found in water and microbial biofilm samples, with special emphasis on acidic and photosynthetic species.
According to RDP Classifier tool the sequences retrieved in the microbial biofilm samples were affiliated with the following taxonomical groups: Acidobacteria, Bacteroidetes, Caldilineae, Chloroflexi, Deinococci, Firmicutes, Alpha-, Beta-, Delta- and Gammaproteobacteria, Nitrospira, Cyanobacteria and chloroplasts of different photosynthetic species (the Classifier tool of RDP includes the 16SrRNA sequence of chloroplasts in a phylum named Cyanobacteria/Chloroplast) and Thermoplasmatales, Sulfolobales, Thermoproteales and Thermoprotei in the domain Archaea.
Among the microorganisms found in the microbial biofilms the photosynthetic species are particularly interesting as they were not detected at all in the water samples (Fig. 2). The phylogenetic distribution of their representative OTUs can be seen in the tree of Fig. 3 and the taxonomical classification according to RDP and their closest BLASTn match can be found in Table S3 of the Supplementary Information section. Among the photosynthetic species detected, those associated to the phylum Cyanobacteria/Chloroplasts (according RDP taxonomical classification tool) were the best represented. The phylogenetically related group taxonomy is the one used by RDP and consequently the one adopted in this work. However, for some authors is believed to be artificial and not reflective of evolutionary relationships, and will likely be revised, or even replaced, as more genetic data become available (Vincent 2009). Recent studies on taxonomy and phylogeny of Cyanobacteria agree that classification should include genetic, phylogenetic, ultrastructural and phenotypic data to create a modern system (Howard-Azzeh et al. 2014; Shi and Falkowski 2008; Komárek 2006; Tomitani et al. 2006; Hoffmann et al. 2005).
Continuing with the description of the photosynthetic species found in the microbial biofilms in the Copahue geothermal ponds, six OTUs were affiliated to the Cyanobacteria class. Three of them, represented by sequences LMa-biof-bac-h7, LVE-biof-bac-a9 and LVE-biof-bac-b10 (Fig. 3; Table S3) could not be further classified. The first two showed 94 % similarity to sequences of Synechococcus elongatus in a BLASTn search. These Cyanobacteria, which belong to the Chroococcales order, are oxygenic phototrophs that can photolyse either H2O or H2S. They are the main source of primary production in oligotrophic aquatic environments and are able to grow at a wide range of light intensities (Scanlan and Nyree 2002). As regards biotechnological applications, Synechococcus species have been used in the production of biofuel and different bioactive compounds (Abed et al. 2009; Machado and Atsumi 2012) The OTU represented by sequence LVE-biof-bac-b10 showed 94 % similarity to sequences of Leptolyngbya antarctica, a species of Cyanobacteria that has been reported in many biodiversity studies done in Antarctica (Taton et al. 2006; Komárek 2007). It is curious to notice in the phylogenetic tree (Fig. 3) that these two sequences clustered together with other Leptolyngbya spp. in a separated and robust clade (99 % bootstrap value) distant from the rest of the known Leptolyngbya species. These three OTUs might point out Copahue’s potential as the habitat of novel uncharacterized Cyanobacteria species which might have possible biotechnological applications.
One of the most abundant OTUs in the microbial biofilms samples, represented by sequence LMa-biof-bact_d8, was affiliated to Cyanobacteria Group I and showed 98–99 % similarity to Mastigocladus/Fischerella like species. The genera Mastigocladus and Fischerella are very similar and have a confused taxonomic history because representatives of both taxa were collected from a hot spring in Czech Republic within 15 years of the description of each other (Kaštovský and Johansen 2008). They belong to the order Stigonemataceae and are filamentous thermophilic Cyanobacteria typical of hot springs. As thermophilic species, Mastigocladus/Fischerella show higher uptake of CO2 than their mesophilic counterparts; such metabolic feature makes them very interesting biotechnological tools in applications such as bioremediation and biofuel production (Kotelev et al. 2013). Other Cyanobacteria detected in the microbial biofilms were classified according to RDP as members of Group V (Table S3) and related by BLASTn search to the genus Leptolyngbya. The OTUs are represented by sequences LMa-biof-bact_d4 and LVE-biof-bact_h9. Notice their position in the phylogenetic tree (Fig. 3) close to other well-recognised Leptolyngbya species. This genus of filamentous Cyanobacteria belongs to the Oscillatoriales order and has been reported in high temperature environments (Castenholz 1996, 2001). As many other Cyanobacteria, Leptolyngbya species have been studied for their potential use in hydrogen and bioactive compound production (Prabaharan et al. 2010; Choi et al. 2010; Thornburg et al. 2011) as well as bioremediation of hydrocarbon contaminated environments (Al-Bader et al. 2013).
