Abstract
The North-Western part of Argentina is particularly rich in wetlands located in the Puna in an altitude between 3,600 and 4,600 m above sea level. Most of these high-altitude Andean lakes are inhospitable areas due to extreme habitat conditions such as high contents of toxic elements, particularly arsenic. Exiguobacterium sp. S17, isolated from stromatolites in Laguna Socompa, exhibited remarkable tolerance to high arsenic concentration, i.e., it tolerated arsenic concentration such as 10 mM of As(III) and 150 mM of As(V). A proteomics approach was conducted to reveal the mechanisms that provide the observed outstanding resistance of Exiguobacterium sp. S17 against arsenic. A comparative analysis of S17, exposed and unexposed to arsenic revealed 25 differentially expressed proteins. Identification of these proteins was performed by MALDI-TOF/MS revealing upregulation of proteins involved in energy metabolism, stress, transport, and in protein synthesis being expressed under arsenic stress. To our knowledge, this work represents the first proteomic study of arsenic tolerance in an Exiguobacterium strain.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Exiguobacterium sp. S17, isolated from a stromatolite from Laguna Socompa, showed a tolerance to high arsenic (As) concentrations. Laguna Socompa is located at the northern part of Salta Province, Argentina (S24°35′34″, W68°12′42″) at 3,800 m of altitude. L. Socompa is one of the high-altitude Andean lakes (HAAL) in the high altitude desert known as Puna (Farías et al. 2011). It is considered an extreme environment due to conditions like high salinity (125 mS), high UV radiation, wide daily thermic amplitudes ranging from 20 to −10 °C in summer and 10 to −40 °C in winter, and a high As content of 32 mg/l (0.42 mM) (Farías et al. 2011, 2012). Stromatolite communities were found along the southern shore of this lake (Farías et al. 2012). Exiguobacterium sp. S17 strain was extracted from one of these stromatolites and belongs to the extremophile culture collection of HAAL described by Ordoñez et al. (unpublished). Several studies have shown that some strains of Exiguobacterium possess unique properties of interest for applications in biotechnology, bioremediation, industry and agriculture (Okeke et al. 2007; Lopez et al. 2005; Pattanapipitpaisal et al. 2002).
Arsenic (As) is a ubiquitous toxic semi-metal or metalloid released in the environment mainly by volcanic activity (Smedley and Kinniburgh 2002; ATSDR 2000; Fergurson and Gavis 1972). The most common oxidation states for soluble arsenic in nature are the pentavalent, arsenate [As(V)], and the trivalent, arsenite [As(III)], present as (AsO −34 ) and (As(OH)3), respectively (Rosen 2002; Anderson and Cook 2004), with As(III) being much more toxic than As(V) (Oremland and Stolz 2003; Mukhopadhyay et al. 2002; Rosen 2002). The toxicity to As(V) is due to its similarity to phosphates (Rosen and Lui 2009), thus leading to the disruption of critical cellular functions or the impairment of synthesis of essential building blocks. In contrast, As(III) toxicity is believed to be due to its ability to covalently bind protein sulfhydryl groups (Parvatiyar et al. 2005).
To survive in extreme environments, cells trigger programs of specific gene expression, which are manifested as an increase or decrease in the amount of a set of proteins synthesized in response to stress (Duché et al. 2002). In the particular case of deleterious effects of As, it was reported that microorganisms have developed several strategies of detoxification including the reduction of As(V) like arsenite methylation, or its oxidation and further extrusion from the cell by a membrane-associated efflux pump, arsB (Lakshmi Sunita et al. 2012; Silver and Phung 2005; Rosen 2002). The genes encoding the arsenite detoxification machinery (ars genes) are widely distributed in bacteria and archaea and can either be found in plasmids or chromosomes (Achour et al. 2007). In this way, proteomics is an important tool to understand the strategies employed by microorganisms under different stress situations. Proteomics together with bioinformatics constitute complementary approaches that can deal with a huge amount of DNA information from genome sequencing projects and can provide new insights into bacterial functional genomics (Champomier-Vergés et al. 2010). This method is an excellent approach to study changes in the proteins expression patterns to elucidate adaptation mechanisms under different conditions. In several of the As removal processes, bacterial activity may play an important catalytic role (Castro de Esparza 2006). Due to the fact that many bacteria are able to modify or reduce the bioavailability of metals, the use of biological based methods appears to be an attractive tool for arsenic removal. Such change in protein concentrations and enzyme activities are best investigated by proteomics which is chosen here as an approach to understand the mechanisms implicated in the tolerance of Exiguobacterium sp. S17 against exposure to As(V) and As(III).
