Abstract
In DNA replication studies, the mechanism for regulation of the various steps from initiation to elongation is a crucial subject to understand cell cycle control. The eukaryotic minichromosome maintenance (MCM) protein complex is recruited to the replication origin by Cdc6 and Cdt1 to form the pre-replication complex, and participates in forming the CMG complex formation with Cdc45 and GINS to work as the active helicase. Intriguingly, Thermoplasma acidophilum, as well as many other archaea, has only one Gins protein homolog, contrary to the heterotetramer of the eukaryotic GINS made of four different proteins. The Gins51 protein reportedly forms a homotetramer (TaGINS) and physically interacts with TaMCM. In addition, TaCdc6-2, one of the two Cdc6/Orc1 homologs in T. acidophilum reportedly stimulates the ATPase and helicase activities of TaMCM in vitro. Here, we found a reaction condition, in which TaGINS stimulated the ATPase and helicase activities of TaMCM in a concentration dependent manner. Furthermore, the stimulation of the TaMCM helicase activity by TaGINS was enhanced by the addition of TaCdc6-2. A gel retardation assay revealed that TaMCM, TaGINS, and TaCdc6-2 form a complex on ssDNA. However, glutaraldehyde-crosslinking was necessary to detect the shifted band, indicating that the ternary complex of TaMCM–TaGINS–TaCdc6-2 is not stable in vitro. Immunoprecipitation experiment supported a weak interaction of these three proteins in vivo. Activation of the replicative helicase by a mechanism including a Cdc6-like protein suggests the divergent evolution after the division into Archaea and Eukarya.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chromosomal DNA replication is essential for cell proliferation. A series of highly regulated processes occurs during DNA replication from initiation to termination to maintain a constant level of genetic material (Masai et al. 2010). The molecular mechanisms of DNA replication have been studied in a variety of organisms from Bacteria, Eukarya, and Archaea (reviewed in Ishino and Ishino 2012; Masai et al. 2010; Mott and Berger 2007). It is very interesting that small circular genomes with defined replication origins are conserved traits in Bacteria and Archaea, while the archaeal proteins involved in DNA replication are not conserved with those in Bacteria, but are highly similar to eukaryotic proteins. The two DNA replication apparatuses are considered to have evolved independently, to the bacterial type and archaeal/eukaryotic type (Leipe et al. 1999). Based on these observations, studies of archaeal DNA replication should help to elucidate the mechanisms common to both the archaeal and eukaryotic systems.
In the eukaryotic replication system, the replicative helicase is recruited to the origin recognition complex (ORC) at the origin by Cdc6 and Cdt1, and the pre-replication complex (pre-RC) is assembled during the G1 phase of the cell cycle. To start the replication process, the replicative helicase unwinds the double-stranded DNA to form the replication forks. Minichromosome maintenance (MCM), a heterohexameric complex (Mcm2–7), is the catalytic component of the replicative helicase, based on evidence that the Mcm4-6-7 subcomplex of HeLa cells exhibited oligonucleotide-displacement activity in vitro (Ishimi 1997). However, the functional configuration of the DNA unwinding machinery during the DNA replication process remained unidentified for a long time.
The Cdc45 protein was found to be involved in the pre-RC with an unknown function in Saccharomyces cerevisiae (Aparicio et al. 1997). The GINS complex, consisting of four different proteins, Sld5, Psf1, Psf2, and Psf3, were also originally identified in S. cerevisiae as essential protein factors for the initiation of DNA replication (Takayama et al. 2003). Immunoprecipitation experiments using Xenopus oocyte extracts detected a physical interaction between MCM, Cdc45, and the GINS complex proteins (Kubota et al. 2003). Subsequently, the complexes containing these three components were isolated from yeast (Gambus et al. 2006), Drosophila (Moyer et al. 2006), and Xenopus oocyte extracts (Pacek et al. 2006). The complex of Cdc45, MCM, and GINS, referred to as the “CMG complex” (Moyer et al. 2006), was immunoaffinity-purified and shown to unwind a 40 nt oligonucleotide annealed to single-stranded DNA in an ATP-dependent manner (Moyer et al. 2006). Cdc45 and GINS are now thought to have structural roles in the replicative helicase core.
In contrast to the heterohexameric MCM complex in Eukarya, most archaeal organisms contain only a single Mcm homolog and the homohexameric complex of the archaeal MCM exhibits distinct DNA helicase activity by itself in vitro (Ishino and Ishino 2012).