Five OTUs were related to the 16S rRNA genes of chloroplast of photosynthetic eukaryotes (class Chloroplast in Table S3 according to the taxonomical classification used by RDP for 16S rRNA gene sequences), two in the genus Bacillariophyta and three in Chlorophyta. Both genera include thermophilic photosynthetic species. The OTUs of chloroplasts from Bacillariophyta, represented by sequences LMi-biof-bact_c1 and B9-biof-bact_h5, were more than 98 % similar to chloroplast sequences found in environments affected by acid mine drainage (González-Toril et al. 2011; unpublished information under NCBI access number AJ536452). The OTUs of chloroplasts of species in the genus Chlorophyta showed low similarities to chloroplast sequences of Scenedesmus obliquus (90 %) and uncultured Chlorella sp. (95 %). The apparent difference to well-characterized species is also reflected in the relative position of the sequences in the phylogenetic tree (Fig. 3). For deeper study of the eukaryotes present in the microbial biofilms we attempted the construction of 18S rRNA gene clone libraries. Unfortunately most of the sequences obtained were of low quality and a comprehensive analysis was not possible. However, we could detect sequences related to Scenedesmus, Chaetophora, Pinnularia, Semispathidium and Platyreta genera (data not shown). It is interesting to notice that no Cyanobacteria species were found in the samples B9-biof and LMi-biof that presented pH values lower than 4 (2.7 and 3.5, respectively) in agreement with the reports of other authors (Brock 1973; Steinberg et al. 1998; González-Toril et al. 2003).
In the phylum Chloroflexi two OTUs were detected. One was related to the genus Roseiflexus, which comprises thermophilic bacteria able to grow photoheterotrophically under anaerobic light conditions and form red mats in natural environments (Hanada et al. 2002). The OTU found in Copahue, represented by sequence LMa-biof-bact_f11 was 98 % similar to a sequence reported in an orange surface microbial mat in El Tatio Geyser Field in Chile (Engel et al. 2013). The other OTU was 98 % similar to Chloroflexus aurantiacus, a photoheterotrophic green non-sulphur bacteria also detected in El Tatio Geyser Field (Engel et al. 2013) as well as in other extreme environments microbial mats (Nübel et al. 2001). Species of the phylum Chloroflexi, especially Chloroflexus, are commonly found in microbial mats, growing under a cyanobacterial layer, taking advantage of the UV light protection and the production of organic compounds (Ruff-Roberts et al. 1994; Boomer et al. 2002); the formation of these photosynthetic multilayer structures has been proven in thermal springs in Yellowstone National Park (Boomer et al. 2009).
Photosynthetic species were also found among Alphaproteobacteria. One OTU was classified within the Rhodospirillaceae, a family composed mainly of photosynthetic purple non-sulphur bacteria. Sequences related to the genus Rhodobacter were also detected in Copahue microbial biofilms. These are purple non-sulphur bacteria that include species with an extensive range of metabolic capabilities, such as lithoautotrophy, photosynthesis, aerobic and anaerobic respiration. As regards microbial diversity, the dominance of Cyanobacteria, Chloroflexus and eventually Proteobacteria has been reported in other microbial mats and biofilms (Coman et al. 2013; Ferris et al. 1996).
Apart from the photosynthetic species described, other bacteria were detected in the microbial biofilms studied. One OTU was classified into genus Meiothermus within the phylum Deinococcus-Thermus and was 99 % similar to Meiothermus hypogaeus, a thermophilic bacterium isolated from a neutrophilic hot spring in Japan (Mori et al. 2012). Another OTU was affiliated to the genus Geothrix in the phylum Acidobacteria and, according to BLASTn, was 99 % similar to an uncultured bacterium detected in acidic Rio Tinto in Spain (Amaral-Zettler et al. 2011). Sequences related to genus Thiomonas, commonly found in natural or related to mining activity acidic environments (Urbieta et al. 2014; Hamamura et al. 2009; Hallberg et al. 2005), were the only sulphur oxidising bacteria detected. The almost absence of sulphur oxidising species is in correlation with the undetection of sulphate ion (the end product of sulphur compounds biooxidation) in the microbial biofilm samples (Table 1). Finally, some sequences were related to species commonly found in soil or aquatic environments, such as Pseudomonas, Aeromonas and Clostridium.
As regards archaea, the 16S rRNA sequences obtained from genomic DNA from the microbial biofilms were affiliated to phyla Euryarchaeota and Crenarchaeota (Table S4). Within Euryarchaeota, the five OTUs detected were classified into the order Thermoplasmatales (Fig. 4a). Two of them could not be further classified and showed over 97 % similarity to sequences reported in a copper mine (Xie et al. 2007) and in different hot springs in Yellowstone National Park (unpublished, information available in NCBI under accession number DQ179028). Another OTU was related to Ferroplasma species, a genus of acidophilic, lithoautotrophic or mixotrophic archaea capable of iron and pyrite oxidation that are very important members of the microbial community in acidic, heavy metal reach environments (Golyshina and Timmis 2005). The Ferroplasma-like sequences found in Copahue microbial biofilms are 99 % similar to sequences detected in Rio Agrio in Copahue and in Rio Tinto in Spain (Urbieta et al. 2012; Amaral-Zettler et al. 2011) (see sequences in purple in Fig. 4a). Other two OTUs were classified in the genus Thermogymnomonas; particularly the one represented by sequence LMi-biof-arch d6 was over 99 % similar to sequences detected in Rio Agrio in Copahue (Urbieta et al. 2012) (Fig. 4a).
