Abstract
Overlapping genes (OGs) draw the focus of recent day’s research. However, the significance of OGs in prokaryotic genomes remained unexplored. As an adaptation to high temperature, thermophiles were shown to eliminate their intergenic regions. Therefore, it could be possible that prokaryotes would increase their OG content to adapt to high temperature. To test this hypothesis, we carried out a comparative study on OG frequency of 256 prokaryotic genomes comprising both thermophiles and non-thermophiles. It was found that thermophiles exhibit higher frequency of overlapping genes than non-thermophiles. Moreover, overlap frequency was found to correlate with optimal growth temperature (OGT) in prokaryotes. Long overlap frequency was found to hold a positive correlation with OGT resulting in an abundance of long overlaps in thermophiles compared to non-thermophiles. On the other hand, short overlap (1–4 nucleotides) frequency (SOF) did not yield any direct correlation with OGT. However, the correlation of SOF with CAIavg (extent of variation of codon usage bias measured as the mean of codon adaptation index of all genes in a given genome) and IG% (proportion of intergenic regions) indicate that they might upregulate the aforementioned factors (CAIavg and IG%) which are already known to be vital forces for thermophilic adaptation. From these evidences, we propose that the OG content bears a strong link to thermophily. Long overlaps are important for their genome compaction and short overlaps are important to uphold high CAIavg. Our findings will surely help in better understanding of the significance of overlapping gene content in prokaryotic genomes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Every gene has its unique and distinct genomic location. However, two adjacent genes are often observed to share their coding sequences with each other, i.e., their coding sequences overlap partially or entirely. For instance, in E. coli enterotoxin gene astA was shown to be completely embedded in transposase-like gene IS1414 (McVeigh et al. 2000). Several genomic analyses have inferred that these overlapping genes are present in a wide range of taxonomic groups including viruses (Chirico et al. 2010; Pavesi 2006; Pavesi et al. 2013; Rancurel et al. 2009; Simon-Loriere et al. 2013) prokaryotes (Cock and Whitworth 2007, 2010; Johnson and Chisholm 2004; Sabath et al. 2008; Sakharkar et al. 2005), plants (Kunova et al. 2012; Quesada et al. 1999) animals, (Makalowska et al. 2007; Sanna et al. 2008; Veeramachaneni et al. 2004) and fungi (Gerads and Ernst 1998; Williams et al. 2005). Moreover, the existence of overlapping genes has been experimentally validated in prokaryotes as well as in eukaryotes (McVeigh et al. 2000; Szklarczyk et al. 2007).
Overlapping genes can be classified broadly into two types: (1) same strand overlapping genes, which are transcribed from the same strand of DNA (also known as parallel or unidirectional overlaps); (2) different strand overlapping genes that are transcribed from two opposite strands of DNA (also known as anti-parallel overlaps) (Sanna et al. 2008). Anti-parallel overlaps are further subdivided into convergent (−><−) and divergent types (<− −>). Convergent type involves the overlapping of 3′ ends and is also denoted as tail-to-tail overlaps, whereas divergent type entails the overlapping of 5′ ends and is often denoted as head-to-head overlap. Different species have different abundance and distributions of parallel and anti-parallel overlaps (Sanna et al. 2008). In bacterial genome, same strand overlaps are more common (Johnson and Chisholm 2004), whereas genomes of higher eukaryotes such as human and mouse configure 90 % of overlapping genes in anti-parallel orientation (Sanna et al. 2008).
There could be varied reasons behind the fixation and maintenance of gene overlaps in different species. The potential role of OGs in gene expression regulation has been demonstrated in Arabidopsis genome (Kunova et al. 2012). An overlapping gene astA that entirely overlapped with IS1414, encode heat-stable enterotoxin 1 (EAST 1) in enteroaggregative E. coli (EAEC). This further emphasizes the role of overlapping genes in bacterial pathogenesis as well (McVeigh et al. 2000). It has already been mentioned that OGs in prokaryotic genomes may cause genome compaction (Sakharkar et al. 2005). However, in viruses, other than genome compaction, OGs may create a new gene that encodes a novel protein necessary for viral adaptation to host by a mechanism, commonly known as “overprinting” (Pavesi 2006). In addition, translational coupling of functionally related genes are favored by gene overlaps in the phage genome (Inokuchi et al. 2000).