The current understanding of the structure–function relationships of the archaeal MCM helicase has been summarized in a review article (Sakakibara et al. 2009). The amino acid sequences of the four subunits, Sld5, Psf1, Psf2, and Psf3, in the eukaryotic GINS complex share some conservation, suggesting that they are ancestral paralogs, and the ORFs encoding the homologs of these Gins proteins were predicted in the archaeal genomes by detailed bioinformatic analyses (Makarova et al. 2005). The sequences of the crenarchaeal homologs are more similar to those of Psf2 and Psf3, while the euryarchaeal homologs are more similar to Sld5 and Psf1 (Makarova et al. 2005). A yeast two-hybrid screen detected an interaction partner of MCM in the crenarchaeon Sulfolobus solfataricus, and it was designated as Gins23. Furthermore, another subunit was identified by the mass spectrometry analysis of an immunoaffinity-purified native GINS complex from a S. solfataricus cell extract, using an anti-Gins23 antibody, and was designated as Gins15, based on its closer similarity to Sld5 and Psf1. Furthermore, a tetrameric structure with a 2:2 molar ratio of Gins23 and Gins15, was identified (Marinsek et al. 2006). We previously characterized the complex of the Gins homologs from Pyrococcus furiosus, including Gins23 and Gins51, and determined a 2:2 ratio in the first report describing a euryarchaeal GINS (Yoshimochi et al. 2008). Our chromatin immunoprecipitation assays revealed that P. furiosus GINS (PfuGINS) is located preferentially at the oriC region of P. furiosus chromosomal DNA during the exponential growth phase, as also reported for P. furiosus MCM (PfuMCM) (Matsunaga et al. 2007; Yoshimochi et al. 2008). The first evidence of the functional interaction between MCM and GINS was the stimulation of the DNA helicase activity of PfuMCM by the addition of the PfuGINS complex (Yoshimochi et al. 2008).
The archaeal equivalent of Cdc45 was only recently identified. An exonuclease, which is stimulated by GINS-association and is called GAN (GINS-associated nuclease), shares similar amino acid sequence and biochemical properties with those of the bacterial RecJ nuclease (Li et al. 2011). A detailed sequence analysis also revealed that GAN, RecJ, and the eukaryotic Cdc45 protein contain a DHH phosphodiesterase domain, supporting the notion that they are homologs (Sanchez-Pulido and Ponting 2011; Makarova et al. 2012).
We have focused on the replicative helicase complex in the thermoacidophilic archaeon, Thermoplasma acidophilum. A single homolog of the Gins protein is found in many euryarchaeal organisms, except for Thermococcales (Yoshimochi et al. 2008), and thus the biochemical properties of the Gins protein from T. acidophilum were analyzed in a previous study (Ogino et al. 2011), where we showed that the T. acidophilum Gins formed a homotetramer complex (TaGINS) and physically interacted with TaMCM. However, TaGINS did not stimulate the helicase activity of TaMCM, even when a 4-fold excess of TaGINS was added to the reaction mixture. In addition, extensive characterizations of the two Cdc6/Orc1 proteins (TaCdc6-1, TaCdc6-2) were reported previously (Haugland et al. 2006; 2008a, b; 2009).
In this study, we performed a more detailed investigation of the interaction between TaMCM and TaGINS to reveal the functional role of TaGINS. A DNA-bound complex of TaMCM–TaGINS–TaCdc6-2 was detected, which may represent a pre-replisome progression complex (pre-RPC) or part of an RPC. Our results suggested the divergence of the replication machinery in Thermoplasma.
Materials and methods
Preparation of recombinant proteins
The recombinant TaMCM, TaGins51, and TaCdc6-2 proteins were prepared according to the previously described procedures, with some modifications (Haugland et al. 2006; Ogino et al. 2011). E. coli BL21-CodonPlus (DE3)-RIL cells (Agilent Technologies) carrying the pET-21a plasmid (Novagen) were cultured at 37 °C in 1 liter of LB medium, containing 50 μg/ml ampicillin and 34 μg/ml chloramphenicol. When the cell density reached an A 600 of 0.50, the gene expression was induced by adding isopropyl β-d-thiogalactopyranoside (IPTG) to a final concentration of 1 mM, followed by further cultivation for 3 h at 37 °C. The cells were harvested by centrifugation (10 min, 5,200×g), and were disrupted by sonication for 10 min in buffer A (50 mM Tris–HCl, pH 8.0, and 10 % glycerol) for TaGins51, and buffer B (50 mM Tris–HCl, pH 8.0, 0.5 M NaCl, 2 mM DTT, and 10 % glycerol) containing 20 mM imidazole for TaMCM and TaCdc6-2. The soluble extracts obtained by centrifugation (10 min, 22,000×g) were heated for 20 min at 60 °C for TaMCM and TaGins51, and 55 °C for TaCdc6-2. The heat-resistant fraction was obtained by centrifugation (10 min, 22,000×g). TaGins51 was then purified as described previously, by hydrophobic and anion-exchange chromatography (Ogino et al. 2011). For TaMCM and TaCdc6-2, the heat-resistant fraction was subjected to chromatography on a 1 ml HisTrap HP column (GE Healthcare), which was developed with a linear gradient of 20–500 mM imidazole. The eluted protein fractions were combined with buffer A, containing 2 mM DTT and 1 M (NH4)2SO4, to a final concentration of 0.5 M (NH4)2SO4, and then loaded to a 5 ml HiTrap Butyl HP column (GE Healthcare), which was developed with a linear gradient of 0.5–0 M (NH4)2SO4 in buffer A. The eluted protein fractions were diluted with buffer A containing 2 mM DTT, and loaded on a Mono Q 5/50 GL column (GE Healthcare), which was developed with a linear gradient of 0.3–0.7 M NaCl in buffer A. The eluted protein fractions were loaded on a HiLoad 16/600 Superdex 200 pg column (GE Healthcare), which was equilibrated with buffer A containing 0.15 M NaCl and 2 mM DTT, and the eluted protein fractions were pooled and concentrated with an Amicon Ultra filter (Millipore). All three proteins were dispensed in small aliquots and stored at −80 °C. The protein concentrations were determined using their absorbances at 280 nm and the extinction coefficients of 49,530 M−1 cm−1 for TaMCM, 11,920 M−1 cm−1 for TaGins51, and 29,465 M−1 cm−1 for TaCdc6-2 as a monomer.
