Abstract
A temperate phage, Psymv2, was isolated from an Antarctic soil bacterium, Psychrobacter sp. MV2. The morphology of Psymv2 was typical of the Siphoviridae, with an isometric head and non-contractile tail. The Psymv2 genome was found to be 35,725 bp in length, had a G + C content of 44.5 %, with 49 protein-coding genes and one tRNA gene predicted. Integration of Psymv2 occurred at an ssrA gene, with the last 27 bases of this gene directly repeated at the prophage ends. The genome was organised in a modular fashion: integration, regulation, packaging, head assembly, tail assembly, host specificity and lysis. While the genome sequence had little similarity on a nucleotide level to previously reported phage sequences, the genome architecture resembled that of Siphoviridae of low G + C Gram-positive bacteria. The closest relatives to Psymv2 were uncharacterized putative prophages within the P. arcticus 273-4 and Acinetobacter baumannii 6013113 genomes. Global alignment of the Psymv2 genome and these prophages revealed significant conservation of the structural modules despite the large spatial divergence of their hosts. A number of unique ORFs were identified in the Psymv2 genome that may contribute to phage and lysogen fitness.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The McMurdo Dry Valleys of eastern Antarctica are one of the most extreme environments on earth; characterised by low temperatures (fluctuating from −55 °C to +25 °C), low water and nutrient availability, high soil salinity, high radiation and strong katabatic winds (Doran et al. 2002; Horowitz et al. 1972; Vincent 1988). These cold desert soils harbour a surprisingly diverse range of microbes, the majority of which are taxonomically distinct and, as yet, uncultured (Pointing et al. 2009; Smith et al. 2006). Microbial activity in this extreme environment is thought to be largely shaped by abiotic factors (Cary et al. 2010). Allen et al. (2010) have, however, recently shown that phages play a crucial role in the regulation of microbial community structure in Arctic soils.
While aquatic Antarctic viral communities are known to be highly diverse (Lisle and Priscu 2004; López-Bueno et al. 2009; Säwström et al. 2008) the abundance and impact of viruses in Antarctic soils has received little to no research attention. In one of the few studies to date, soils collected from the rims of Antarctic ephemeral ponds were shown to contain only 16-fold less extracellular virus like particles than moist organic rich soils (Williamson et al. 2007). The incidence of lysogeny within these soils was additionally estimated by Williamson et al. (2007) and a correlation between water content and lysogeny observed; with a potential increase in lysogeny in drier soils.
The cultivation and genomic characterisation of cold-adapted viruses from this unique environment is of great interest as these organisms potentially represent important elements of the Antarctic desert soil trophic structure, and may play key roles in critical processes such as lateral gene transfer. In this study we screened cultured psychrophilic Dry Valley soil isolates for prophages and report on the isolation of phage Psymv2 from a Psychrobacter spp. To our knowledge this is the first report of the isolation and complete genome sequencing of a bacteriophage specific for a cold-adapted terrestrial microbe.
Materials and methods
Sampling, enrichment and isolation of bacterial strains
Soil samples were collected aseptically from beneath hypoliths in Miers Valley (78°05′S, 163°45′E) in the McMurdo Dry Valleys, South Victoria Land, Antarctica. Samples were stored at −80 °C and defrosted at 4 °C prior to use. Bacterial enrichment cultures were established by suspending soil samples (ca. 1–1.5 g) in 6 ml half-strength liquid Nutrient Broth (Merck) and incubated at 16 °C for 2.5 h with intermittent shaking. Dilution series of the enrichment cultures were plated onto half-strength Nutrient Agar (Merck) and isolates grown at 16 °C for 2–7 days. Isolated colonies were picked and subjected to additional rounds of purification by inoculation onto fresh NA plates. Isolates capable of growth at 4 °C were selected for induction experiments.
The 16S rRNA gene was amplified by PCR from genomic DNA using the universal bacterial primers E9F and U1510R (Hansen et al. 1998; Reysenbach and Pace 1995). The PCR product was cloned using the pGem-T plasmid cloning system (Promega) according to standard procedures. Plasmids were sequenced with M13 primers using an ABI PRISM Big Dye Terminator Cycle Sequencing Ready Reaction kit on an ABI PRISM 3130xl Genetic Analyzer (Perkin Elmer). Sequence identity was analysed by comparison to the GenBank nr/nt database using the BLASTN program (Altschul et al. 1990).