Prokaryotes are often subjected to environmental changes. Changes in habitat temperature is one such environmental challenges that prokaryotes are frequently exposed to. High temperature often denatures protein (Lepock et al. 1988), disrupts membrane structure and integrity (Baatout et al. 2005) and hampers cell homeostasis. Therefore, high temperatures could pose severe threat to cell survival. In order to combat this adverse or stressful condition, some prokaryotes have evolved several molecular strategies. These include: increase in purine content of mRNA (Lao and Forsdyke 2000; Das et al. 2006; Lambros et al. 2003), rRNA secondary structure stability (Galtier and Lobry 1997), reduction of protein structural disorder (Burra et al. 2010) and more importantly, the reduction of genome size that facilitates thermophilic adaptation of bacteria (Sabath et al. 2013). Bacterial overlapping genes originate through simple extension or elongation of one coding frame into another due to the absence of stop codon in the coding region (Fukuda et al. 1999). Recently, it was proposed that prokaryotes could reduce their genome size by elimination of intergenic regions (genomic streamlining) in response to rise in growth temperature (Sabath et al. 2013). It has already been mentioned that OGs facilitate genome compaction (Sakharkar et al. 2005). However, no studies have ever linked OG content of prokaryotic genome to their growth temperatures. Hence, we intended to investigate whether there is an abundance of OGs in thermophilic prokaryotes compared to non-thermophilic ones. An in-depth comparison of OGs between 50 thermophilic and 206 non-thermophilic genomes revealed that OGs have significant contribution in thermophilic stress tolerance of prokaryotic genomes which leads to their enrichment in thermophilic genomes. Moreover, we have also illustrated the different mechanisms of long (7–50 nucleotides) and short (1–4 nucleotides) overlaps in acclimatizing higher temperature. Thus our study would surely help in better understanding of the mechanisms of thermophilic stress tolerance.
Materials and methods
Retrieval of genomic information
The dataset of this study comprises 256 unique prokaryotes for which, gene overlap data as well as Optimal Growth Temperature (OGT) data were available (Additional file 1). OGT data were obtained from NCBI database (ftp://www.ncbi.nlm.nih.gov/genomes/genomeprj_archive/) and supplementary dataset of Wang et al. (2006). Overlapping gene data were retrieved from Pairwise neighbours database (Palleja et al. 2009). Overlap frequency was calculated as the total number of adjacent gene pairs that overlap divided by total number of adjacent gene pairs in that genome (Sabath et al. 2008). Long overlaps were defined as overlaps spanning a region of 7–50 nucleotides, whereas short overlaps are defined as overlaps spanning a region of 1–4 nucleotides (Fonseca et al. 2014). Spacer length data were extracted from pairwise neighbours database. Proportion of the intergenic region (IG %) was calculated as sum of all spacer length between all adjacent gene pairs divided by genome size.
Genome size information for each genome was obtained from (ftp://www.ncbi.nlm.nih.gov/genomes/genomeprj_archive/lproks_0.txt).
Average Codon Adaptation Index (CAIavg) values of each prokaryotic genome were taken from the supplementary material of Botzman and Margalit (2011).
Calculation of overlap formation frequency
For the calculation of overlap formation frequency in response to change in optimal growth temperature, we considered nine thermophilic–mesophilic pairs that have been previously studied by McDonald (2010). Members of these nine thermophilic–mesophilic pairs were phylogenetically close, exhibited similar GC content but differ in their habitat temperature (Table 2). Additionally, we chose one mesophilic–psychrophilic pair to study the changes in overlap frequency during cold stress. One-to-one orthologous genes between the members of those meso-thermo and meso-psychro pairs were retrieved from OMA genome browser (Altenhoff et al. 2011). For each genome we first estimated the number of OGs that have detectable orthologs in the paired genome. Next, we calculated the number of newly formed overlapping gene in a genome as the number of genes that rendered overlapping in that genome but whose orthologs rendered non-overlapping in the paired genome. Overlap formation frequency of each genome in each pair was estimated as the ratio of number of newly formed OGs divided by the number of OGs that have detectable orthologs in its counter genome. Overlap formation frequency in thermophilic, mesophilic and psychrophilic were denoted as OGthermo, OGmeso, OGpsychro, respectively.