Materials and methods
Arsenic tolerance
The strain Exiguobacterium sp. S17 was identified by sequencing of 16S DNA (Ordoñez 2012). The strain was routinely grown in Luria-Bertani broth (LB, Britania) at 30 °C. To determine the tolerance of arsenic [As(V) and As(III)], tubes with 3 ml of LB50 (LB diluted to 50 % final concentration) used as control and LB50 containing different concentrations of arsenic: As(III), 2.5, 5, 10 mM and As(V), 20, 50, 100, 150 mM were inoculated to an initial OD600nm between 0.05 and 0.09. The tubes were incubated at 30 °C with constant agitation (150 rpm). Samples were taken during 24 h and OD600nm were determined.
Bacterial strain and culture conditions
A single colony of Exiguobacterium sp. S17 was grown in LB50 broth at 30 °C for 24 h and this culture was used for further inoculation of 100 ml of LB (control), 100 ml of LB supplemented with As(V) (100 mM) and 100 ml of LB supplemented with As(III) (7.5 mM) at initial OD600nm 0.1. Erlenmeyer flasks were incubated up to the mid-exponential growth phase (OD600nm 0.55 ± 0.05), the cells were harvested by centrifugation (7,000 rpm, 10 min at 18–20 °C) and washed twice with Tris–HCl buffer (0.1 M) pH 7.5. The pellets were then resuspended in 5 ml of the same buffer and were passed through a French press at 2,000 psi (SLM Instruments, Inc., Haverhill, MA, USA). Unbroken cells and other debris were removed by centrifugation at 14,000 rpm for 10 min at 4 °C (Beckman, USA). The total protein concentration was determined by Bradford method using BioRad reagents (Biorad, Richmond, CA), using bovine serum albumin (Sigma) as the standard. Aliquots of 400 μg of protein were stored at −80 °C for the isoelectrofocusing assay. Three independent assays for each condition were performed.
Two-dimensional electrophoresis (2DE)
Sample preparation and 2DE gels were carried out according to Borja Sánchez et al. (2005) with some modifications. If required, nucleic acids were removed by treatment with 1 μl of benzonase (Novagen®) in the presence of 1 μl of 1 M MgSO4 for 30 min at 37 °C. Total proteins were precipitated with 3 volumes of cold acetone, and after incubation at −20 °C for 16 h, samples were centrifuged (14,000 rpm, 10 min). The protein pellets were air-dried and solubilized in 40 μl of solubilization mixture. The suspension was centrifugated at 3,500 rpm for 10 min and loaded onto immobilized pH gradient strips (pH 4.0–7.0, 18 cm, GE Health Care, Sweden). Gels were passively rehydrated for 20 h. The isoelectrofocusing assay was performed using IPGphor (GE, HealthCare, Sweden) at 53,500 V/h and the focusing strips were stored at −20 °C until separation in second dimension was performed. The second dimension was performed by SDS-PAGE on gels containing 12.5 % (w/v) polyacrylamide and carried out in a Bio-Rad Protean II xi cell (Biorad, Richmond, CA). Proteins were resolved overnight at a constant current of 11 mA/gel at 4 °C. Gels were stained with Biosafe colloidal Coomassie blue (Biorad, Richmond, CA, USA) according to the manufacturer’s instructions, scanned with Image Scanner III and analyzed with Prodigy SameSpots (Nonlinear Dynamic Group, UK).
Protein identification using peptide mass fingerprinting
For each independent assay, the respective 2DE gels were carried out. Briefly, prominent spots were used to manually assign vectors in each gel image and the automatic vectors feature of the software was used to add additional vectors, which were manually verified. These vectors were used to warp the images and align the spot position to a common reference gel. Spot detection performed according to this reference gel was edited and artifacts removed. To correct the variability due to staining and to reflect the quantitative variation in intensity of protein spots, the spot volumes were normalized as a percentage of the total volume in all spots in the gel. A spot was considered significant when its resulting normalized volume showed more than 1.2-fold variation with respect to the control (LB medium) at the level of p < 0.05.