Prophage induction and purification
Isolates were screened for prophages through the addition of various concentrations (0.001–6 μg/ml) of Mitomycin C (MC; Sigma–Aldrich) to mid-log phase cultures for 1 h at 16 °C. Bacterial cells and debris were removed from induced cultures by low speed centrifugation and filter sterilisation using 0.45 μm pore size syringe filters (Nalgene). Bacteriophage suspensions for electron microscopy analyses and DNA isolation were prepared using the method of Ackermann (2009). Briefly, bacteriophages were collected from filtered suspensions by centrifugation at 25,000 g for 60 min in a fixed-angle rotor. Phages were then washed at least twice with 0.1 M ammonium acetate (pH 7) and sedimented at each wash step by centrifugation as described. Sedimented phages were gently resuspended in 0.1 M ammonium acetate (pH 7).
Transmission electron microscopy
For negative staining, bacteriophage suspensions were spotted onto carbon-coated copper grids for 1 min, stained for a further minute with 2 % (wt/vol) uranyl acetate, pH 4 and air-dried (Ackermann 2009). Phages were visualised using a Jeol 200CX electron microscope, at 60 kV and magnifications ranging from 50,000× to 100,000×.
Phage DNA isolation
Viral genomic DNA was extracted using a standard proteinase K and SDS protocol (Sambrook and Russell 2001). Briefly, phage preparations were incubated with both DNase (5 mg/ml) and RNase (10 mg/ml) for 2 h at 37 °C. Samples were then treated with Proteinase K (100 μg/ml) in the presence of 0.5 % SDS for 1 h at 65 °C, extracted with phenol/chloroform, and the DNA recovered from the aqueous phase by ethanol precipitation.
Cloning, sequencing and analysis
Genomic DNA from Psychrobacter sp. MV2 was cloned into the pCC1FOS™ vector (Epicentre) using the manufacturer’s recommended protocol. Fosmid clones harbouring the phage genome were identified by Southern blot using Psymv2 DNA, labelled with the DIG-DNA labelling kit (Roche), as probe. Four fosmid clones, with an average insert size of approximately 30 kb, were sequenced by Inqaba Biotechnical Industries (Pty) Ltd (Pretoria, SA) using a 454 GS-FLX platform (Roche Diagnostics). The genome sequencing of phage Psymv2 was completed by PCR amplification directly from genomic DNA. The chromosomal insertion site of the bacteriophage was sequenced by genome walking from an adaptor ligated Psychrobacter sp. MV2 genomic library using the method of Siebert et al. (1995).
Sequence analysis and assembly was carried out using the CLC Genomics Workbench package (CLC bio, Katrinebjerg, Denmark). The accuracy of the genome sequence assembly was verified by amplification by PCR. Ab initio gene predictions were performed using GeneMark.hmm 2.0 (http://www.exon.biology.gatech.edu/heuristic_hmm2.cgi) (Besemer and Borodovsky 1999), fgenesv and fgenesb (http://www.linux1.softberry.com/berry.phtml) (Softberry, Mount Kisco, NY, USA) using the generic bacterial genetic code. tRNA and tmRNA genes were predicted using Aragorn (http://www.130.235.46.10/ARAGORN/) (Laslett and Canback 2004) and BRUCE v1.0 (Laslett et al. 2002). Sequences were compared to the GenBank_nr database using the Blastp and/or tblastx algorithms with an E-value cut-off of 10−5. Annotations were additionally based on results from RPS-BLAST searches, with an E-value threshold of 10−2, against the NCBI conserved domains database or CDD (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) (Marchler-Bauer et al. 2009) and Interpro scan searches (http://www.ebi.ac.uk/Tools/InterProScan/) (Hunter et al. 2009). Transmembrane domains were predicted using the Transmembrane hidden Markov model TMHMM v2.0 (Krogh et al. 2001) and protein localisation predicted using PSORTb v3.0.2 (Yu et al. 2010). Genome alignments with the closest prophages were performed using the progressive Mauve algorithm (Darling et al. 2010).