Statistical analyses
All statistical analyses were conducted using the software SPSS version 13. All types of overlap frequencies were found to be non-parametric in Shapiro–Wilk test (P < 0.05). Following non-parametric distribution we used Mann–Whitney U test to detect significant difference in the distribution of different variables between two groups. Spearman’s correlation tests were performed to analyze the correlations between different variables.
Results
Thermophiles have higher OG frequency compared to non-thermophiles
Here, we found a strong negative correlation between OG frequency and the percentage of intergenic DNA in a genome (IG%) (Spearman’s ρ = −0.328; P = 10−6; N = 256). Hence, it is logical to hypothesize that the presence of OGs could be a critical feature of thermophilic prokaryotes. To validate our hypothesis, we conducted a study on 256 distinct prokaryotes (Additional file 1) out of which 50 were thermophiles (optimal growth temperature ≥45 °C) and 206 non-thermophiles (temperature ranges from 16.5 to 42 °C). We calculated overlap frequency for each genome mentioned above and compared the mean overlap frequency of thermophiles with that of non-thermophiles. The results of Mann–Whitney U test including statistical parameters, P value and mean overlap frequency of both the groups (thermophiles and non-thermophiles) were enlisted in Table 1. Our results show that thermophiles have an elevated frequency of gene overlaps than non-thermophiles. We also observed a weak but significant correlation between optimal growth temperature and gene overlap frequency (Spearman’s ρ = 0.197; P = 1.3 × 10−3; N = 256). This observation prompted us to perform further investigation on the role of OGs in acclimatizing thermal stress.
In order to study how shift in growth temperature modulates overlap frequency in prokaryotic genomes, we estimated gene overlap formation frequencies in nine thermophilic–mesophilic genome pairs that have been previously studied by McDonald (2010) (details given in “Materials and methods” section). Additionally, we chose one meso-psychro pair to study the changes in gene overlap formation frequency during cold stress. Our detailed analysis revealed that in eight out of nine meso-thermo pairs overlap formation frequency in thermophilic genomes (OGthermo) was significantly higher than overlap formation frequency in mesophilic genomes (OGmeso) (Table 2). For the meso-psychro pair, OGmeso was also found to be significantly higher than OGpsychro (overlap formation frequency in psychrophilic genomes) (Table 2).
Effect of long and short overlaps in regulating thermophilic stress
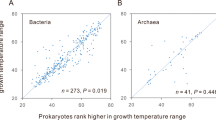
We compared the distribution of short (1–4 nucleotides) and long overlap (7–50 nucleotides) frequency between non-thermophiles and thermophiles in our dataset. We obtained a very weak, but significant difference of short overlap frequency between non-thermophiles and thermophiles, whereas the difference of long overlap frequency was quite pronounced in thermophiles compared to non-thermophiles (Table 3). Hence, it would be interesting to examine whether short and long overlaps differ in their impact on the process of acclimatization to a higher temperature range. Long overlap frequency (7–50 nucleotides) was found to yield a significant and robust correlation with optimal growth temperature (Spearman’s ρ = 0.489; P = 10−6; N = 256) (Fig. 1) while short overlap frequency (1–4 nucleotides) did not hold any significant correlation with optimal growth temperature (Spearman’s ρ = 0.062; P = 0.263; N = 256) (Fig. 2). In order to test how LOF is associated with the degree of thermophilicity, we considered their correlation in thermophilic and non-thermophilic groups separately. LOF was found to correlate with OGT in both thermophilic and non-thermophilic group (Spearman’s ρ Thermo = 0.704; P = 10−6; N = 50 and Spearman’s ρ Non-thermo = 0.320; P = 3 × 10−6; N = 206) (Fig S1 and S2, additional file2). Here, we also noticed that LOF and OGT shares significant correlation in archae (37 genomes) (Spearman’s ρ archae = 0.754; P = 10−6) and eubacterial domains (219 genomes) (Spearman’s ρ eubacteria = 0.404; P = 10−6). In our dataset, many overlaps were detected to exceed 50 nucleotides in length. So, we were also curious to study the relationship of these types of overlaps (>50 nucleotides) with temperature. We have also found a significant correlation between very long overlap frequency (>50 nucleotides) and OGT (Spearman’s ρ = 0.276; P = 8 × 10−5; N = 256). Interestingly, comparison of the correlation coefficients between long (7–50 nucleotides) as well as very long overlaps (>50 nucleotides) with OGT, using Steiger’s Z test has shown that long overlap frequency has a more profound effect over temperature compared to very long overlaps (>50 nucleotides) (Z = 3.92; P < 0.01).