Individual spots were excised from the gels and subjected to mass spectrometry analyses that were carried out by CEQUIBIEM (Centro de Estudios Químicos y Biológicos de Espectrometría de Masa), Facultad de Ciencias Exactas y Naturales, UBA, Argentina. Protein identity from peptide mass fingerprints was determined by the MASCOT program (Matrix Science Inc., Boston, MA; http://www.matrixsciende.com/search-form-select.html). Fragmentation was carried out with more intense MS peaks (MS/MS). When possible, MS and MS/MS information was combined for one or more peptide searches.
Results and discussion
Growth of Exiguobacterium sp. S17 under arsenic stress
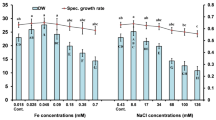
To evaluate quantitatively the S17 response to arsenic exposure, we determined the pattern of growth by measuring OD600nm at different times in the presence of increasing concentrations of the arsenic. Figure 1 shows the growth values obtained at 12 h and the tolerance was measured both in arsenate as sodium arsenite. Exiguobacterium sp. S17 was able to grow in all concentrations of arsenate assayed, showed similar growth rates, being the concentrations of 150 mM tested in this work (Fig. 1a). With regard to the presence of arsenite, this strain also showed tolerance to all concentrations tested for this metalloid, being 10 mM the highest evaluated concentration (Fig. 1b). The MIC was realized for all conditions. The maximum value was 15 mM of As(III) and 250 mM of As(V). The values selected for proteomic assays [As(V) = 100 mM and As(III) = 7.5 mM] were made to guarantee a good cellular growth.
In contrast with our result, Lakshmi Sunita et al. (2012) reported a strain of Exiguobacterium, isolated from estuary of Goa, with a maximum tolerance of 0.5 mM As(III), twenty times lower than the maximum concentration observed in Exiguobacterium sp. S17. These results may be due to the fact that Exiguobacterium sp. S17 came from Laguna Socompa, which has an arsenic concentration of about 32 mg/l (Farías et al. 2011). The response of a bacterium to a particular stress is related to the environment where it was isolated. Cai et al. (2009) suggest that soils with long-term arsenic contamination may result in the selection of highly diverse arsenic resistance mechanisms that could be probably spread throughout horizontal gene transfer events. Fernández Zenoff et al. (2006) also demonstrated this effect in bacteria isolated from HAAL while investigating UV radiation stress.
Proteomic analysis of Exiguobacterium sp. S17
The mechanisms of response to the presence of arsenic were investigated by a proteomics approach. Cells were grown in LB (control), LB + As(V) (100 mM) and LB + As(III) (10 mM), and were harvested at the mid log phase of growth (OD600nm ~ 0.55). These samples were subjected to 2DE, which led to successful identification of proteins involved in the arsenic response. Identification of changes is based on a spot-by-spot comparison of gels containing samples extracted under different conditions, always in comparison to a reference (standard gels). Two-dimensional protein patterns revealed a total of 113 spots differentially expressed under the assayed conditions. Twenty-five statistically significant spots, by a factor greater than 1.2, were identified by MALDI-TOF/MS, as shown in Table 1. Among them, eight spots (positions 8, 9, 13, 15, 26, 34, 62 and 67) were over-expressed in the presence of As(V) compared with the control medium and were not detected in the presence of As(III), while nine spots (positions 1, 12, 33, 37, 75, 96, 97, 98 and 100) were detected only in the presence of As(V). Four other positions (27, 28, 42 and 48) were detected in the presence of either As(V) or As(III) but not in the control medium (Figs. 2, 3). On the other hand, under control conditions, one spot (44) corresponding to an over-expressed protein was identified. Finally, three spots (20, 53, 71) were detected in all tested conditions; nevertheless, the spots’ intensity was higher in the presence of arsenate.
Exiguobacterium sp. S17 showed over-expression of a superoxide dismutase “SOD” (spot 71) in the presence of As(V) and As(III) compared to the control. It is known that exposure to arsenic induces response against oxidative stress in bacterial cells (Ciprandi et al. 2012; Rodriguez-Gabriel and Russell 2005; Moore et al. 2005). SOD is an enzyme that catalyzes the dismutation of superoxide ion into oxygen and hydrogen peroxide, which then is reduced to H2O by peroxidases, constituting an important defense strategy against reactive oxygen species (ROS), responsible for oxidative stress in cells. Ciprandi et al. (2012) described the same effect in Chromobacterium violaceum in response to As(III), which increased SOD expression quite comparable to oxidative stress conditions. Furthermore, Kramer and Ames (1988) reported that SOD is an important protein in providing resistance to selenite toxicity in Salmonella typhimurium. The over-expression of SOD in Exiguobacterium sp. S17 under As exposition is thus considered to be directly involved in the cellular response to reduce the alterations, such as DNA damage and protein oxidation, caused by the presence of this metalloid.