Preparation of DNA substrates
To prepare the DNA substrate for the gel-retardation assay, the 54-mer oligonucleotide HJ-3-54mer (5′-dTCACTCCGCATCTGCCGATTCTGGCTGTGGCGTGTTTCTGGTGGTTCCTAGGTC-3′), synthesized by Sigma-Aldrich Japan K. K., was labeled at the 5′-terminus with 32P, using [γ-32P]ATP and T4 polynucleotide kinase (New England Biolabs). The labeled oligonucleotide was separated from the free nucleotides with a Microspin G-50 column (GE Healthcare). For the helicase assay, the HJ-3-54mer, with a DyLight 800-labeled 5′-terminus, was synthesized by Takara Bio Inc. To prepare the splayed-arm DNA substrate, the labeled HJ-3-54mer and HJ-4 (5′-dGACCTAGGAACCACCAGAAACACGCCACAGCCAGGAAGCCGATTGCGAGGCCGTCCTACCATCCTGCAGG-3′) were mixed in 20 mM Tris–HCl, pH 8.0, and 50 mM NaCl, heated at 95 °C for 3 min, and cooled to 25 °C for 1 h. The splayed-arm DNA was fractionated on a Superdex 200 5/150 GL column (GE Healthcare), which was equilibrated with 20 mM Tris–HCl, pH 8.0, and 0.15 M NaCl, concentrated with an Amicon Ultra filter, and used in this study.
Gel-retardation assay
The DNA-binding activity of TaMCM was measured in reaction mixtures (20 µl), containing 20 mM HEPES–NaOH, pH 7.5, 10 mM MgCl2, 0.1 mg/ml BSA, 50 fmol of the DNA substrate, 0.01 % Tween 20, and proteins (TaMCM, TaGins51, TaCdc6-2) in the presence of 1 mM ATPγS (a very slowly hydrolyzed substrate for ATPases), which were incubated at 59 °C for 10 min. Glutaraldehyde was then added to a final concentration of 0.01 %, and the reaction mixtures were incubated at room temperature for 10 min to crosslink the DNA substrate and the DNA-bound proteins. Loading buffer (5 ×), containing 20 mM HEPES–NaOH, pH 7.5, 12.5 % Ficoll, and 0.1 % Orange G, was added to the reaction mixtures. An aliquot (10 µl) was fractionated on a 4 % polyacrylamide gel in 1 × TAE by electrophoresis at 15 mA for 50 min and visualized with an image analyzer, Typhoon Trio + (GE Healthcare).
Immunoprecipitation assay
T. acidophilum cells were cultured in 200 ml of medium at 56 °C with shaking, as described previously (Yasuda et al. 1995), and were harvested at the exponential growth phase (A 600 = 0.25) by centrifugation (10 min, 6,000×g). The cells (1 × 1011) were suspended and disrupted in lysis buffer (50 mM Tris–HCl, pH 8.0, 10 mM MgCl2, 0.5 % Triton X-100) by sonication for 1 min (5 s on–5 s off). A portion (10 μl) of Dynabeads Protein G (Novex) was washed twice with TBS–T (20 mM Tris–HCl, pH 7.5, 500 mM NaCl, 0.1 % Tween 20), mixed with TBS–T containing 10 μl of anti-TaMcm, anti-TaCdc6-2, or anti-TaGins51 antiserum (prepared by injecting the purified recombinant proteins into rabbits), and incubated at room temperature for 1 h on a rotary shaker. Each mixture was washed twice with TBS–T, and then twice with 0.2 M triethanolamine, pH 8.0. The antibody was cross-linked to the Dynabeads Protein G with dimethyl pimelimidate 2 HCl (DMP, Thermo Scientific Pierce), according to the manufacturer’s protocol. Pre-immune serum was used for negative control experiments. After equilibration of the antibody-conjugated Dynabeads Protein G with lysis buffer, an aliquot of the cell extract (500 μl, 1.7 × 1010 cells) was added, and the mixture was incubated at room temperature for 1 h on the rotary shaker. The precipitates were washed twice with lysis buffer, and the immunoprecipitated proteins were eluted from the beads with 40 μl of gel loading solution (50 mM Tris–HCl, pH 6.8, 10 % glycerol, 100 mM DTT, 0.2 mg/ml bromophenol blue, 2 % SDS) at 98 °C for 3 min. Five microliter portions of the eluates were subjected to 10–20 % SDS-PAGE, followed by western blot analysis.
Western blot analysis
The proteins on the gel were electroblotted onto a polyvinylidene difluoride (PVDF) membrane (Bio-Rad) using a Trans-Blot Turbo Transfer System (Bio-Rad), and reacted with the anti-TaMCM, anti-TaCdc6-2 and anti-TaGins51 antisera, respectively. Rabbit TrueBlot: Anti-Rabbit IgG HRP (Rockland Immunochemicals, Inc.) was used as the secondary antibody. The proteins were visualized by ECL prime (GE Healthcare), and images were obtained with an LAS-3000 image analyzer (Fujifilm).