Next, we analyzed the role of long and short overlap frequencies in the process acclimatization to high temperature. Since, genome size compaction is an exclusive phenomenon of thermophilic stress tolerance, we decided to focus on the impact of long and short overlap frequency over genome size. We have performed a non-parametric Spearman’s correlation test between genome size and long overlap frequency (LOF) as well as short overlap frequency (SOF). We found a significant negative correlation between LOF and genome size (Spearman’s ρ = −0.548; P = 10−6; N = 256) which indicates that LOF has a significant contribution in genome compaction. Surprisingly, we obtained a positive correlation between SOF and genome size (Spearman’s ρ = 0.168; P = 0.007; N = 256). It was also proposed that genome size in prokaryotes associates positively with genomic GC content (Hildebrand et al. 2010). Furthermore, the short overlap frequency in prokaryotic genomes increases gradually with genomic GC content (Fonseca et al. 2014). Hence, we found it essential to investigate the correlation between genome size and both long and short overlap frequency after controlling for genomic GC content. In both the cases, i.e., LOF and SOF yielded a negative correlation with genome size after controlling GC content (Spearman’s ρ LOF = −0.424; P = 10−5; r 2 = 0.180 Spearman’s ρ SOF = −0.277; P = 1 × 10−4; r 2 = 0.077; N = 256). It is apparent from the r 2 values of the two correlations that LOF accounts for 18 % variation of genome size and SOF accounts for 7.7 % variation of genome size. Therefore, our results indicate that both short and long overlaps participate in genome compaction but the effect of long overlaps on genome size were far more pronounced compared to short overlaps.
OG frequency and other salient factors influencing optimum growth temperature
According to Sabath et al. (2008) the variability of codon usage may influence OG frequency in prokaryotic genomes (Sabath et al. 2008). Consequently, Botzman and Margalit (2011) pointed out that growth temperature has a robust impact over codon usage bias of prokaryotic genomes (Botzman and Margalit 2011). In the previous section, OG frequency was found to be correlated with both intergenic regions (IG%) and growth temperature (OGT). Hence, it would be interesting to explore whether CAIavg and IG% has any influence over both long and short overlaps, respectively. Table 4 clearly shows that both SOF and LOF hold significant correlation with CAIavg and IG%. Abundance of short overlap frequency in thermophilic genomes even in the absence of any direct correlation with growth temperature suggests that the short overlap frequency may have guided by any other confounding factors. Therefore, high CAIavg and low IG% may instigate the rise SOF in thermophilic genomes. To quantitate and evaluate the contribution of each factor related to growth temperature in our study, we did a multivariate linear regression analysis. Table 5 delineates the result of this multivariate regression analysis. The result indicates that the genomic factors studied here guide growth temperature in the following order: long overlap frequency (LOF) > CAIavg > genome size. We found no significant effect of IG% and SOF on optimal growth temperature. So, from our study it is evident that the long overlap frequency is the strongest potential factor regulating OGT.