Nucleotide sequence accession numbers
The 16S rRNA (1,496 bp) and ssrA gene sequences of Psychrobacter sp. MV2 and the phage Psymv2 genome were deposited in GenBank under accession numbers HQ610925, JF270479 and JF270478, respectively.
Results and discussion
Isolation and morphology of bacteriophage Psymv2
Twenty-two psychrophilic microbial strains were cultured from soil collected from the Miers Valley, Antarctica, and screened for prophages by treatment with Mitomycin C (MC). Phage Psymv2 was induced from an aerobic non-motile Gram-negative coccobacillus, with optimal induction achieved at 1 μg/ml MC. The 16S rRNA gene sequence of the bacterial isolate had 99.06 % identity to that of the strict psychrophile Psychrobacter frigidicola type strain DSM 12411T (Bowman et al. 1996; GenBank accession number AJ609556). The isolate, termed Psychrobacter sp. MV2, had an optimal growth temperature of 16 °C and was incapable of growth above 27 °C. Members of the genus Psychrobacter, within the class Gammaproteobacteria, have been isolated from a wide variety of low temperature marine and terrestrial environments, as well as various other sources such as food products, human tissues and body fluids. The widespread distribution of members of this genus reflects their capacity for reproduction within a wide range of temperatures (−10 to 42 °C) (Bowman et al. 1996; Maruyama et al. 2000; Romanenko et al. 2004; Bowman 2006; Bakerman et al. 2006). Several psychrotrophic strains, such as the recently sequenced Psychrobacter arcticus 273-4 (Ayala-del-Río et al. 2010) isolated from Siberian permafrost, are seen as models for understanding cold-adaptation in microorganisms. No further characterisation of Psychrobacter sp. MV2 has been carried out to date.
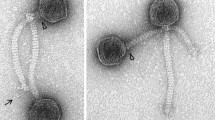
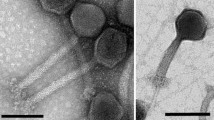
The morphology of Psymv2 was examined by electron microscopic analysis of phage particles (Fig. 1). Psymv2 had an isometric head with an average diameter of approximately 53 nm and an approximately 188 nm long non-contractile tail. Terminal tail fibres were additionally visible. This is the characteristic morphology of the B1 morphotype group of the Siphoviridae family, order Caudovirales (Ackermann 2007).
General features of the Psymv2 genome
The Psymv2 genome was sequenced and annotated as outlined in the Materials and Methods. The double stranded (ds) DNA genome was 35,725 base pairs (bp) in length, with an average guanine-plus-cytosine (G + C) content of 44.5 %. This is similar to the G + C composition of the currently sequenced Psychrobacter strains: P. arcticus 273-4 (42.8 %; Ayala-del-Río et al. 2010), P. cryohalolentis strain K5 (42 %; Genbank NC_007969) and Psychrobacter sp. PRwf1 (44 %; Genbank NC_009524). Segments within the phage genome with lower G + C contents, that may delineate functional clusters and/or regions of lateral gene transfer, were identified and are discussed later.
A total of 49 putative open reading frames (ORFs), with a minimum of 50 amino acids (aa) coding length, were identified. The start codon usage for the ORFs was 75.5 % ATG (37/49) and 24.5 % GTG (12/49) and nine of these ORFs are transcribed from the complementary strand. The deduced amino acid sequence of each ORF was compared to the NCBI nr database using Blastp software with an E-value cut-off of e-5. Of the 49 Psymv2 coding sequences (CDS), 34 (71.4 %) had homologues in the Genbank database, with top hits to proteins encoded by uncharacterised functional or cryptic prophages (76.5 % or 26/34), characterised phages (11.7 % or 4/34) and bacterial genomes (11.7 % or 4/34). The latter may, however, be present in as yet unrecognised prophage and prophage-like elements. Predicted functions could be assigned to 51 % (25/49) of the phage Psymv2 proteins, based on sequence homology, conserved motifs, HMM profiles and other sequence signatures. The remaining 24 (49 %) protein products could not be assigned a putative function. A single tRNA gene specific for Lys/Asn at bp 11,082–11,160 was predicted with Aragorn (Laslett and Canback 2004). The genetic map of the Psymv2 prophage is given in Fig. 2. The positions, putative functions and top Blastp hits of the proteins encoded by Psymv2 are given in Table 1.