In addition, the differential proteome of Exiguobacterium sp. S17 revealed the expression of heat shock proteins (Hsps), e.g., DnaK chaperone (spot 20), GroEL (spot 15) and cpn10 (or GroEs) (spot 62). All these stress proteins are over-expressed in the presence of arsenate. These proteins are highly conserved within species and carry out essential functions such as folding, translocation, and assembly in stressed and non-stressed cells (Hartl 1996; Parsell and Lindquist 1993). The Hsps have an important function against environmental stress conferring tolerance to high temperatures, high salt and heavy metal concentrations (Tkáčová and Angelovičová 2012; Ron 2006; Kiriyama et al. 2001; Tedengren et al. 1999; Parsell and Lindquist 1993). With respect to a direct As exposition effect, Zhang et al. (2007) observed an increase of DnaK chaperone in Comamonas sp. under arsenate stress. Similar results were reported for Ferroplasma acidarmanus Fer1 by Baker-Austin et al. (2007) and for Pseudomonas aeruginosa by Parvatiyar et al. (2005) when this bacterium was exposed to As(III). On the other hand, Rodrigues et al. (2008) observed that Exiguobacterium sibiricum 255–15 increased the expression of genes dnaK, groES and groEL upon elevated temperature (39 °C). Apparently high temperatures produce denaturation of proteins in E. sibiricum, and to overcome this problem, the expression of genes encoding Hsp proteins is induced. Clearly, the finding of an increase of these proteins in Exiguobacterium sp. S17 in this study identifies them as an important component of the stress response induced by As exposition apparently requiring the increased recovery of misfolded proteins caused by the presence of this metalloid (Nguyen et al. 2009; Mogk et al. 2003). Interestingly, one universal stress protein, UspA (spot 75) was expressed only in the presence of As(V). In contrast, Cleiss-Arnold et al. (2010) reported UpsA induction by As(III). Multiple members of the UspA family of proteins are found in the genomes of bacteria, archaea and fungi, and it has been reported that this protein is expressed in response to a large variety of stress conditions (Kvint et al. 2003), However, the biological and biochemical functions of the UspA protein family are still not known (Siegele 2005). Curiously, the Hsps proteins (cold-shock protein, GroEL, Cpn10 and UspA), were not detected in the presence of arsenite, probably due to the fact that the cell removes the arsenite and the defense machinery in the presence of this metalloid may not be activated. Ordoñez (2012) reported that the genome of Exiguobacterium sp. S17 carries a copy of an ArsB gene encoding a protein that acts as an efflux pump of cytoplasmic arsenite. Additionally, in the same strain, an As(III) resistance protein (ARC3) has been reported that also acts as an efflux pump. The presence of this gene in the Exiguobacterium genus has not been reported before. The combined work of both efflux pumps (arsB and ACR3) might explain the enhanced tolerance of this strain to As(III) (Ordoñez et al. unpublished). Proteomic analysis did not allow detecting differential expression of the protein responsible for the transport of arsenic. This is probably due to the fact that these transport proteins are expressed under all the studied conditions as constitutive proteins, and thus are not detected as differential spots. It should also be kept in mind that ArsB, the protein responsible to the arsenite resistance, is a membrane protein which often is highly insoluble and, therefore, is not readily resolved by 2D-PAGE (Baker-Austin et al. 2007). However, two proteins involved in transport were detected only in the presence of As(V), and ABC transporter (spot 97) and a protein member of the toxic anion resistance family protein (spot 33). It has been reported that the inward transport of arsenate is carried out by phosphate transport membrane systems as Pit (phosphate transport) and Pst (phosphate specific transporter) (Paez-Espino et al. 2009). This latter protein is an ABC transporter that binds and uses the energy of ATP to transport substances across membranes (Couoh Uicab et al. 2010). In Exiguobacterium sp. S17, this protein is probably involved in the transport of As(V) into the cell, since its presence under As(III) expositions and in the control medium was not observed. In relation with the other identified protein (spot 33), a toxic anion resistance family protein, it might be assumed that the cells promote the elimination of toxic anion arsenite as mechanism of arsenic resistance.