Discussion
This study reveals a robust association between overlapping gene content and optimal growth temperature of prokaryotic genomes. The probability of gene overlapping is markedly increased when two neighboring genes are brought closer to each other with minimal intergenic region. This is the reason why overlapping genes are very common in operons where genes are arranged co-directionally, separated by short intergenic spacers (Moreno-Hagelsieb and Collado-Vides 2002). Previously, it has been observed that thermophiles have more structured genomic architecture, where genes are very frequently organized into operons (Yoon et al. 2011). These operons are seldom disrupted by genetic rearrangements in thermophiles (Yoon et al. 2011). These observations prompted us to study whether there is any association between OG content and OGT in prokaryotes. We found a significant difference of overlap frequency between thermophiles and non-thermophiles. In addition, a significant strong correlation between overlap frequency and OGT was also observed. Moreover, multivariate regression analysis revealed a robust impact of LOF over OGT. This further implied that like many other genomic features, OG content is determined by the environmental factor like OGT and increase in OG content could be an effective strategy of prokaryotes in combating thermal stress.
Many theoretical studies as well as experimental evidences suggest that the last universal common ancestor (LUCA) of both archae and bacteria lived in (hyper) thermophilic environment (Akanuma et al. 2013; Brooks et al. 2004; Di Giulio 2003). Therefore, we find it interesting to investigate whether during the evolution of thermophilic ancestor to mesophilic or psychrophilic successor; there exist a selection against the trait of gene overlapping. In order to address this issue, we have chosen ten prokaryotic pair, members of nine of these pairs were one mesophilic and one thermophilic and the tenth pair had one mesophilic and one psychrophilic member. We took orthologous genes between members of each pairs and conducted an in-depth analysis on the conservation of overlapping relationship between the members of each pair. Here, it was noticed that in the course of evolution of prokaryotes from high growth temperature to relatively colder one, frequency of overlapping gene is markedly reduced. Moreover, our results revealed that members of mesophilic and psychrophilic genomes have consistently lower overlap formation frequency than their thermophilic counterpart. Thus, these results imply that in comparison to the genomes of higher growth temperature, evolutionary pressure for overlap formation is generally relieved in the genomes of low optimal growth temperature.
Length of gene overlap shows a wide variation in prokaryotes (Fonseca et al. 2014). Earlier, Fonseca et al. 2014 reported that, in prokaryotes, selection acts against long overlaps while short overlaps are profusely present in the prokaryotic genomes. Another interesting observation in our study with respect to overlap length is the association of long overlap frequency with OGT which is stronger and more robust than total overlap frequency. It was reported that mechanism of thermophilic adaptation differs between archae and eubacteria (Mizuguchi et al. 2007). Hence, we also investigated whether association between OGT and LOF varies between archae and eubacteria. It was found that association between LOF and OGT were significant in both domains of archae and eubacteria. This further shows that although different mechanisms of thermal stress tolerance exist between archae and eubacteria, an elevated overlap frequency was common between the two superkingdoms. Moreover, our results revealed that frequency of long overlaps consistently increases with increase in optimal growth temperature. Thus, it suggests that long overlap frequency changes with the degree in thermophilicity in prokaryotic genomes. But, strikingly short overlap frequency yielded no such direct correlation with OGT. Therefore, we wondered for an explanation of such an observation. Studies on viral system revealed that the overlapping region encoding simultaneously for two protein products are under stronger selective constraints (Simon-Loriere et al. 2013). Moreover, it has been shown that genes that overlap through their entire length (internal overlaps) are evolutionarily more conserved than the genes that overlap partially (terminal overlaps) (Simon-Loriere et al. 2013). Thus, the length of gene overlap could be regarded as an important factor to modulate the selective constraints on overlapping genes. It is evident that mutations that are neutral or nearly neutral under optimal physiological conditions could become deleterious at high temperature and they are commonly called temperature-sensitive mutations (Drake 2009). For this reason, number of studies have reported that coding regions of thermophilic genomes undergo lower rate of base substitution than the coding regions of non-thermophilic genomes (Drake 2009; Friedman et al. 2004). Therefore, it is logical to hypothesize that the increased selective pressure on the overlapping region may favor overlapping genes to be more abundant in thermophilic genomes compared to non-thermophilic genomes. Moreover, due to more stringent selection on long overlaps, these types of overlaps may have been favored in thermophilic genomes as compared to short overlaps. However, further studies are required to assess the effect of overlap length on overall base substitution rate of a given gene in prokaryotic genomes. In our study short overlaps are found to hold a strong correlation with CAIavg. Since, CAIavg has a broad role in thermophilic adaptation (Botzman and Margalit 2011), it might be possible that along with long overlaps, short overlaps helps in survival at higher temperature through increasing CAIavg. Long overlaps (7–50 nucleotides) has a more pronounced effect over genome size compaction than short overlaps and for this reason they may also be more abundant in thermophiles compared to short overlaps and share a robust correlation with OGT.