ATPase assay
TaMCM (1 pmol as the hexamer) was incubated in a 15 μl reaction mixture, containing 20 mM Tris–HCl, pH 8.0, 10 mM MgCl2, 1 mM DTT, 0.1 mg/ml BSA, 0.01 % Tween 20, 40 μM ATP, 0.33 pmol of [γ-32P] ATP and increasing amounts of TaGINS, in the presence or absence of 5 µM nucleotides (base pairs) M13 single-stranded or double-stranded DNA. After incubations at 59 °C for 15, 30, 45, and 60 min, an aliquot (1 μl) was spotted onto a polyethyleneimine-cellulose thin layer chromatography plate (Merck), to separate the ATP and released Pi, using a solvent containing 1 M formic acid and 0.5 M LiCl. The autoradiograms were obtained with an image analyzer, Typhoon Trio+, and the ATPase activity was quantified using the ImageQuant TL analysis software (GE Healthcare). The amount of Pi in the reaction without the enzyme was deducted from that of each reaction, to correct for the spontaneous hydrolysis at high temperature. The standard error of the mean was calculated from three independent experiments.
Helicase assay
The helicase activity of TaMCM was measured in a 15 μl reaction mixture, containing 20 mM Tris–HCl, pH 8.0, 10 mM magnesium acetate, 2 mM DTT, 0.1 mg/ml BSA, 0.1 % Triton X-100, 3.3 mM ATP, 0.5 or 1 pmol of the DNA substrate, 0.5 or 1 pmol of the trap DNA (5′-dGACCTAGGAACCACCAGAAACACGCCACAGCCAG-3′) and TaMCM, with increasing amounts of TaGINS and/or TaCdc6-2. After an incubation at 50 °C for 15 min, the samples were immediately transferred to ice, and 5 μl of 4 × stop buffer (100 mM EDTA, 4 % SDS, 10 % Ficoll, and 0.1 % Orange G) was added. An aliquot (5 µl) was loaded onto a 10 % polyacrylamide gel in 0.5 × TBE (45 mM Tris–borate, pH 8.5, and 1 mM EDTA) and electrophoresed at 15 mA for 50 min. The gel image was obtained with an Odyssey Infrared Imaging System (LI-COR), and the helicase activity was quantified using the application software (LI-COR). The standard error of the mean was calculated from three independent experiments.
Results
Purification of TaCdc6-2
To investigate the activation of TaMCM, we purified TaCdc6-2 as a recombinant protein with a C-terminal His-tag, as reported previously (Haugland et al. 2006), but with a slight modification. The absorbance curve in the UV region (210–320 nm) indicated that the protein fraction, purified by a HisTrap HP column, contained a significant amount of nucleic acids, which are probably tightly bound to TaCdc6-2. Therefore, the protein fractions were subjected to hydrophobic interaction chromatography, to remove the TaCdc6-2-bound nucleotides. The fraction eluted from the HiTrap Phenyl HP column had a 280 nm to 260 nm absorbance ratio above 1.8, suggesting that almost all of the nucleotides were removed from TaCdc6-2. The TaCdc6-2 protein was then concentrated, using an anion exchange column. A SDS-PAGE image of the purified TaCdc6-2 protein is shown in Fig. S1 together with the TaGins51 and TaMCM proteins, which were also purified reproducibly, according to our previous study (Ogino et al. 2011). Starting from 1 l culture, 7.1 mg of homogeneous TaCdc6-2 was obtained. Since TaCdc6-2 became denatured when it was stored at 4 °C or at −25 °C, the solution was promptly frozen and stored at −80 °C. The insolubility of TaCdc6-2 at low temperatures is a similar property to the Cdc6 protein from P. furiosus (Akita et al. 2010).
TaGINS stimulates the ATPase and helicase activities of TaMCM
We previously showed that TaGINS forms a homotetramer and physically interacts with TaMCM. However, the stimulation of the helicase activity by GINS as observed for P. furiosus GINS and MCM (Yoshimochi et al. 2008), was not detected (Ogino et al. 2011). To understand the physiological function of the TaMCM–TaGINS interaction, we investigated the helicase activity of TaMCM in more detail, in the presence of TaGINS in vitro. The reaction conditions for the ATPase assay and the helicase assay were re-examined, and the molar ratio of TaGINS/TaMCM was increased, as compared to our previous study. The reaction solution was also re-examined, and some detergents were found to be effective. The ATPase activity of TaMCM was stimulated by about 1.5-fold in the presence of single-stranded and double-stranded DNA, as observed previously (Ogino et al. 2011) (Fig. 1a). We examined the effect of TaGINS on the ATPase activity, and only a minimal response was observed until a five-fold excess amount of TaGINS to TaMCM was used. However, this DNA-dependent ATPase activity became stronger with increasing amounts of the TaGins51 protein, and was stimulated by about 2-fold when TaGINS was present in a 20-fold excess over TaMCM in the reaction mixtures (Fig. 1a). Since the stimulation was observed even without DNA, the increased ATPase activity is partly caused by a direct interaction between TaMCM and TaGINS. To examine whether this increased ATPase activity affects the helicase activity of TaMCM, an in vitro helicase assay was performed. A fluorescently labeled (5′-DyLight 800) DNA was used, and a trap DNA was added to prevent re-annealing of the unwound DNA. As shown in Fig. 1b, unwound DNA was re-annealed with the trap DNA. The helicase activity of TaMCM was clearly stimulated in the presence of TaGINS (Fig. 1b). The stimulation of unwinding by TaGINS was quantified and plotted (Fig. 1b, c). The results from the ATPase and helicase activity measurements suggested that TaMCM undergoes a conformational change by the interaction with TaGINS.