Additionally, as shown in Table 1, sigma 54 modulation protein/ribosomal protein S30EA (spot 28), ribosomal protein S2 (spot 96), ribosome recycling factor (spot 100), prolyl-tRNA synthetase and elongation factor TS (spot 53) are all overexpressed in the presence of As(V). All of these proteins are involved in transcriptional and translational processes. With respect to the EF-Ts, it is responsible for mediating the regeneration of EF-Tu–GDP complex, catalyzing the addition of aminoacyl tRNA into the ribosome enabling protein synthesis. According to Caldas et al. (1998), the EF-Tu has a dual function in protein synthesis and stress response. The results obtained here are in agreement with this finding as spot corresponding to EF-Ts were observed in the three study conditions, and in fact, it was the most intense spot under arsenate presence. Similar results were also described by Pandey et al. (2011), who observed an over-expression of EF-Tu in Anabaena sp. PCC7120 under arsenate stress. Baker-Austin et al. (2007) suggested that the amount of ribosomal sub-units increases due to the requirement of increased protein biosynthesis to combat the cellular stress caused by the presence of metalloids. Based on these constraints, the results presented here reflect the ability of Exiguobacterium sp. S17 to adapt and survive in the presence of arsenic.
Screening of response against arsenic stress by 2DE yielded also proteins that belong to the general metabolism pathways (carbohydrate and amino acid metabolism) and identified their differentially expression in Exiguobacterium sp. S17. In the presence of As(V), a differential up-regulation, glyceraldehyde-3-phosphate dehydrogenase (spot 13) and phosphopyruvate hydratase (spot 48), enzymes involved in the glycolytic pathway were detected. Both proteins are involved in the second stage of glycolysis, and thus in energy (ATP-) generation. In addition, the results showed that, in the presence of arsenate, proteins involved in amino acid metabolism were expressed. Up-regulation was found for: 3-methyl-2-oxobutanoate dehydrogenase (spot 12), involved in the metabolism of leucine, valine and isoleucine, delta-1-pyrroline-5-carboxylate dehydrogenase (spot 1), implicated in arginine and proline metabolism, and finally serine hydroxymethyl transferase (spot 67), performing conversion of serine to glycine. Identification of enzyme involved in the amino acid metabolism is again indicative of an increased energy generation, as deaminated amino acids (after conversion into alpha-keto acids) are fed into the tricarboxylic acid cycle (TCA). TCA cycle is the final pathway for the oxidation of molecules such as amino acids, fatty acids and sugars, and also provides intermediates for biosynthesis. In accordance to this finding, a change was also detected in fumarate hydratase concentration (spot 37), another enzyme of the TCA cycle. These results concur with reports by TCA cycle is the final pathway for the oxidation of molecules such as amino acids, fatty acids and sugars, and also provides intermediates for biosynthesis. Baker-Austin et al. (2007) reported an up-regulation of enzymes that feed into or are part of TCA cycle in Ferroplasma acidarmanus Fer1 under As(III). Taking together reports from the literature and findings from this study, the results could suggest that in Exiguobacterium sp. S17 upregulates significantly the increment of energy-providing metabolic pathways to overcome the cellular stress caused by the exposition may be necessary to manage the cellular stress caused by the addition of arsenic.
Again directing in the energy-providing pathways in the identification of two enzymes involved in purine metabolism was also detected in 2DE gels. Inosine 5-monophosphate dehydrogenase (spot 34) becomes over-expressed in the presence of As(V), as compared with the control, but it is not detected in the presence of arsenite. Purine nucleoside phosphorylase (spot 44) was found over-expressed in control conditions and downregulated in cells under stress. These enzymes have an essential role in providing two precursors (adenine and guanine) for RNA and DNA synthesis. Overexpression is thus understandable as it is known that bacteria have evolved salvage pathways of purine and pyrimidine, which can be used as a source of energy, and for providing carbon and nitrogen (Middelhoven et al. 1984). On the other hand, the down-regulation under arsenic presence suggests that the cells by strictly regulating the purine synthesis, thus insuring bacterial survival under harmful conditions.