From our study, it is evident that genome compaction is the primary reason of association of overlapping gene content to thermophily. However, genome size reduction does not necessarily involve an increase in OG frequency. Genomes of endosymbionts often are of small size but contain large intergenic regions due to their functional constraints (Degnan et al. 2011), and hence, contain limited repertoire of gene overlap. Previous studies (Kelkar and Ochman 2013; Sakharkar and Chow 2005; Sakharkar et al. 2004) showed that loss of genes could result in shortening of genome length. Thermophiles, in contrast increases frequency of gene overlapping in their genomes and shorten intergenic region to reduce their genome sizes. In connection to this, here, we would like to draw an important example of cell size reduction in response to high temperature in marine planktonic bacteria (Chrzanowski et al. 1988), where gradual shrinkage of cell volume was found to be an intrinsic property of the cells in response to rise in OGT. More importantly, genome streamlining is also observed in these groups of marine planktonic bacteria (Swan et al. 2013). Hence, further studies are necessary to explore whether OGT has any impact over cell size reduction in overall prokaryotic world, and if it is then the genome compaction (through rise in OG) could be a primary prerequisite to accommodate the genome into a smaller cellular space.
In summary, our observations and interpretations shed light into a relatively unrecognized facet of genomic adaptation of prokaryotes to extreme temperature, where we find an essential and nontrivial connection between overlapping gene content and thermophily. Our study will surely pave inroads to future research on prokaryotic adaptation to extreme temperature.
Abbreviations
- OG:
-
Overlapping gene
- OGT:
-
Optimal growth temperature
- LOF:
-
Long overlap frequency
- SOF:
-
Short overlap frequency
- CAIavg :
-
Average of codon adaptation index
- IG %:
-
Proportion of intergenic region in a given bacterial genome
References
Akanuma S, Nakajima Y, Yokobori S, Kimura M, Nemoto N, Mase T, Miyazono K, Tanokura M, Yamagishi A (2013) Experimental evidence for the thermophilicity of ancestral life. Proc Natl Acad Sci USA 110:11067–11072
Altenhoff AM, Schneider A, Gonnet GH, Dessimoz C (2011) OMA 2011: orthology inference among 1000 complete genomes. Nucleic Acids Res 39:D289–D294
Baatout S, De Boever P, Mergeay M (2005) Temperature-induced changes in bacterial physiology as determined by flow cytometry. Ann Microbiol 55:73–80
Botzman M, Margalit H (2011) Variation in global codon usage bias among prokaryotic organisms is associated with their lifestyles. Genome Biol 12:R109
Brooks DJ, Fresco JR, Singh M (2004) A novel method for estimating ancestral amino acid composition and its application to proteins of the last universal ancestor. Bioinformatics 20:2251–2257
Burra PV, Kalmar L, Tompa P (2010) Reduction in structural disorder and functional complexity in the thermal adaptation of prokaryotes. PloS One 5:e12069
Chirico N, Vianelli A, Belshaw R (2010) Why genes overlap in viruses. Proc Biol Sci 277:3809–3817
Chrzanowski TH, Crotty RD, Hubbard GJ (1988) Seasonal-variation in cell volume of epilimnetic bacteria. Microb Ecol 16:155–163
Cock PJA, Whitworth DE (2007) Evolution of gene overlaps: relative reading frame bias in prokaryotic two-component system genes. J Mol Evol 64:457–462
Cock PJA, Whitworth DE (2010) Evolution of relative reading frame bias in unidirectional prokaryotic gene overlaps. Mol Biol Evol 27:753–756