Phylogenetic representation of the archaea found in the water and microbial biofilms in Copahue geothermal area: a Euryarchaeota, b Crenarchaeota. Sequences in purple were detected in Rio Agrio (Urbieta et al. 2012)
The sequences in the phylum Crenarchaeota were all affiliated to the class Thermoprotei (Table S4; Fig. 4b). The OTU represented by sequence LMi-biof-arch-b11 could not be further classified and was distantly related to an archaeal clone from Norris Geyser Basin in Yellowstone National Park (unpublished, information available under NCBI accession number DQ924756). The other crenarchaeal OTUs were related to thermophilic species like Sulfolobus, Pyrobaculum and Vulcanisaeta. The genus Sulfolobus has been described long ago by Brock (Brock et al. 1972) as strict aerobic, acidophilic, thermophilic, sulphur oxidizing archaea, typical of geothermal environments. Pyrobaculum species are neutrophilic and extreme thermophiles, capable of growing up to 104 °C; the species are either facultative aerobic or strictly anaerobic and they can grow either chemolithoautotrophically by sulphur reduction or organotrophically by sulphur respiration or by fermentation (Volkl et al. 1993). Species from the genus Vulcanisaeta are anaerobic, hyperthermophilic and moderately acidophilic, they grow heterotrophically and are able to use thiosulphate as electron acceptor (Itoh et al. 2002). Considering the temperature measured in the microbial biofilms samples probably the 16S rRNA gene sequences of these extreme thermophilic archaea corresponds to species that are not metabolically active in the microbial biofilms but develop in high temperature environments nearby, such as the hot springs waters (see the following section).
Biodiversity in water of ponds and hot springs
Prokaryotic biodiversity assessment in water samples was approached parallel by two techniques: bacterial and archaeal 16S rRNA cloning and sequencing and in situ hybridisation. The OTUs found in water samples, together with the number of clones of each OTU and their closest BLAST hit and their accession number, are listed in Table S5 in the Supplementary Information section. Table S6 presents absolute counts and hybridisation percentages of the probes used. Figure 5 shows hybridisation percentages of total microorganisms stained with DAPI for probes EUB338 (Bacteria domain) and ARQ915 (Archaea domain) for LMa, LVE, B9 and LMi water samples. Figure 6 shows epifluorescence microscope images of these hybridisations and their corresponding DAPI stains to provide a quantitative idea of the abundances of the two domains in the different ponds studied. These results show that LMa and LVE were dominated by bacteria, with 84 and 86 % of hybridisation with the bacterial probe EUB338, respectively. On the other hand, B9 and LMi samples were dominated by archaea (88 and 91 % of hybridisation with ARQ915 probe, respectively). Considering that FISH reveals only viable cells, as labile RNAs are the probes target, we used hybridisations counts to guide the biodiversity analysis in the four water ponds studied.
Epifluorescense microscopic images showing the prevailing domain in water samples of the Copahue geothermal ponds. On the left side, DAPI stained all the microorganisms present. On the right side, hybridisations with bacteria general probe (EUB338) or archaea general probe (ARQ915). EUB338 probes in LMa FISH hybridisation and in LMi CARD-FISH hybridisation are labelled with the green fluorochrome Alexa 488 (green emission). The other FISH probes used are labelled with red CY3 flourochrome (red emission)
In LMa and LVE, bacterial community was more deeply analysed (Fig. 2; Table S7). In these two acidic, moderate temperature ponds, the bacterial 16S rRNA gene clones libraries were similar. In LMa1, the majority of the clones were approximately 99 % similar to species of Thiomonas. These are moderately acidophilic, facultative chemolithotrophic or mixotrophic bacteria, able to grow on some metal sulphides or sulphur (Moreira and Amils 1997) and have been reported in other acidic environments, including those related with mining activity (Hallberg et al. 2005). The second better represented bacteria in LMa waters were related to Acidithiobacillus species. This genus is composed of aerobic, obligate acidophilic and lithoautotrophic bacteria. Acidithiobacillus species are able to obtain energy from the oxidation of sulphur compounds, and some of them from the oxidation of ferrous iron (Kelly and Wood 2000). In the waters of LVE, Thiomonas- and Acidithiobacillus-related species were also important members of the pond’s bacterial community. Two other species associated with acidic environments appeared in this pond: Acidiphilium and Hydrogenobaculum. The former are aerobic, acidophilic, obligate chemoorganotrophic bacteria that oxidise sulphur compounds and are generally found accompanying Acidithiobacillus, as they metabolise organic compounds which are toxic for lithoautotrophs (Johnson and Hallberg 2003). Hydrogenobaculum are aerobic, acidophilic and thermophilic bacteria that obtain carbon from carbon dioxide fixation and energy from oxidising hydrogen or reduced sulphur compounds (Stohr et al. 2001). Conversely to microbial biofilms, in LMa and LVE water samples the detection of a high proportion of sulphur oxidising species is in good correlation with the high concentrations of sulphate ion measured (Table 1). The bacterial clone library in LVE water sample was completed by the detection of some heterotrophic species, such as Acinetobacter and Pseudomonas, not related with acidic or geothermal environments. Their presence could be explained by the fact that LVE pond is located very near the Copahue thermal health centre, and it has a higher anthropogenic influence.