Schematic illustration of phage Psymv2 in the Psychrobacter sp. MV2 genome. The nucleotide sequence numbering is shown above and predicted open reading frames (ORFs) illustrated by arrows indicating the direction of transcription. ORFs with functional annotations are labelled, the abbreviations terS and terL are for the small and large terminase subunits respectively. HP indicates hypothetical proteins and H-T, head-to-tail. The location of the phage att sites are shown
The genome of phage Psymv2 displayed a compact modular arrangement of functional gene clusters for integration, regulation, packaging, head morphogenesis, head–tail joining, tail morphogenesis and lysis (Fig. 2). While the order of functional modules is similar to that of the Lambda-like group of Siphoviridae phages (reviewed by Casjens 2005), the position of the genes responsible for host cell lysis downstream of the tail morphogenesis gene cluster is atypical. A similar arrangement has been noted in other Siphoviridae phages (Casjens 2008), including the well-studied Pseudomonas phage D3 (Kropinski 2000), the recently described Burkholderia phage phi644-2 (Ronning et al. 2010) and several phages infecting low G + C Gram-positive bacteria (Canchaya et al. 2003).
The closest related elements to Psymv2, based on the greatest number of shared homologous proteins, were uncharacterised prophages within the genomes of members of the Pseudomonadales. An uncharacterised lambda-like prophage between bp 551715 and 585095 in the P. arcticus 273-4 genome (Genbank accession number: NC_007204; Ayala-del-Río et al. 2010) shared the greatest number of homologous proteins with the highest identity (35–93 %). These clustered within the tail morphogenesis modules of both prophages. Proteins within the packaging and head morphogenesis module of Psymv2 had the highest identity (62–74 %) to proteins encoded by an uncharacterised prophage in the whole genome shotgun sequence of Acinetobacter baumannii 6013113 (Genbank accession number: NZ_ACYR01000154.1). For the purposes of this paper these putative prophages will be termed Psyar273-4/1 and Acba6013113/1, respectively.
Host interaction and phage regulation modules
The product of Psymv2 ORF1 is a putative integrase protein with conserved motifs of the tyrosine site-specific recombinase family. The Psymv2 integrase had 65 % identity to the predicted integrase encoded by Acba6013113/1 and 41 % to that encoded by Psyar273-4/1. The integrase had 25–30 % identity to numerous phage integrase proteins, with the highest identity (30 %) to the integrase (Bbp50) of the podovirus Bordetella BPP-1. Several proteins involved in phage regulation were identified, with Psymv2 ORFs 9, 10 and 11 encoding lambda CI-, Cro- and CII-like proteins, respectively. The CI and Cro repressor proteins had the highest identity to the cognate proteins of Pseudomonas phage D3 (32 and 50 % identity, respectively), while the CII homolog had 38 % identity to gp14 of Burkholderia phage Bcep176. None of these proteins showed any sequence identity to those ascribed the same function in Psyar273-4/1 or Acba6013113/1. The regulatory region additionally contains a DNA-binding protein (encoded by ORF 13) that may function in a manner similar to the Roi protein, providing superinfection immunity.
In the majority of characterised dsDNA bacteriophages, host cell lysis is a result of the combined action of two proteins; endolysin and holin (Wang et al. 2000). Typical bacteriophage endolysins have an N-terminal catalytic domain and a C-terminal cell wall binding domain. While the protein encoded by Psymv2 ORF 49 did not show significant similarity to characterised bacteriophage endolysins, the presence of these two conserved domains within the protein facilitated functional assignment. The product of ORF 49 is a 164 aa polypeptide with an N-terminal domain (aa 6–88) that shows significant sequence similarity (E-value 5.85e-20) to DUF847 super family members, predicted to be related to lysozyme enzymes. The C-terminal region (aa 91–154) contains a potential peptidoglycan binding domain (E-value 2.17e-15) (Table 1). Interestingly, the closest Blastp match to the Psymv2 putative lysin was to the Bordetella phage BPP-1Bbp2 protein (56 % identity) of the Podoviridae family (Table 1).