Das S, Paul S, Bag SK et al (2006) Analysis of Nanoarchaeum equitans genome and proteome composition: indications for hyperthermophilic and parasitic adaptation. BMC Genomic 7:186
Degnan PH, Ochman H, Moran NA (2011) Sequence conservation and functional constraints in intergenic spacers of reduced genomes of obligate symbiont Buchnera. PloS Genet 7(9):e1002252
Di Giulio M (2003) The universal ancestor was a thermophile or a hyperthermophile: tests and further evidence. J Theor Biol 221:425–436
Drake JW (2009) Avoiding dangerous missense thermophiles display especially low mutation rates. PloS Genet 5:e1000520
Fonseca MM, James Harris D, Posada D (2014) Origin and length distribution of unidirectional prokaryotic overlapping genes. G3 Bethesda 4:19–27
Friedman R, Drake JW, Hughes AL (2004) Genome-wide patterns of nucleotide substitution reveal stringent functional constraints on the protein sequences of thermophiles. Genetics 167:1507–1512
Galtier N, Lobry JR (1997) Relationships between genomic G+C content, RNA secondary structures, and optimal growth temperature in prokaryotes. J Mol Evol 44:632–636
Gerads M, Ernst JF (1998) Overlapping coding regions and transcriptional units of two essential chromosomal genes (CCT8, TRP1) in the fungal pathogen Candida albicans. Nucleic Acid Res 26:5061–5066
Fukuda Y, Washio T, Tomita M (1999) Comparative study of overlapping genes in the genomes of Mycoplasma genitalium and Mycoplasma pneumoniae. Nucleic Acid Res 27:1847–1853
Hildebrand F, Meyer A, Eyre-Walker A (2010) Evidence of selection upon genomic GC content in bacteria. PLoS Genet 6:e1001107
Inokuchi Y, Hirashima A, Sekine Y et al (2000) Role of ribosome recycling factor (RRF) in translational coupling. EMBO J 19:3788–3898
Johnson ZI, Chisholm SW (2004) Properties of overlapping genes are conserved across microbial genomes. Genome Res 14:2268–2272
Kelkar YD, Ochman H (2013) Genome reduction promotes increase in protein functional complexity in bacteria. Genetics 193:303–307
Kunova A, Zubko E, Meyer P (2012) A pair of partially overlapping Arabidopsis genes with antagonistic circadian expression. Int J Plant Genomic 2012:349527
Lambros RJ, Mortimer JR, Forsdyke DR (2003) Optimum growth temperature and the base composition of open reading frames in prokaryotes. Extremophiles 7:443–450
Lao PJ, Forsdyke DR (2000) Thermophilic bacteria strictly obey Szybalski’s transcription direction rule and politely purine-load RNAs with both adenine and guanine. Genome Res 10:228–236
Lepock JR, Frey HE, Rodahl AM, Kruuv J (1988) Thermal-analysis of CHL V79 cells using differential scanning calorimetry—implications for hyperthermic cell killing and the heat shock response. J Cell Physiol 137:14–24
Makalowska I, Lin C-F, Hernandez K (2007) Birth and death of gene overlaps in vertebrates. BMC Evol Biol 7:193
McDonald JH (2010) Temperature at homologous sites in proteins from nine thermophile-mesophiles species pairs. Genome Biol Evol 2:267–276
McVeigh A, Fasano A, Scott DA et al (2000) IS1414, an insertion sequence with a heat-stable enterotoxin gene embedded in a transposase-like gene. Infect Immun 68:5710–5715
Mizuguchi K, Sele M, Cubellis MV (2007) Environment specific substitution tables for thermophilic proteins. BMC Bioinform 8:S15
Moreno-Hagelsieb G and Collado-Vides J (2002) A powerful non-homology method for the prediction of operons in prokaryotes. Bioinformatics: S329–36
Palleja A, Reverter T, Garcia-Vallve S, Romeu A (2009) Pairwise neighbours database: overlaps and spacers among prokaryote genomes. BMC Genomics 10:281