In the analysis of the prokaryotic community of the other ponds, B9 and LMi, FISH showed that archaea were the dominant species (Figs. 5, 6). The classification of these archaeal OTUs according to RDP is presented in Table S8 in the Supplementary Information section. The water of the pond B9 and chiefly the hot spring LMi presented the highest temperature values and the most acidic pH conditions (Table 1). An interesting fact about the archaea found in the high-temperature ponds in Copahue is that, while the majority of the bacteria sequences were more than 97 % similar to known and cultivated microorganisms, almost all the archaea sequences were less than 97 % similar to any cultivated species, and in some cases even to uncultured clones (Tables S5; S8). According to these results, the Copahue geothermal field could be the habitat of potentially novel, possibly autochthonous, archaea. The archaeal community in B9 waters was dominated by sequences affiliated to the Thermoplasmatales family in the Euryarchaeota phylum represented by sequences B9-arch-f6 and B9-arch h12 (Fig. 4a; Table S8). A minor fraction, represented by sequence B9-ach-d12, was affiliated to the phylum Crenarchaeota, specifically to the genus Sulfolobus. It is interesting to notice in the phylogenetic trees of Fig. 4 that the archaeal sequences found in the waters of B9 form long branches within well-established classes, but distant from other reported cultured or uncultured species. In LMi, the extreme physical conditions of this hot spring seem to be reflected in its archaeal population, as the 95 clones analysed belonged to only one OTU affiliated with the Sulfolobus genus.
The species related to the sequences found in Copahue water ponds and hot springs are mainly related to the sulphur cycle. The geochemical conditions of the area, particularly the acidity, the reported abundance of sulphur compounds produced by the sustained volcanic activity (Mas et al. 1996; Varekamp et al. 2009; Urbieta et al. 2014) and the measured concentrations of ion sulphate (the final product or sulphur compounds biooxidation) suggest that the sulphur cycle plays a key role in the geochemistry of this environment.
Comparison of microbial species in water and microbial biofilms in Copahue geothermal springs and in other geothermal environments
In spite of some cases presenting similar pH and temperature conditions, microbial biofilms and the water of the ponds and hot springs are different environments within the Copahue geothermal area, which is reflected in their different prokaryotic community composition. The samples B9-biof, LMi-biof, B9 and LMi, have significant difference in temperature and at lesser extent in pH values, with more extreme conditions in the water samples. These marked differences in physicochemical conditions help to explain better their microbial community composition: B9 and LMi water samples are dominated by archaea, with a high proportion of Sulfolobales, particularly in LMi; while the biofilm samples show a quite diverse microbial community that includes photosynthetic eukaryotic species such as Chlorophyta and Bacillariophyta.
In general, the different nature of both environments analysed in this study have a marked impact on their microbial biodiversity. In microbial biofilms, prokaryotic communities are more diverse and photosynthetic species are present while acidophilic sulphur oxidising bacteria, the main prokaryotes found in the water samples become far less important. That could be related with another important difference in physicochemical parameters between microbial biofilms and water samples; in the former no ion sulphate (the final product of sulphur compounds oxidation) was detected while in water samples the concentration was in all cases (except LMa) higher than the normal values for fresh water (between 10 and 250 mg/L). The sequences related to the genus Thiomonas were the only species of acidophilic sulphur oxidising bacteria present in the microbial biofilms. In fact, Thiomonas were the only bacterial species detected in both habitats analysed in this work. This result could be considered as an indication of the importance of Thiomonas in the microbial community and biogeochemistry of Copahue. On the other hand, no photosynthetic species of any kind were detected in the water of any of the ponds or hot springs studied.
The archaeal community was similar in the water and microbial biofilms samples studied. The species found in both environments are similar to sequences associated with Thermoplasmatales and Sulfolobales (Fig. 2). It is particularly interesting to notice in the phylogenetic tree of the Euryarchaeota phylum (Fig. 4a) that many of the sequences detected in the microbial biofilms clustered together with sequences found in Rio Agrio (coloured in purple), also part of the Copahue geothermal area. Similarly, the Crenarchaeota phylum tree (Fig. 4b) shows that the only one archaeal OTU found in the waters of LMi hot spring (LMi87-arch-f1) is highly related to one of the OTUs found in the microbial biofilms (LMi-biof-arch-f8). These findings support the idea that Copahue geothermal area posses a common autochthonous and apparently biogeographically determined archaeal community.