Effect of TaGINS on the ATPase and helicase activities of TaMCM. a The ATPase activity is expressed as the amount of Pi released by a constant amount of the TaMCM protein (as a hexamer) after 60-min incubation in the absence or presence of M13 ss or dsDNA. The standard error of the mean was calculated from three independent experiments. b The DyLight 800-labeled splayed-arm DNA substrate (0.5 pmol) was incubated with 1 pmol TaMCM and TaGINS. The helicase activity is expressed at the bottom of the panel, as the relative amount of unwound DNA (%) in each reaction condition with or without TaGINS. Single-stranded and trapped DNAs were loaded in parallel as controls for the unwinding reaction. c The averages of the helicase activities in three independent experiments, with the standard error of the mean, are shown
TaCdc6-2 synergistically stimulates the helicase activity of TaMCM with TaGINS
We used the 5′-DyLight 800-labeled DNA substrates to confirm that the TaMCM activation by TaCdc6-2 is reproducible in the current fluorescently labeled helicase assay. As shown in Fig. 2a and b, the helicase activity of TaMCM was enhanced with increasing amounts of TaCdc6-2, consistent with the results reported previously (Haugland et al. 2006, 2008b). We then repeated the helicase assay under the same reaction conditions as performed for TaCdc6-2 (1 pmol DNA, 0.5 pmol TaMCM, but not the condition shown in Fig. 1) to assess the effect of TaGINS. As shown in Fig. 2c, a stimulation of the helicase activity was observed, but it was clearly less than that caused by TaCdc6-2 (Fig. 2a). We then performed the helicase assay in the presence of both TaGINS and TaCdc6-2, to determine whether a synergistic activation occurs in vitro. The activation of TaMCM by TaCdc6-2 (Fig. 3a, lanes a–e) and TaGINS (Fig. 3a, lanes a, f) individually in this experiment was reproducible, as compared with Fig. 2. The increased unwinding efficiency in the presence of TaGINS was further enhanced with increasing amounts of TaCdc6-2 (Fig. 3a, lanes g–j), demonstrating a synergistic effect of the two proteins. The unwinding efficiencies in this experiment were quantified and the stimulation of the helicase activity by Cdc6-2 in the presence and absence of TaGINS was plotted in Fig. 3b. All standard error of the means were less than 1.6 %, indicating that the synergistic effect of the two proteins should be significant.
Comparison of the stimulating ability of TaCdc6-2 and TaGINS on the helicase activity of TaMCM. a The DyLight 800-labeled splayed-arm DNA substrate (1 pmol) was incubated with TaMCM and TaCdc6-2, as described in the Materials and methods. The helicase activity is indicated at the bottom of the panel, as the relative amount of unwound DNA (%). Single-stranded and trapped DNAs were loaded in parallel, as controls for the unwinding reaction. b The averages of the helicase activities in three independent experiments, with the standard error of the mean, are shown. c The helicase activity was measured with TaMCM and TaGINS under the same reaction conditions as in a
The helicase activity of TaMCM in the presence of TaCdc6-2 and TaGINS. a TaCdc6-2 and TaGINS were added to the assay mixtures, containing 1 pmol of the DyLight 800-labeled splayed-arm DNA substrate. The amounts of the proteins are indicated at the top of each lane. The helicase activity is indicated at the bottom of the panel, as the relative amount of unwound DNA (%). Single-stranded and trapped DNAs were loaded in parallel, as controls of the unwinding reaction. b The averages of the helicase activities in three independent experiments, with the standard error of the mean, are shown. All standard errors of the means were less than 1.6 %
TaCdc6-2 and TaGINS form a complex with TaMCM on DNA
We performed a gel-retardation assay, using 5′-32P-labeled single-stranded DNA as a probe, to evaluate the interactions between TaMCM, TaCdc6-2 and TaGINS on DNA. The DNA band was shifted in the presence of TaMCM (Fig. 4a, lane e). On the other hand, neither TaGINS nor TaCdc6-2 shifted the DNA band (Fig. 4a, lanes b–d). However, the TaMCM-shifted band was shifted further by the addition of TaCdc6-2 (lane f), TaGINS (lane g), or both of these proteins (lane h).The shifted band by the three proteins was slightly moved from the well. This result indicates that TaGINS and TaCdc6-2 can load onto TaMCM-bound DNA. Glutaraldehyde treatment was needed to detect the shifted band in this experiment, due to the rapid dissociation of TaMCM from DNA (Haugland et al. 2009; Ogino et al. 2011). Some DNA remained in the wells of lanes e, f, and g perhaps due to crosslinked large molecules. This gel shift experiment suggested that TaMCM remains more stably bound on the DNA in the presence of TaGINS and TaCdc6-2, and that TaMCM can be activated not only by the stimulation of its ATPase activity but also by the enhancement of its DNA-binding ability. We previously reported that TaCdc6-2 does not affect DNA binding of TaMCM (Haugland et al. 2008b). However, the present gel shift image showed that the affinity of the TaMCM-TaCdc6-2 complex to DNA remarkably increased from that of TaMCM alone (Fig. 4a, lanes e, f). These results, as well as the results in our previous report (Ogino et al. 2011) were, however strictly ATP-dependent as no band-shift could be observed without ATPγS addition (data not shown). To confirm whether TaMCM, TaGINS, and TaCdc6-2 form a complex in T. acidophilum cells, immunoprecipitation experiment was performed using extracts from exponentially growing cells and antibodies against TaMcm, TaGins51, and TaCdc6-2. As shown in Fig. 4b, TaMcm and TaGins51 co-precipitated with anti-TaMcm or anti-TaGins51 antibody, respectively, as reported previously (Ogino et al. 2011). However, TaCdc6-2 was not detected clearly from either anti-TaMcm or anti-TaGins51. These results suggest that the ternary complex of TaMCM–TaGINS–TaCdc6-2 is fragile, and especially the interaction between TaCdc6-2 and other two proteins is too weak to be detected.