Finally, one other down-regulated protein, transketolase (spot 26), was detected under arsenic stress compared with the control. This enzyme participates in the formation of xylulose 5-phosphate in the pentose phosphate pathway for NADPH production. The same effect was observed in Anabaena under arsenic stress by Pandey et al. (2011). According to Kaiser (1979), this down-regulation may be due to high sensitivity toward accumulated H2O2 in the cells after As treatment. Alternatively, one might speculate that this diminution is due directly to the arsenate presence, which acts as competitive inhibitor of the reaction catalyzed by transketolase (Gorbach et al. 1981). These results constitute evidence of the adaptive response of Exiguobacterium sp. 17 against arsenic stress conditions, which thereby guarantee its survival.
Concluding remarks
In conclusion, 25 proteins were described that are differentially expressed under As(V) or As(III) exposition; all (100 %) of these proteins were expressed or over-expressed in the presence of As(V), while 32 and 48 % of these were detected in the As(III)-supplemented medium with As(III) and or in the control, respectively. Probably, most of them are synthesized by the cells but in lower concentrations than in medium with As(V) and were undetected. The lower expression of proteins detected in the presence of As(III) could be explained by the finding that Exiguobacterium sp. S17 is able to remove the As(III) through ACR3 and ArsB pumps, while the As(V) has to must be reduced and recently expelled prior excretion. In fact, the genes responsible for the reduction and transport of arsenic were found in the strain under study (unpublished). It is highly probable that the greater permanence and higher concentration of metalloid inside within the cells could be responsible for triggering the defense machinery to insure adaptation and survival, exactly as reflected in the increased expression of stress-related proteins, described in this work (Fig. 3). The current findings are the basis for future studies, e.g., as the construct generation of deletion mutants for of the arc3 gene, one of the most prominent proteins identified in this survey might help understand if this gene in Exiguobacterium sp. S17 is responsible for the ability to adapt and survive under arsenate stress conditions (Fig. 4).
Schematic representation of the proteins involved in the transport of arsenic (adapted from Paez-Espino et al. 2009)
This report, representing the first proteomic study of arsenic tolerance in an Exiguobacterium strain isolated from the HAAL, constitutes an important contribution to future studies about the potential application of this strain in bioremediation processes.
References
Achour AR, Bauda P, Billard P (2007) Diversity of arsenite transporter genes from arsenic-resistant soil bacteria. Res Microbiol 158:128–137
Anderson CR, Cook GM (2004) Isolation and characterization of arsenate-reducing bacteria from arsenic contaminated sites in New Zealand. Curr Microbiol 48:341–347
ATSDR—Agency for Toxic Substances and Disease Registry (2000) Toxicological profile for arsenic september prepared for the US department of health and human services by syracuse research corporation
Baker-Austin G, Dopson M, Wexler M, Sawers RG, Stemmler A, Rosen BP, Bond PL (2007) Extreme arsenic resistance by the acidophilic archaeon ‘Ferroplasma acidarmanus’ Fer1. Extremophiles 11:425–434
Borja Sánchez B, Champomier-Vergès MC, Anglade P, Baraige F, de los Reyes-Gavilán CG, Margolles A, Zagorec M (2005) Proteomic analysis of global changes in protein expression during bile salt exposure of bifidobacterium longum NCIMB 8809. J Bacteriol 187(16):5799–5808
Cai L, Lui G, Rensing C, Wang G (2009) Genes involved in arsenic transformation and resistance associated with different levels of arsenic-contaminated soils. BMC Microbiol 9:4
Caldas TD, Yaagaubia AE, Richarme G (1998) Chaperone properties of bacterial elongation factor EF-Tu. J Biol Chem 273:11478–11482