Holins represent one of the most diverse groups of proteins, with greater than 50 divergent gene families. The lack of sequence similarity between members of this group is probably a reflection of the evolutionary pressures related to their function as regulators of the duration of the lytic infection cycle (Wang et al. 2000). Holins are typically small proteins of between 71 and 161 aa, with two to three transmembrane domains, and may possess a dual translation initiation site. In most phages the ORF encoding holin immediately precedes or overlaps the endolysin gene (Wang et al. 2000). Psymv2 ORF 48, immediately upstream of the ORF encoding the predicted Psymv2 lysin, encodes a small protein of 108 aa. This protein contains two predicted transmembrane domains (TMDs) (aa 10–32 and aa 45–67) and may be localised to the cytoplasmic membrane (PSORTb localisation score 10.00). The first two codons of ORF48 are the initiation codons GTG and ATG. Therefore, this protein may function as a class II holin.
Packaging and structural modules in Psymv2
The packaging module of phage Psymv2 contains ORFs 23 and 24, which are predicted to encode the small and large subunits of the phage terminase, respectively (Table 1). The small subunit is responsible for recognising and binding the cos site in concatemeric phage DNA, the endonuclease and ATPase activities of the large subunit are then responsible for cleaving the DNA for packaging into the phage head (Rao and Feiss 2008). Based on sequence similarity and the presence of conserved domains, ORFs 25, 26 and 27 are predicted to encode the portal protein, the phage maturation protease and the major capsid protein, respectively (Table 1). The packaging and head morphogenesis proteins of phage Psymv2 have 62–68 % identity to related proteins of Acba6013113/1 and show little to no identity to those encoded by Psyar273-4/1. The Psymv2 portal, protease and capsid show homology to cognate proteins of the Enterobacteria phage HK97 and Pseudomonas phage D3.
In the majority of Siphoviridae the packaging and head genes are followed by a cluster of head or tail completion and tail encoding genes. A similar arrangement was seen in the Psymv2 genome (Fig. 2; Table 1). Psymv2 ORFs 29 and 30, with overlapping stop and start codons, encode proteins that share conserved motifs found in the head (or tail) completion proteins of long-tailed phages such as HK97 (Cardarelli et al. 2010). Head (or tail) completion proteins are specialised proteins of the phage head that serve as an attachment point for the tail and allow for the movement of the DNA out of the capsid during infection.
The tail morphogenesis module of Psymv2 spans ORFs 37–45. A number of putative minor tail components and tail assembly proteins are encoded by ORFs 37, 39, 41-44. The protein encoded by ORF 38 had only limited homology to any known or hypothetical phage proteins but was identified as the phage tape measure protein (tmp) based on the presence of four predicted TMDs (aa 391–413, 437–459, 519–541 and 548–570), and a conserved core region of a family of phage tail proteins, which includes the TP901 tmp. Additionally, the size of the encoded protein (1,107 aa) corresponds well with the measured Psymv2 tail length of approximately 188 nm. It is well established that the tmp determines tail length (Katsura and Hendrix 1984). Psymv2 ORF 43 encodes a tail-associated protein with an N-terminal metallo-endopeptidase motif (aa 6–109) and a C-terminal domain (aa 124–226) with predicted cell-wall hydrolase activity. This tail-associated protein may then function during cell adsorption and penetration as a peptidoglycan-degrading enzyme. The enzymatic activity of the protein would cleave both the peptide cross-links and polysaccharide chains within the peptidoglycan cell wall, facilitating entry of the dsDNA genome through the phage tail and into the host cell. Psymv2 ORF45 encodes a large protein of 1,503 aa, the N-terminal region (aa 1–896) showed significant sequence identity (84 %, E-value 0.0) to the cognate protein of Psyar273-4/1. This protein has similarity to the tail fibres of a number of lambda-like phages, including HK97 and HK022, and is the putative phage tail fibre involved in determining host specificity.
Embedded within the Psymv2 structural cluster were a number ORFs encoding proteins of unknown function but conserved sequence, including ORF 33 and 34, which encode members of the DUF646 and DUF3168 families, respectively (Table 1). ORFs 31 and 32 may be involved in superinfection exclusion, and are discussed later.