Physical interaction of TaMCM, TaCdc6-2, and TaGINS. a The DNA binding activity of TaMCM, TaCdc6-2, and TaGINS is shown. The TaMcm, TaCdc6-2, and TaGins51 proteins were incubated with a 54 nt 32P-labeled ssDNA at 59 °C for 10 min, crosslinked with glutaraldehyde, and analyzed by 4 % PAGE. The amounts of the proteins are shown at the top of each lane. The assignment of each band was indicated by arrows, 1 well, 2 TaMCM–TaCdc6-2–TaGINS–DNA, 3 TaMCM–TaGINS–DNA, 4 TaMCM–TaCdc6-2–DNA, 5 TaMCM–DNA, 6 ssDNA. b Immunoprecipitation analyses were performed to confirm the formation of a complex including MCM, GINS, and Cdc6-2 in the T. acidophilum cell extract. The immunocomplexes were captured with anti-TaCdc6-2, anti-TaMcm, and anti-TaGins51 antibodies, respectively, from the whole cell extract (as shown on the top), and were subjected to 10–20 % SDS-PAGE, followed by western blot analyses using these antibodies (shown on the right side). The whole cell extracts without immunoprecipitation (input) or precipitated with DynaBeads Protein G treated with pre-immune serum were also loaded as positive and negative controls, respectively
Discussion
The protein forming the helicase complex in the archaeal DNA replication process has not been disclosed yet. We investigated the effect of TaGINS on TaMCM in more detail, based on the results from our previous study and found that TaGINS clearly stimulated the helicase activity of TaMCM in vitro. This is the third investigation to show experimentally that GINS stimulates the helicase activity of MCM in Archaea, as previously demonstrated for P. furiosus (Yoshimochi et al. 2008) and T. kodakarensis (Ishino et al. 2011), and the first evidence to show that a hometetramer GINS, composed of a single protein (TaGins51), stimulates their cognate MCM helicases. We found here a more suitable reaction condition for the helicase assay than the previous study. Some detergents, such as 0.1 % Triton X-100 or 0.01 % Tween 20, were effective for the enzyme assays. The detergents probably help to maintain the active forms in solution. In addition, the stimulation became more obvious with increasing molar ratio of TaGINS/TaMCM in the reaction mixture. The two proteins do not interact sufficiently at low concentrations, and higher amounts of TaGINS are thus required to detect the effect on TaMCM. Since TaMcm and TaGins51 proteins are abundantly produced in vivo (Haugland et al. 2009; Ogino et al. 2011), the stimulation of the TaMCM helicase activity by TaGINS in vitro should be physiologically relevant in T. acidophilum cells.
T. acidophilum has two Cdc6/Orc1-like proteins and one of them, Cdc6-2, stimulated the MCM helicase in our previous study as a unique phenomenon in Archaea (Haugland et al. 2006). We attempted to evaluate the activation of the MCM helicase by two proteins, Cdc6-2 and GINS from T. acidophilum, and demonstrated that the helicase activity of TaMCM was enhanced more effectively in the presence of both TaCdc6-2 and TaGINS than in the presence of either protein alone. The results of the gel-retardation assay showed that DNA-bound TaMCM forms a complex with TaCdc6-2 and TaGINS. Furthermore, TaMCM forms a more stable complex on DNA in the presence of these two proteins.
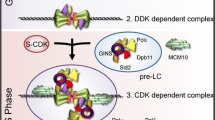
In vitro experiments showed that a stable eukaryotic CMG complex is formed on single-stranded DNA as compared with Mcm2–7 alone (Ilves et al. 2010). Furthermore, the structures of Mcm2–7 and the CMG complex from Drosophila, determined by electron microscopy and single particle image analysis, suggested that Cdc45 and GINS bridge the gap between Mcm2 and Mcm5 in the gapped-ring structure of the Mcm2–7 hexamer (Costa et al. 2011). All of the archaeal Mcm proteins characterized to date form homomultimers, and it is unclear whether the archaeal MCMs have gapped ring-structures when they are loaded onto DNA, although it is known that archaeal MCMs can form a spiral state (Chen et al. 2005; Costa et al. 2008; Fu et al. 2013; Slaymaker et al. 2013). TaCdc6-2 and TaGINS are probably loaded on the TaMCM-bound DNA and form, at least, a temporary TaMCM–TaGINS–TaCdc6-2 complex. Possible models for the TaMCM activation in T. acidophilum are presented in Fig. 5. An important question is whether TaCdc6-2 participates in the RPC or leaves the complex after activating the MCM conformation. Another consideration is that TaMCM, TaGINS, and TaCdc6-2 should assemble at the oriC region to form the active helicase complex. Our analyses showed that TaMCM interacts with TaGINS and TaCdc6-2 without the oriC region. If TaMCM continuously interacts with TaGINS and TaCdc6-2, then the replicative helicase should be constantly active. The regulatory mechanism underlying the stage-specific activation of the replicative helicase in the T. acidophilum cell cycle should be elucidated. The detailed structural basis for the MCM activation in Archaea remains enigmatic, and the determination of the actual shape of the helicase complex in T. acidophilum is an interesting target for structural biology.