Castro de Esparza ML (2006) Remocion del arsenico en el agua para bebida y bioremediacion de suelos from http://www.bvsde.paho.org/bvsacd/cd51/remocion-agua.pdf International Congress Mexico City, Natural arsenic in groundwater of Latin America
Champomier-Vergés MC, Zagorec M, Fadda S (2010) Bacteria adaptation to streeful environments. In: Mozzi F, Raya R, Vignolo G (eds) Biotecnology of lactic acid bacteria. Novel applications. Wiley, USA, pp 57–71
Ciprandi A, Azevedo Barauna R, Valadares Santos A, Costa Goncalves E, Peixe Carepo MS, Cruz Schneider MP, Silva A (2012) Proteomic response to arsenic stress in chromobacterium violaceum. J I Omics 2:69–73
Cleiss-Arnold J, Koechler S, Proux C, Fardeau ML, Dillies MA, Coppee JY, Arsène-Ploetze F, Bertin PN (2010) Temporal transcriptomic response during arsenic stress in Herminiimonas arsenicoxydans. BMC Genomics 11:709
Couoh Uicab YL, Canto Canche BB, Islas Flores I (2010) Review of characteristics of ABC transporters involved in fungal pathogenesis. Tecnociencia Chihuahua 4(2):87–96
Duché O, Trémoulet F, Glaser P, Labadie J (2002) Salt stress proteins induced in Listeria monocytogenes. Appl Environ Microbiol 68:1491–1498
Farías ME, Revale S, Mancini E, Ordoñez O, Turjanski A, Cortez N, Vazquez MP (2011) Genome sequence of Sphingomonas sp. S17, isolated from an alkaline, hiperarsenic, and hypersaline volcano-associated lake at high altitud in the Argentinean Puna. J Bacteriol 193:3686–3687
Farías ME, Rascovan N, Toneatti DM, Albarracín VH, Flores MR, Poire DG, Collavino MM, Aguilar OM, Vazquez MP, Polerecky L (2012) The discovery of stromatolites developing at 3570 m above sea level in a high-altitude volcanic lake Socompa, Argentinean Andes. PLOS ONE (accepted to publish)
Ferguson JF, Gavis J (1972) A review of arsenic cycle in natural waters. Water Res 6(11):1259–1274
Fernández Zenoff V, Siñeriz F, Faria ME (2006) Diverse responses to UV-B radiation and repair mechanisms of bacteria isolated from high-altitude aquatic environments. Appl Environ Microbiol 72(12):7857–7863
Gorbach ZV, Maglysh SS, Kubyshin VL, Ostrovskii IM (1981) Purification and properties of rat liver transketolase. Biokhimiia 46:1963–1969
Hartl FU (1996) Molecular chaperones in cellular protein folding. Nature 381:571–580
Kaiser W (1979) Reversible inhibition of the Calvin cycle and activation of oxidative pentose phosphate cycle in isolated intact chloroplasts by hydrogen peroxide. Planta 145:377–382
Kiriyama MT, Oka M, Takehana M, Kobayashi S (2001) Expression of a small heat shock protein 27 (HSP27) in mouse skin tumors induced by UVB-irradiation. Biol Pharm Bull 24:197–200
Kramer GF, Ames BM (1988) Isolation and characterization of a selenium metabolism mutant of Salmonella typhimurium. J Bacteriol 170:736–743
Kvint K, Nachin L, Diez A, Nystrom T (2003) The bacterial universal stress protein: function and regulation. Curr Opin Microbiol 6(2):140–145
Lakshmi Sunita MS, Prashant S, Bramha Chari PV, Nageswara Rao S, Padma Balaravi PB, Kishor K (2012) Molecular identification of arsenic-resistant estuarine bacteria and characterization of their ars genotype. Ecotoxicology 21(1):202–212
Lopez L, Pozo C, Rodelas B, Calvo C, Juarez B, Martinez-Toledo MV, Gonzalez-Lopez J (2005) Identification of bacteria isolated from an oligotrophic lake with pesticide removal capacities. Ecotoxicology 14(3):299–312
Middelhoven WJ, Hoogkramer-Te Niet MC, Kreger-Van Rij NJW (1984) Trichosporon adeninivorans sp. nov., a yeast species utilizing adenine, xanthine, uric acid, putrescine and primary n-alkalymines as the sole source of carbon, nitrogen and energy. Antonie Van Leeuwenhoek 50:369–378
Mogk A, Deurling E, Voderwulbecke S, Vierling E, Bukau B (2003) Small heat shock proteins, ClpB and the DnaL system from a functional triade in reversing protein aggregation. Mol Bio 50(2):585–595
Moore LE, Pfeiffer R, Warner M, Clark M, Skibola C, Steinmous C, Alguacil J, Rothman N, Smith MT, Smith AH (2005) Identification of biomarkers of arsenic exposure and metabolism in urine using SELDI technology. J Biochem Mol Tox 19:176
Mukhopadhyay R, Rosen BP, Phung LT, Silver S (2002) Microbial arsenic: from geocycles to genes to enzymes. FEMS Microbiol Rev 26:311–325