Comparison of Psymv2 with related phages: identification of unique regions
A global genome alignment of Psymv2 with the closest known elements Psyar273-4/1 and Acba6013113/1 revealed the mosaic arrangement of the functional modules within the Psymv2 genome (Fig. 3a). Clearly defined regions of nucleotide sequence homology or local collinear blocks (LCB), indicated by similarly coloured blocks in Fig. 3a, were apparent. The first such region spans the ORF encoding the integrase in all three phages. The packaging and head module (ORF 21–30) of Psymv2 showed clear homology to the same region in Acba6013113/1. Part of the Psymv2 tail morphogenesis module (ORFs 39–45), including the host specificity protein, showed homology to the module within the Psyar273-4/1 genome. The abrupt cessation of the homologous regions between the phages suggests modular exchanges or rearrangements and insertions. These results support the current view that the genomes of many tailed phages are mosaics that arise through genetic exchange from a diverse pool of phages (Casjens 2005). While large sections of the three phage genomes share little nucleotide sequence similarity, the overall order of functional modules is conserved (Fig. 3a). As if often the case, this is a reflection of the conserved regulatory and transcriptional control patterns governing phage life cycles.
Comparison of phage Psymv2 nucleotide and protein sequences to related phages. a Whole genome alignment of phage Psymv2 and putative prophages within the P. arcticus 273-4 and A. baumanni 6013113 genomes. Similarly coloured regions indicate homology or local collinear blocks (LCB) between nucleotide sequences, with the level of similarity indicated by the height of the bars within each LCB. Genome alignments were performed using Mauve. b and c Blastp hits for Psymv2 proteins against the nr database (b) and against proteins of viral origin (virus[Orgn]) (c), with an E-value of less than 1e-5. Coloured stars indicate the top hits for each coding sequence and circles other hits with similarity. The percentage identity between the Psymv2 proteins and the Blastp hits is indicated, with red above 90 %, pink 70–90 %, green 50–70 %, blue 30–50 % and black less than 30 % identity. The abbreviations γP and βP designate Gammaproteobacteria and Betaproteobacteria respectively. The grey blocks indicate regions with little to no similarity to known phages. The % G + C composition of the Psymv2 genome is indicated by the bar at the bottom of the figure. A sliding window length of 75 bp was used with % G + C composition scaled from black (0 %) to white (100 %) (colour figure online)
To gain a greater understanding of the homology of Psymv2 to other bacteriophages, and to identify proteins that are unique to Psymv2, the Psymv2 predicted proteins were compared to both the NCBI nr database and proteins of viral origin using Blastp (Fig. 3b, c, respectively). From this it was clear that both the head and tail structural proteins of Psymv2 show the highest levels of homology to cognate proteins in prophages (Fig. 3b) but also have detectable homologues in a number of Siphoviridae phages, including the well-studied HK97, HK022 and D3 (Fig. 3c). The structural proteins, particularly the terminase and procapsid, are often the most conserved proteins of tailed phages (Casjens 2008). The notable exception within the structural morphogenesis region was the tmp (Box in Fig. 3b, c). While no single phage shared homologues to all or even a majority of the proteins involved in integration, regulation and lysis, detectable homologues were identified amongst a number of phages in the Siphoviridae, Podoviridae and Myoviridae families (Fig. 3c).
The Psymv2 genome contains four regions that appear to encode proteins specific to Psymv2 with no homologues encoded by known phages and with lower G + C contents (Grey shading in Fig. 3b, c). The novelty of these regions was confirmed by tBlastx analysis of the nucleotide sequence (results not shown). These regions may have been acquired through horizontal gene transfer. These regions span ORFs 2–5, 15–20, 31–32 and 46–47. The first of these regions includes four genes of unknown function (ORFs 2–4) and ORF 5, which encodes a predicted dksA (DnaK suppressor) transcriptional regulator. The protein encoded by ORF 4 showed 85 % identity to a hypothetical protein encoded by a gene in the P. arcticus 273-4 genome that lies outside of the predicted Psyar273-4/1 prophage region.