Possible models for the activation and progression of the replicative helicase in T. acidophilum, based on this study. The predicted dynamics of replication fork activation is indicated. TaCdc6-1 or TaCdc6-2 recognizes the replication origin, and TaCdc6-2, TaMCM, and TaGINS form the “CMG like” complex at the origin to activate the TaMCM helicase. The [TaCdc6-2–TaMCM–TaGINS] or [TaMCM–TaGINS] complex unwinds the duplex DNA, as a replication fork progression complex. It is also possible that GAN (Cdc45) participates as a part of the active helicase
Regarding functions of the two Cdc6 proteins in Thermoplasma, one may be a recruiter for MCM to the oriC region, as in the case of P. furiosus (Akita et al. 2010). The oriC region in the T. acidophilum genome must be identified to clarify this issue. The possibility of the TaCdc6-1-TaCdc6-2 heterodimer formation should also be analyzed. In the case of S. solfataricus, the Cdc6-1 and Cdc6-3 proteins form a heterodimer binding to ori2, one of the three origins in this organism (Dueber et al. 2007). Phosphorylation may be involved in the initiation of DNA replication in Archaea, as in the CDK and DDK-dependent activation in Eukarya. TaCdc6-1 is autophosphorylated more efficiently as compared with TaCdc6-2, and this may indicate a functional difference between the two Cdc6 proteins (Haugland et al. 2008a). TaCdc6-1 and TaCdc6-2 may share their functions in initiation regulation and in fork progression, respectively.
T. acidophilum has two GAN (Cdc45)-like ORFs, whose gene products might interact with TaGINS forming a eukaryotic-like CMG complex. An important future task will be to investigate whether the GAN proteins affect the helicase activity of TaMCM, and the TaMCM–TaGINS, or TaMCM–TaGINS–TaCdc6-2 complexes.
Searching for the genome sequences of the available three genuses in Thermoplasmatales, Thermoplasma, Picrophilus, and Ferroplasma, revealed that one for each mcm, gins, and cdc6-2 genes are commonly predicted with more than 49 % similarity. Therefore, the activation mechanism of the replicative helicase in T. acidophilum presented here may be expanded to Thermoplasmatales.
The mechanism of DNA replication in Archaea is a very interesting subject. Comparison of the experimental data among the three domains of life will provide extensive amounts of information for understanding the evolution and diversification of the DNA replication machinery.
References
Akita M, Adachi A, Takemura K, Yamagami T, Matsunaga F, Ishino Y (2010) Cdc6/Orc1 from Pyrococcus furiosus may act as the origin recognition protein and Mcm helicase recruiter. Genes Cells 15:537–552
Aparicio OM, Weinstein DM, Bell SP (1997) Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell 91:59–69
Chen YJ, Yu X, Kasiviswanathan R, Shin JH, Kelman Z, Egelman EH (2005) Structural polymorphism of Methanothermobacter thermautotrophicus MCM. J Mol Biol 346:389–394
Costa A, van Duinen G, Medagli B, Chong J, Sakakibara N, Kelman Z, Nair SK, Patwardhan A, Onesti S (2008) Cryo-electron microscopy reveals a novel DNA-binding site on the MCM helicase. EMBO J 27:2250–2258
Costa A, Ilves I, Tamberg N, Petojevic T, Nogales E, Botchan MR, Berger JM (2011) The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat Struct Mol Biol 18:471–477
Dueber EL, Corn JE, Bell SD, Berger JM (2007) Replication origin recognition and deformation by a heterodimeric archaeal Orc1 complex. Science 317:1210–1213
Fu Y, Slaymaker IM, Wang J, Wang G, Chen XS (2013) The 1.8-Å crystal structure of the N-terminal domain of an Archaeal MCM as a right-handed filament. J Mol Biol 426:1512–1523
Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K (2006) GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol 8:358–366
Haugland GT, Shin JH, Birkeland NK, Kelman Z (2006) Stimulation of MCM helicase activity by a Cdc6 protein in the archaeon Thermoplasma acidophilum. Nucleic Acids Res 34:6337–6344
Haugland GT, Innselset M, Madern D, Birkeland NK (2008a) Characterization of the Cdc6 Homologues from the Euryarchaeon Thermoplasma acidophilum. Open Biochem J 2:129–134
Haugland GT, Sakakibara N, Pey AL, Rollor CR, Birkeland NK, Kelman Z (2008b) Thermoplasma acidophilum Cdc6 protein stimulates MCM helicase activity by regulating its ATPase activity. Nucleic Acids Res 36:5602–5609
Haugland GT, Rollor CR, Birkeland NK, Kelman Z (2009) Biochemical characterization of the minichromosome maintenance protein from the archaeon Thermoplasma acidophilum. Extremophiles 13:81–88