The microbial diversity of the Copahue geothermal system can be compared with that of other similar systems, natural or artificial, around the globe. Rio Tinto, in southern Spain is one of the most studied natural acidic environments. The prokaryotic biodiversity in its water column as well as in the macroscopic filaments found in still water areas is dominated by iron oxidising species such as At. ferrooxidans, L. ferrooxidans and Acidiphilium spp. (García-Moyano et al. 2007; González-Toril et al. 2003). The difference in prokaryotic community composition between Rio Tinto and Copahue is mainly related with the fact that the Spanish river has very high iron concentrations (over 1000 mg/L) compared with the ones measured in Copahue. In algal photosynthetic biofilms in Rio Tinto, species of the eukaryotes Euglena, Pinnularia and Chlorella have been reported but no Cyanobacteria, probably due to the low pH of the river (Souza-Egipsy et al. 2008; Amaral-Zettler et al. 2011). As regards archaeal community, all the mentioned studies on Rio Tinto coincide that Thermoplasmata, particularly Ferroplasma species similar to those detected in Copahue are dominant, but there are no records of Crenarchaeota from Rio Tinto. When considering acidic environments of anthropogenic origin, a study done on macroscopic biofilms in acidic, metal-rich waters in an abandoned copper mine and a chalybeate spa in North Wales showed that the biodiversity was more related to the one described in Copahue moderate temperature water samples than in microbial biofilms, although the Welsh biofilms showed a higher presence of iron oxidising species such as At. ferrooxidans, Acidiphilium, Ferromicrobium, Thiomonas and no detection of archaea or photosynthetic species (Hallberg et al. 2005). Similar biodiversity, with a higher proportion of Leptospirillum, was detected in a biofilm in an acid mine drainage site (Bond et al. 2000). Another very well studied geothermal area is the Uzon Caldera at Kamchatka in Russia; there the microbial community presented some similarities with Copahue, with a marked presence of chemolithotrophic species such as Hydrogenobaculum in the most acidic sites (as the area is rich in As) and dominance of Chloroflexus, Deltaproteobacteria and Clostridia in the less acidic pools (Burgess et al. 2012). A study done specifically in microbial mats revealed the presence of many photosynthetic species similar to those found in Copahue (Ward et al. 1994). A similar study done in sulphur and non-sulphur microbial mats in solfataric fields in south-western Iceland showed similar results to those presented in this study, where the sulphur reach samples were dominated by chemolithotrophic sulphur oxidising species and there were no photosynthetic species reported, whereas the non-sulphur mats were dominated by Chloroflexus, photoheterotrophic green non-sulphur bacteria, with very little influence of sulphur oxidisers (Skirnisdottir et al. 2000). In a study of bacterial and archaeal biodiversity of microbial mats from a slightly alkaline geothermal region in Romania, similar Cyanobacteria and Chloroflexi species were found, however, the other bacteria and chiefly the archaeal community were completely different, more related to methanogenic species such as Methanomassiliicoccus and Methanococcus (Coman et al. 2013). Recently, a study has been published describing the morphological and phylogenetic diversity of Cyanobacteria in Algerian hot springs (temperatures ranging from 39 to 93 °C and pH from 6 to 7) where the hottest springs were dominated by Leptolyngbya, Synechococcus-like Cyanobacteria and Gloeomargarita, whereas Oscillatoriales Chroococcales and Stigonematales dominated lower temperature springs (Amarouche-Yala et al. 2014). It is curious to notice that two of the Cyanobacteria species that dominate the very high temperature mats were also present in the moderate temperature microbial mats in Copahue.
Conclusion
The comparison between the prokaryotic biodiversity of the water of ponds and hot springs and the microbial biofilms found in Copahue geothermal area showed two different ecological niches dominated by different species. In the water samples, the dominant prokaryotes were chemolithoautotrophic or mixotrophic, mainly sulphur oxidising, bacteria in the moderate temperature ponds and archaea in the high-temperature hot springs; while in microbial biofilms photosynthetic species, such as Cyanobacteria and the eukaryotes Bacilariophyta and Chlorophyta were the most important primary producers.
References
Abed RM, Dobretsov S, Sudesh K (2009) Applications of Cyanobacteria in biotechnology. J Appl Microbiol 106:1–12
Achenbach L, Woese C (1999) 16S and 23S rRNA-like primers. In: Sower KR, Schreier HJ (eds) Archaea: a laboratory manual. Cold Spring Harbor Laboratory Press, New York, pp 521–523
Al-Bader D, Eliyas M, Rayan R, Radwan S (2013) Air–dust-borne associations of phototrophic and hydrocarbon-utilizing microorganisms: promising consortia in volatile hydrocarbon bioremediation. Environ Sci Pollut Res 19:3997–4005