The second region, from ORF 15 to 20, encodes hypothetical proteins of unknown function, includes a tRNA gene and is flanked by a homing endonuclease gene. The protein encoded by ORF 16 had 42 % identity to a protein encoded by A. baumannii SDF, while ORF 18 encoded a protein with 51 % identity to a protein encoded by an uncharacterised Mu-like prophage in the P. arcticus 273-4 genome. A predicted tRNA gene specific for Lys/Asn at bp 11,082–11,160 was identified in this region. Analysis of the codon usage in Psymv2 revealed a significantly higher number of codons for Lys (3.584 %) compared to the average codon usage of between 0.675 and 2.45 %. The presence of the tRNA Lys/Asn gene in the Psymv2 genome therefore appears to correspond to the high usage of the Lys codon in the Psymv2 genes. Interestingly, Ayala-del-Río et al. (2010) recently showed that cold-adapted proteins encoded by P. arcticus 273-4 have an increased Lys content. Amino acid substitutions that increase the structural flexibility of proteins, such as Lys for Arg, have been proposed to be necessary for activity at low temperatures and are thought to be indicators of cold-adaptation (Ayala-del-Río et al. 2010). Flanking this region is ORF 21, which encodes a homing endonuclease containing a conserved active site core motif of the HNHc family (Table 1). Homing endonuclease genes are known as selfish genetic elements and are commonly found in phages and bacteria (reviewed by Marcaida et al. 2010). A homing mechanism is used to copy the sequence, and often surrounding sequences, to the same site at different loci, such as in related phages. In phage T4 the SegB homing endonuclease has been shown to be responsible for the transfer of the flanking T4 tRNA region to related phages during co-infection (Brok-Volchanskaya et al. 2008). The HNH endonuclease, and any surrounding co-transferred sequences, may then confer a selective advantage to the phage and/or its lysogen.
The region from ORF 31 to 32 lies within the Psymv2 structural module and may be involved in superinfection exclusion. Both encoded proteins have a TMD and are predicted to be membrane proteins, with the ORF 31 gene product being significantly related to a family of lipoproteins (Table 1). Many phages use membrane targeted proteins that prevent the entry of DNA from superinfecting phage into the cell. The protein encoded by ORF 32 had 84 % identity (57 % query coverage) to a hypothetical protein encoded by P. arcticus 273-4. The last region, from ORF 46 to 47, lies between the structural and lysis modules of Psymv2 and the encoded proteins had no known function or significant homologues.
Psymv2 bacterial integration site
The Psymv2 chromosomal integration site was sequenced by genome walking outwards from both ends of the integrated prophage on the Psychrobacter sp. MV2 genome. Analysis of the sequence upstream of the prophage integrase (ORF 1) using Aragorn (Laslett and Canback 2004) identified an ssrA gene as the Psymv2 integration site. The ssrA gene encodes a small stable tmRNA, with both tRNA- and mRNA-like functions, that is known to be involved in the recovery of stalled ribosomes on degraded mRNA. The tmRNA encodes a short peptide tag which is added to partially synthesised proteins targeting them for degradation (Karzai et al. 2000). Immediately upstream of the ssrA gene in the Psychrobacter sp. MV2 genome was an ORF encoding a putative phospholipase/carboxylesterase (Fig. 4a) with 90 % identity (E-value 5e-116) to a cognate protein encoded by P. arcticus 273-4.
Integration of phage Psymv2 into the T-loop of the tRNA region of the Psychrobacter sp. MV2 ssrA gene. a The integrated Psymv2 prophage is flanked by an exact 27 bp direct repeat that duplicates the last 27 bp of the 3′ end of the Psychrobacter sp. MV2 ssrA gene. b The tRNA-like domain of the Psychrobacter sp. MV2 tmRNA predicted using Aragorn (Laslett and Canback 2004). The Psymv2 phage att site is indicated by the arrow
The last 27 bp of the ssrA gene were found to be directly repeated at the opposite prophage end (Fig. 4a). This 27 bp identical repeat presumably represents the conserved core sequence of the Psymv2 attachment site (attB, attP, attL and attR) and functions as the recognition site for Psymv2 integration and excision. The att core sequence represents the T-loop of the tRNA region of the Psychrobacter sp. MV2 tmRNA (Fig. 4b). The exact repetition of this sequence in the phage genome upstream of the integrase (attL) reconstitutes the ssrA gene following integration. The ssrA gene was found to be highly conserved in the P. arcticus 273-4 genome with 97 % nucleotide identity. Neither Psyar273-4/1 nor Acba6013113/1 were found to be integrated at an ssrA gene. The 27 bp Psymv2 att core sequence was, however, found in numerous Psychrobacter and Acinetobacter genomes (including A. baumannii 6013113) with 100 % identity, in many cases flanking an integrase or transposase gene (results not shown). Prophage insertion at ssrA genes is estimated to occur for 8 % of predicted prophages (Fouts 2006).