Pavesi A (2006) Origin and evolution of overlapping genes in the family Microviridae. J Gen Virol 87:1013–1017
Pavesi A, Magiorkinis G, Karlin DG (2013) Viral proteins originated de novo by overprinting can be identified by codon usage: application to the “gene nursery” of deltaretroviruses. PloS Comput Biol 9:e1003162
Quesada V, Ponce MR, Micol JL (1999) OTC and AUL1, two convergent and overlapping genes in the nuclear genome of Arabidopsis thaliana. FEBS Lett 461:101–106
Rancurel C, Khosravi M, Dunker AK, Romero PR, Karlin D (2009) Overlapping genes produce proteins with unusual sequence properties and offer insight into de novo protein creation. J Virol 83:10719–10736
Sabath N, Graur D, Landan G (2008) Same-strand overlapping genes in bacteria: compositional determinants of phase bias. Biol Direct 3:36
Sabath N, Ferrada E, Barve A, Wagner A (2013) Growth temperature and genome size in bacteria are negatively correlated, suggesting genomic streamlining during thermal adaptation. Genome Biol Evol 5:966–977
Sakharkar KR, Dhar PK, Chow VTK (2004) Genome reduction in prokaryotic obligatory intracellular parasites of humans: a comparative analysis. Int J Syst Evol Microbiol 54:1937–1941
Sakharkar KR, Chow VTK (2005) Strategies for genome reduction in microbial genomes. Genome Inform 16:69–75
Sakharkar KR, Sakharkar MK, Verma C et al (2005) Comparative study of overlapping genes in bacteria, with special reference to Rickettsia prowazekii and Rickettsia conorii. Int J Syst Evol Microbiol 55:1205–1209
Sanna CR, Li WH, Zhang L (2008) Overlapping genes in the human and mouse genomes. BMC Genomics 9:169
Simon-Loriere E, Holmes EC, Pagan I (2013) The Effect of gene overlapping on the rate of RNA virus evolution. Mol Biol Evol 30:1916–1928
Swan BK, Tupper B, Sczyrba A, Lauro FM, Martinez-Garcia M, Gonzalez JM, Luo H, Wright JJ, Landry ZC, Hanson NW, Thompson BP, Poulton NJ, Schwientek P, Acinas SG, Giovannoni SJ, Moran MA, Hallam SJ, Cavicchioli R, Woyke T, Stepanauskas R (2013) Prevalent genome streamlining and latitudinal divergence of planktonic bacteria in the surface ocean. Proc Natl Acad Sci USA 110:11463–11468
Szklarczyk R, Heringa J, Pond SK, Nekrutenko A (2007) Rapid asymmetric evolution of a dual-coding tumor suppressor INK4a/ARF locus contradicts its function. Proc Natl Acad Sci USA 104:12807–12812
Veeramachaneni V, Makalowski W, Galdzicki M et al (2004) Mammalian overlapping genes: the comparative perspective. Genome Res 14:280–286
Wang HC, Susko E, Roger AJ (2006) On the correlation between genomic G + C content and optimal growth temperature in prokaryotes: data quality and confounding factors. Biochem Biophys Res Commun 342:681–684
Williams BAP, Slamovits CH, Patron NJ et al (2005) A high frequency of overlapping gene expression in compacted eukaryotic genomes. Proc Natl Acad Sci USA 102:10936–10941
Yoon SH, Reiss DJ, Bare JC et al (2011) Parallel evolution of transcriptome architecture during genome reorganization. Genome Res 21:1892–1904
Acknowledgments
We thank two anonymous referees for their helpful suggestions and critical comments. We acknowledge Department of Biotechnology, Govt. of India, for financial help. We also thank Mr. S.K. Gupta for his technical helps.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by H. Atomi.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Saha, D., Panda, A., Podder, S. et al. Overlapping genes: a new strategy of thermophilic stress tolerance in prokaryotes. Extremophiles 19, 345–353 (2015). https://doi.org/10.1007/s00792-014-0720-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-014-0720-3