Amann RI (1995) In: van Elsas JD, de Bruijn FJ (eds) situ identification of microorganisms by whole cell hybridization with rRNA-targeted nucleic acid probes, In: Akkermans ADL. Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, pp 1–15
Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA (1990) Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol 56:1919–1925
Amaral-Zettler LA, Zettler ER, Theroux SM, Palacios C, Aguilera A, Amils R (2011) Microbial community structure across the tree of life in the extreme Rio Tinto. ISME J 5:42–50
Amarouche-Yala S, Benouadah A, Bentabet AEO, López-García P (2014) Morphological and phylogenetic diversity of thermophilic Cyanobacteria in Algerian hot springs. Extremophiles. doi:10.1007/s00792-014-0680-7
Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ (2006) New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl Environ Microbiol 72:5734–5741
Bond PL, Banfield JF (2001) Design and performance of rRNA targeted oligonucleotide probes for in situ detection and phylogenetic identification of microorganisms inhabiting acid mine drainage environments. Microb Ecol 41:149–161
Bond PL, Smriga SP, Banfield JF (2000) Phylogeny of microorganisms populating a thick, subaerial, predominantly lithotrophic biofilm at an extreme acid mine drainage site. Appl Environ Microbiol 66:3842–3849
Boomer SM, Lodge DP, Dutton BE, Pierson B (2002) Molecular characterization of novel red green nonsulfur bacteria from five distinct hot spring communities in Yellowstone National Park. Appl Environ Microbiol 68:346–355
Boomer SM, Noll KL, Geesey GG, Dutton BE (2009) Formation of multilayered photosynthetic biofilms in an alkaline thermal spring in Yellowstone National Park, Wyoming. Appl Environ Microbiol 75:2464–2475
Brock TD (1973) Lower pH limit for the existence of blue-green algae: evolutionary and ecological implications. Science 179:480–483
Brock TD, Brock KM, Belly RT, Weiss RL (1972) Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Archiv für Mikrobiologie 84:54–68
Burgess AE, Unrine JM, Mills GL, Romanek CS, Wiegel J (2012) Comparative geochemical and microbiological characterization of two thermal ponds in the Uzon Caldera, Kamchatka, Russia. Microb Ecol 63:471–489
Castenholz RW (1996) Endemism and biodiversity of thermophilic Cyanobacteria. Nova Hedwig 112:33–47
Castenholz RW (2001) Phylum BX. Cyanobacteria: oxygenic photosynthetic bacteria. In: Garrity GM, Boone DR, Castenholz RW (eds) Bergey’s manual of systematic bacteriology. Springer, New York, pp 473–600
Chiacchiarini P, Lavalle L, Giaveno A, Donati E (2010) First assessment of acidophilic microorganisms from geothermal Copahue–Caviahue system. Hydrometallurgy 104:334–341
Choi H, Engene N, Smith JE, Preskitt LB, Gerwick WH (2010) Crossbyanols A-D, toxic brominated polyphenyl ethers from the Hawaiian bloom-forming cyanobacterium Leptolyngbya crossbyana. J Nat Prod 73:517–522
Cohen Y, Gurevitz M (1992) The Cyanobacteria: ecology, physiology and molecular genetics. In: Balows A, Trüper HG, Dworkin M, Harder W, Schleifer KH (eds) The Prokaryotes, 2nd edn. Springer, New York, pp 2079–2104
Coman C, Drugă B, Hegedus A, Sicora C, Dragoş N (2013) Archaeal and bacterial diversity in two hot spring microbial mats from a geothermal region in Romania. Extremophiles 17:523–534
Daims H, Bruhl A, Amann R, Schleifer KH, Wagner M (1999) The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol 22:434–444
Engel AS, Johnson LR, Porter ML (2013) Arsenite oxidase gene diversity among Chloroflexi and Proteobacteria from El Tatio Geyser Field, Chile. FEMS Microbiol Ecol 83:745–756
Ferris MJ, Muyzer G, Ward DM (1996) Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl Environ Microbiol 62:340–346
García-Moyano A, González-Toril E, Aguilera A, Amils R (2007) Prokaryotic community composition and ecology of floating macroscopic filaments from an extreme acidic environment, Rio Tinto (SW, Spain). Syst Appl Microbiol 30:601–614
Giaveno MA, Urbieta MS, Ulloa R, González-Toril E, Donati ER (2013) Physiologic versatility and growth flexibility as the main characteristics of a novel thermoacidophilic Acidianus strain isolated from Copahue geothermal area in Argentina. Microb Ecol 65:336–346
Golyshina OV, Timmis KN (2005) Ferroplasma and relatives, recently discovered cell wall-lacking archaea making a living in extremely acid, heavy metal-rich environments. Environ Microbiol 7:1277–1288
González-Toril E, Llobet-Brossa E, Casamayor EO, Amann R, Amils R (2003) Microbial ecology of an extreme acidic environment, the Tinto River. Appl Environ Microbiol 69:4853–4865
González-Toril E, Aguilera A, Souza-Egipsy V, Pamo EL, España JS, Amils R (2011) Geomicrobiology of La Zarza-Perrunal acid mine effluent (Iberian Pyritic Belt, Spain). Appl Env Microbiol 77:2685–2694
Hallberg KB, Coupland K, Kimura S, Johnson DB (2005) Macroscopic streamer growths in acidic, metal rich mine waters in North Wales consist of novel and remarkably simple bacterial communities. Appl Environ Microbiol 72:2022–2030