Ilves I, Petojevic T, Pesavento JJ, Botchan MR (2010) Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell 37:247–258
Ishimi Y (1997) A DNA helicase activity is associated with an MCM4, -6, and -7 protein complex. J Biol Chem 272:24508–24513
Ishino Y, Ishino S (2012) Rapid progress of DNA replication studies in Archaea, the third domain of life. Sci China Ser-C Life Sci 55:1–18
Ishino S, Fujino S, Tomita H, Ogino H, Takao K, Daiyasu H, Kanai T, Atomi H, Ishino Y (2011) Biochemical and genetical analyses of the three mcm genes from the hyperthermophilic archaeon, Thermococcus kodakarensis. Genes Cells 16:1176–1189
Kubota Y, Takase Y, Komori Y, Hashimoto Y, Arata T, Kamimura Y, Araki H, Takisawa H (2003) A novel ring-like complex of Xenopus proteins essential for the initiation of DNA replication. Genes Dev 17:1141–1152
Leipe DD, Aravind L, Koonin EV (1999) Did DNA replication evolve twice independently? Nucleic Acids Res 27:3389–3401
Li Z, Pan M, Santangelo TJ, Chemnitz W, Yuan W, Edwards JL, Hurwitz J, Reeve JN, Kelman Z (2011) A novel DNA nuclease is stimulated by association with the GINS complex. Nucleic Acids Res 39:6114–6123
Makarova KS, Wolf YI, Mekhedov SL, Mirkin BG, Koonin EV (2005) Ancestral paralogs and pseudoparalogs and their role in the emergence of the eukaryotic cell. Nucleic Acids Res 33:4626–4638
Makarova KS, Koonin EV, Kelman Z (2012) The CMG (CDC45/RecJ, MCM, GINS) complex is a conserved component of the DNA replication system in all archaea and eukaryotes. Biol Direct 7:7
Marinsek N, Barry ER, Makarova KS, Dionne I, Koonin EV, Bell SD (2006) GINS, a central nexus in the archaeal DNA replication fork. EMBO Rep 7:539–545
Masai H, Matsumoto S, You Z, Yoshizawa-Sugata N, Oda M (2010) Eukaryotic chromosome DNA replication: where, when, and how? Annu Rev Biochem 79:89–130
Matsunaga F, Glatigny A, Mucchielli-Giorgi MH, Agier N, Delacroix H, Marisa L, Durosay P, Ishino Y, Aggerbeck L, Forterre P (2007) Genomewide and biochemical analyses of DNA-binding activity of Cdc6/Orc1 and Mcm proteins in Pyrococcus sp. Nucleic Acids Res 35:3214–3222
Mott ML, Berger JM (2007) DNA replication initiation: mechanisms and regulation in bacteria. Nat Rev Microbiol 5:343–354
Moyer SE, Lewis PW, Botchan MR (2006) Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci USA 103:10236–10241
Ogino H, Ishino S, Mayanagi K, Haugland GH, Birkeland N-K, Yamagishi A, Ishino Y (2011) The GINS complex from the thermoacidophilic archaeon, Thermoplasma acidophilum may function as a homotetramer in DNA replication. Extremophiles 15:529–539
Pacek M, Tutter AV, Kubota Y, Takisawa H, Walter JC (2006) Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol Cell 21:581–587
Sakakibara N, Kelman LM, Kelman Z (2009) Unwinding the structure and function of the archaeal MCM helicase. Mol Microbiol 72:286–296
Sanchez-Pulido L, Ponting CP (2011) Cdc45: the missing RecJ ortholog in eukaryotes? Bioinformatics 27:1885–1888
Slaymaker IM, Fu Y, Toso DB, Ranatunga N, Brewster A, Forsburg SL, Zhou ZH, Chen XS (2013) Mini-chromosome maintenance complexes form a filament to remodel DNA structure and topology. Nucleic Acids Res 41:3446–3456
Takayama Y, Kamimura Y, Okawa M, Muramatsu S, Sugino A, Araki H (2003) GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes Dev 17:1153–1165
Yasuda M, Oyaizu H, Yamagishi A, Oshima T (1995) Morphological variation of new Thermoplasma acidophilum isolates from Japanese hot springs. Appl Environ Microbiol 61:3482–3485
Yoshimochi T, Fujikane R, Kawanami M, Matsunaga F, Ishino Y (2008) The GINS complex from Pyrococcus furiosus stimulates the MCM helicase activity. J Biol Chem 283:1601–1609
Acknowledgments
This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (Grant numbers 21113005, 23310152, and 26242075 to Y. I.). H. O. is supported by Grant-in-Aid for Japan Society for the Promotion of Science (JSPS) Fellows. This manuscript was written during N-K. Birkeland’s visit to Ishino’s laboratory by the Invitation Fellowship Programs for Research in Japan of JSPS.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. W. W. Adams.
This article is part of a special issue based on the 10th International Congress on Extremophiles held in Saint Petersburg, Russia, September 7–11, 2014.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ogino, H., Ishino, S., Haugland, G.T. et al. Activation of the MCM helicase from the thermophilic archaeon, Thermoplasma acidophilum by interactions with GINS and Cdc6-2. Extremophiles 18, 915–924 (2014). https://doi.org/10.1007/s00792-014-0673-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-014-0673-6