Nguyen TTA, Michaud D, Cloutier C (2009) A proteomic analysis of the aphid Macrosiphum euphorbiae under heat and radiation stress. Insect Biochem Mol Biol 39:20–30
Okeke BC, Laymon J, Oji C, Crenshaw S (2007) Rapid bioreduction of hexavalent chromium in water by Exiguobacterium sp. GS1. In: ASM general 107th meeting, ASM Press, Toronto, pp Q-199
Ordoñez OF (2012) Factores ambientales extremos en ecosistemas microbianos de humedales altoandinos: mecanismos de adaptacion. PhD thesis from Universidad Nacional de Tucumán, Tucumán, Argentina
Oremland RS, Stolz JF (2003) The ecology of arsenic. Science 300:939–944
Paez-Espino D, Tamames J, de Lorenzo V, Canovas D (2009) Microbial responses to environmental arsenic. Biometals 22:117–130
Pandey S, Rai R, Lal Chand Rai (2011) Proteomics combines morphological, physiological and biochemical attributes to unravel the survival strategy of Anabaena sp. PGG7120 under arsenic stress. J Proteomics 75:921–937
Parsell DA, Lindquist S (1993) The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Annu Rev Genet 27:437–496
Parvatiyar K, Alsabbagh EM, Ochsner UA, Stegemeyer MA, Smulian AG, Hei Hwang S, Jackson CR, McDermott T, Hassett DJ (2005) Global analysis of cellular factors and responses involved in Pseudomonas aeruginosa resistance to arsenite. J Bacteriol 187(14):4853–4864
Pattanapipitpaisal P, Mabbett AN, Finlay JA, Beswick AJ, Paterson-Beedle M, Essa A, Wright J, Tolley MR, Badar U, Ahmed N, Hobman JL, Brown NL, MAcaskie LE (2002) Reduction of Cr(VI) and bioaccumulation of chromium by gram positive and gram negative microorganisms not previously exposed to CR-stress. Environ Technol 23:731–745
Rodrigues FD, Ivanova N, He Z, Huebner M, Zhou J, Tiedje JM (2008) Architecture of thermal adaptation in an Exiguobaterium sibiricum strain isolated from 3 million year old permafrost: a genome and transcriptome approach. BMC Genomics 9:547
Rodriguez-Gabriel MA, Russell P (2005) Distinct signaling pathways respond to arsenite and reactive oxygen species in Schizosaccharomyces pombe. Eukaryot Cell 4(8):1396–1402
Ron EZ (2006) Bacterial stress response. In: Dworkin M, Falkow S, Resenberg E, Schleifer KH, Stackebrandt E (eds) The Prokaryotes. Springer, Singapore, pp 1012–1027
Rosen BP (2002) Biochemistry of arsenic detoxification. FEBS Lett 529:86–92
Rosen BP, Lui Z (2009) Transport pathways for arsenic and selenium: a miniriew. Environ Int 35(3):512–515
Siegele D (2005) Universal stress proteins in Escherichia coli. J Bacteriol 187(18):6253–6254
Silver S, Phung LT (2005) Genes and enzymes involved in bacterial oxidation and reduction of inorganic arsenic. Appl Environ Microbiol 71:599–608
Smedley PL, Kinniburgh DG (2002) A review of source, behavior and distribution of arsenic in natural waters. Appl Geochem 17:517–568
Tedengren M, Olsson B, Bradley B, Zhou LZ (1999) Heavy metal uptake, physiological response and survival of the blue mussel (Mytilus edulis) from marine and brackish waters in relation to the induction of heat-shock protein 70. Hydrobiologia 393:261–269
Tkáčová J, Angelovičová M (2012) Heat shock proteins (Hsps): a review. Anim Sci Biotechnol 45(1):349–353
Zhang Y, Ma YF, Qi SW, Meng B, Tausif Chaudhry M, Liu SQ and Liu SJ (2007) Responses to arsenate stress by Comamonas sp. strain CNB-1 at genetic and proteomic levels. Microbiology 153:3713–372
Acknowledgments
This work was supported by Préstamo BID PICT 2010 N°1788-Agencia Nacional de Promoción Científica y Tecnológica. Carolina Belfiore and Omar F. Ordoñez are recipients of a CONICET fellowship. We thank Dra. Silvia de Moreno for their technical assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by H. Atomi.
Rights and permissions
About this article
Cite this article
Belfiore, C., Ordoñez, O.F. & Farías, M.E. Proteomic approach of adaptive response to arsenic stress in Exiguobacterium sp. S17, an extremophile strain isolated from a high-altitude Andean Lake stromatolite. Extremophiles 17, 421–431 (2013). https://doi.org/10.1007/s00792-013-0523-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-013-0523-y