Hamamura N, Macur RE, Korf S, Ackerman G, Taylor WP, Kozuba M, Inskeep WP (2009) Linking microbial oxidation of arsenic with detection and phylogenetic analysis of arsenite oxidase genes in diverse geothermal environments. Environ Microbiol 11:421–431
Hanada S, Takaichi S, Matsuura K, Nakamura K (2002) Roseiflexus castenholzii gen. nov., sp. nov., a thermophilic, filamentous, photosynthetic bacterium that lacks chlorosomes. Int J Syst Evol Microbiol 52:187–193
Herrera A, Cockell CS (2007) Exploring microbial diversity in volcanic environments: a review of methods in DNA extraction. J Microbiol Methods 70:1–12
Hoffmann L, Komárek J, Kaštovský J (2005) System of cyanoprokaryotes (Cyanobacteria)—state in 2004. Algol Stud 117:95–115
Howard-Azzeh M, Shamseer L, Schellhorn HE, Gupta RS (2014) Phylogenetic analysis and molecular signatures defining a monophyletic clade of heterocystous Cyanobacteria and identifying its closest relatives. Photosynth Res 122:171–185
Itoh T, Suzuki KI, Nakase T (2002) Vulcanisaeta distributa gen. nov., sp. nov., and Vulcanisaeta souniana sp. nov., novel hyperthermophilic, rod-shaped crenarchaeotes isolated from hot springs in Japan. Int J Syst Evol Microbiol 52:1097–1104
Johnson DB, Hallberg K (2003) The microbiology of acidic mine waters. Res Microbiol 154:466–473
Kaštovský J, Johansen JR (2008) Mastigocladus laminosus (Stigonematales, Cyanobacteria): phylogenetic relationship of strains from thermal springs to soil-inhabiting genera of the order and taxonomic implications for the genus. Phycology 47:307–320
Kelly DP, Wood AP (2000) Reclassification of some species of Thiobacillus to the newly designated genera Acidithiobacillus gen. nov., Halothiobacillus gen. nov. and Thermithiobacillus gen. nov. Int J Syst Evol Microbiol 50:511–516
Komárek J (2006) Cyanobacterial taxonomy: current problems and prospects for the integration of traditional and molecular approaches. Algae 21:349–375
Komárek J (2007) Phenotype diversity of the cyanobacterial genus Leptolyngbya in maritime Antarctica. Pol Polar Res 28:211–231
Kotelev MS, Antonov IA, Beskorovainyi AV, Vinokurov VA (2013) Photobioreactor operation condition optimization for high-energy cyanobacterial biomass synthesis to produce third-generation biofuels. Chem Tech Fuels Oils 49:1–4
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematic. Wiley, Chichester, pp 115–175
Lavalle L, Chiacchiarini P, Pogliani C, Donati E (2005) Isolation and characterization of acidophilic bacteria from Patagonia, Argentina. Process Biochem 40:1095–1099
Ludwig W, Strunk O, Westram R, Richter L, Meier H, Buchner A, Schleifer KH (2004) ARB: a software environment for sequence data. Nucleic Acids Res 32:1363–1371
Machado IMP, Atsumi S (2012) Cyanobacterial biofuel production. J Biotechnol 162:50–56
Manz W, Amann R, Ludwig W, Wagner M, Schleifer KH (1992) Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst Appl Microbiol 15:593–600
Mas G, Mas L, Bengochea L (1996) Hydrothermal, surface alteration in the Copahue geothermal field (Argentina). In: Proceedings twenty-fifth workshop on geothermal reservoir engineering, Stanford University. Stanford, California, pp 22–24
Massana R, Murray AE, Preston CM, Delong EF (1997) Vertical distribution and phylogenetic characterization of marine planktonic Archaea in the Santa Barbara Channel. Appl Environ Microbiol 63:50–56
Moreira D, Amils R (1997) Phylogeny of Thiobacillus cuprinus and other mixotrophic Thiobacilli: proposal for Thiomonas gen. nov. Int J Syst Evol Microbiol 47:522–528
Mori K, Iino T, Ishibashi J, Kimura H, Hamada M, Suzuki K (2012) Meiothermus hypogaeus sp. nov., a moderately thermophilic bacterium isolated from a hot spring. Int J Syst Evol Microbiol 62:112–117
Neef A (1997) Anwendung der in situ-Einzelzell-Identifizierung von Bakterien zur Populations analyse in komplexen mikrobiellen Biozonösen. Ph.D. thesis. Technical University of Munich, Munich, Germany
Nübel U, Bateson MM, Madigan MT, Kühl M, Ward DM (2001) Diversity and distribution in hypersaline microbial mats of bacteria related to Chloroflexus spp. Appl Environ Microbiol 67:4365–4371
Parker SR, Gammons CH, Pedrozo FL, Wood SA (2008) Diel changes in metal concentrations in a geogenically acidic river: Rio Agrio, Argentina. J Volcanol Geoth Res 178:213–223
Pernthaler A, Pernthaler J, Amann R (2002) Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl Environ Microbiol 68:3094–3101
Prabaharan D, Arun-Kumar D, Uma L, Subramanian G (2010) Dark hydrogen production in nitrogen atmosphere, an approach for sustainability by marine cyanobacterium Leptolyngbya valderiana BDU 20041. Int J Hydrog Energ 35:10725–10730
Roeselers G, Norris TB, Castenholz RW, Rysgaard S, Glud RN, Kühl M, Muyzer G (2007) Diversity of phototrophic bacteria in microbial mats from Arctic hot springs (Greenland). Environ Microbiol 9:26–38
Ruff-Roberts AL, Kuenen JG, Ward DM (1994) Distribution of cultivated and uncultivated Cyanobacteria and Chloroflexus-like bacteria in hot spring microbial mats. Appl Environ Microbiol 60:425–468
Rusch A, Amend JP (2004) Order-specific 16S rRNA-targeted oligonucleotide probes for (hyper)thermophilic archaea and bacteria. Extremophiles 8:357–366
Scanlan DJ, Nyree J (2002) Molecular ecology of the marine cyanobacterial genera Prochlorococcus and Synechococcus. FEMS Microbiol Ecol 40:1–12
Schloss PD (2005) Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol 71:1501–1506
Shi T, Falkowski PG (2008) Genome evolution in cyanobacteria: the stable core and the variable shell. Proc Natl Acad Sci USA 105:2510–2515
Skirnisdottir S, Hreggvidsson GO, Hjörleifsdottir S, Marteinsson VT, Petursdottir SK, Holst O, Kristjansson JK (2000) Influence of sulfide and temperature on species composition and community structure of hot spring microbial mats. Appl Environ Microbiol 66:2835–2841
Souza-Egipsy V, González-Toril E, Zettler E, Amaral-Zettler LA, Aguilera A, Amils R (2008) Prokaryotic community structure in algal photosynthetic biofilms from extreme acidic streams in Río Tinto (Huelva, Spain). Int Microbiol 11:251–260
Stal LJ (2000) Cyanobacterial mats and stromatolites. In: Whitton BA, Potts M (eds) The ecology of cyanobacteria: their diversity in time and space. Kluwer Academic Publishers, Dordrecht, pp 61–120
Steinberg CEW, Schäfer H, Beisker W (1998) Do acid-tolerant Cyanobacteria exist? Acta Hydroch Hydrob 26:13–19
Stohr R, Waberski A, Volker H, Tindall BJ, Thomm M (2001) Hydrogenothermus marinus gen. nov., sp. nov., a novel thermophilic hydrogen-oxidizing bacterium, recognition of Calderobacterium hydrogenophilum as a member of the genus Hydrogenobacter and proposal of the reclassification of Hydrogenobacter acidophilus as Hydrogenobaculum acidophilum gen. nov., comb. nov., in the phylum ‘Hydrogenobacter/Aquifex’. Int J Syst Evol Microbiol 51:1853–1862
Taton A, Grubisic S, Balthasart P, Hodgson DA, Laybourn-Parry J, Wilmotte A (2006) Biogeographical distribution and ecological ranges of benthic Cyanobacteria in East Antarctic lakes. FEMS Microbiol Ecol 57:272–289
Thornburg CC, Thimmaiah M, Shaala LA, Hau AM, Malmo JM, Ishmael JE, McPhail KL (2011) Cyclic depsipeptides, grassypeptolides D and E and Ibu-epidemethoxylyngbyastatin 3 from a red sea Leptolyngbya cyanobacterium. J Nat Prod 74:1677–1685
Tomitani A, Knoll AH, Cavanaugh CM, Ohno T (2006) The evolutionary diversification of Cyanobacteria: molecular–phylogenetic and paleontological perspectives. Proc Natl Acad Sci USA 103:5442–5447
Urbieta MS, González-Toril E, Giaveno MA, Aguilera A, Donati E (2012) First prokaryotic biodiversity assessment using molecular techniques of an acidic river in Neuquén, Argentina. Microb Ecol 64:91–104
Urbieta MS, González-Toril E, Aguilera A, Giaveno MA, Donati E (2014) Archaeal and bacterial diversity in five different hydrothermal ponds in the Copahue region in Argentina. Syst Appl Microbiol 37:429–441
Varekamp JC, Ouimette A, Herman S, Flynn KS, Bermúdez AH, Delpino DH (2009) Naturally acid waters from Copahue volcano, Argentina. Appl Geochem 24:208–220
Vincent WF (2009) Cyanobacteria. In: Likens GE (ed) Plankton of Inland Waters. Elsevier, London, pp 226–231
Volkl P, Huber R, Drobner E, Rachel R, Burggraf S, Trincone A, Stetter KO (1993) Pyrobaculum aerophilum sp. nov., a novel nitrate-reducing hyperthermophilic archaeum. Appl Environ Microbiol 59:2918–2926
Ward DM, Weller R, Shiea J, Castenholz RW, Cohen Y (1989) Hot spring microbial mats: anoxygenic and oxygenic mats of possible evolutionary significance. In: Cohen Y, Rosenburg E (eds) Microbial mats: physiological ecology of benthic microbial communities. American Society for Micriobiology, Washington, DC, pp 2–15
Ward DM, Ferris MJ, Nold SC, Bateson MM, Kopczynski ED, Ruff-Roberts AL (1994) Species diversity in hot spring microbial mats as revealed by both molecular and enrichment culture approaches—relationship between biodiversity and community structure. In: Stal LJ, Caumette P (eds) Microbial mats: structure, development and environmental significance. Springer, Heidelberg, pp 33–44
Ward DM, Ferris MJ, Nold SC, Bateson MM (1998) A natural view of microbial biodiversity within hot spring cyanobacterial mat communities. Microbiol Mol Biol Rev 62:1353–1370
Xie X, Xiao S, He Z, Liu J, Qiu G (2007) Microbial populations in acid mineral bioleaching systems of Tong Shankou Copper Mine, China. J Appl Microbiol 103:1227–1238
Acknowledgments
This work was partially supported by Grants PIP 0348 (CONICET), PICT-2012-0623 (ANPCYT) and MICINN-Spain (CGL2011-22540).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Oren.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Urbieta, M.S., González-Toril, E., Bazán, Á.A. et al. Comparison of the microbial communities of hot springs waters and the microbial biofilms in the acidic geothermal area of Copahue (Neuquén, Argentina). Extremophiles 19, 437–450 (2015). https://doi.org/10.1007/s00792-015-0729-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-015-0729-2