Concluding remarks
In this paper we present the first phage isolated from a member of the genus Psychrobacter. Morphologically, the phage can be classified as a Siphoviridae. This classification was supported by the fact that many of the phage structural proteins have homology to cognate proteins of several members of this family. The closest relatives in terms of gene order, nucleotide and predicted amino acid sequence similarity were uncharacterised prophages within the genomes of Psychrobacter arcticus 273-4 and Acinetobacter baumannii 6013113. A more detailed understanding of the phylogeny of Psymv2 was hampered, as is the case with most phages, by the mosaic nature of the genome and the limited number of complete genome sequences of related characterised phages. The availability of additional genomic data from phages infecting bacteria within the family Moraxellaceae should no doubt provide greater insight.
A number of interesting and unique ORFs were identified within the Psymv2 genome. These will be the subject of future investigations as they are of interest for their potential involvement in the environmental fitness of the lysogen, Psychrobacter sp. MV2. Recent studies have shown that, at least in marine environments, lytic infection appears to be prevalent in highly productive habitats, while lysogeny is more common under unfavourable environmental conditions. Here prophages may support the survival of their host by encoding repressors and transcriptional regulators that inhibit host genes controlling superfluous metabolic processes (Paul 2008).
Viral control of microorganisms is now considered to be an important component of ecological and geochemical processes in most environments. Several bacteriophages infecting deep-sea hydrothermal vent bacteria have been characterised, including the well-studied Siphoviridae GVE2; a lytic phage of the thermophile Geobacillus sp. E263 (Liu et al. 2006; Liu and Zhang 2008). An analysis of the effects of GVE2 infection on the host cellular proteome revealed phage-induced differential expression of host proteins involved in energy metabolism, replication and transportation of energy and matter (Wei and Zhang 2010). A similar investigation of the response of Pscyhrobacter sp. MV2 to infection with Psymv2 would contribute to our understanding of virus-host interactions at low temperature.
Abbreviations
- aa:
-
Amino acid
- CDS:
-
Coding sequence
- LCB:
-
Local collinear block
- MC:
-
Mitomycin C
- ORF:
-
Open reading frame
- TMD:
-
Transmembrane domain
References
Ackermann H-W (2007) 5500 Phages examined in the electron microscope. Arch Virol 152:227–243
Ackermann H-W (2009) Basic phage electron microscopy. In: Clokie MRJ, Kropinski AM (eds) Bacteriophages. Humana Press Inc, USA, pp 113–126
Allen B, Willner D, Oechel WC, Lipson D (2010) Top-down control of microbial activity and biomass in an Arctic soil ecosystem. Environ Microbiol 12:642–648
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Ayala-del-Río HL, Chain PS, Grzymski JJ, Ponder MA, Ivanova N, Bergholz PW, Di Bartolo G, Hauser L, Land M, Bakermans C, Rodrigues D, Klappenbach J, Zarka D, Larimer F, Richardson P, Murray A, Thomashow M, Tiedje JM (2010) The genome sequence of Psychrobacter arcticus 273-4, a psychroactive siberian permafrost bacterium, reveals mechanisms for adaptation to low-temperature growth. Appl Environ Microbiol 76:2304–2312
Bakerman C, Ayala-del-Río HL, Ponder MA, Vishnivetskaya T, Gilichinsky D, Thomashow MF, Tiedje JM (2006) Psychrobacter cryohalolentis sp. nov. and Psychrobacter arcticus sp. nov., isolated from Siberian permafrost. Int J Syst Evol Microbiol 56:1285–1291
Besemer J, Borodovsky M (1999) Heuristic approach to deriving models for gene finding. Nucleic Acid Res 27:3911–3920
Bowman J (2006) The genus Psychrobacter. In: Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, Dworkin M (eds) The prokaryotes. Springer-Verlag, New York, pp 920–930
Bowman JP, Cavanagh J, Austin JJ, Sanderson K (1996) Novel Psychrobacter species from Antarctic ornithogenic soils. Int J Syst Bacteriol 46:841–848
Brok-Volchanskaya VS, Kadyrov FA, Sivogrivov DE, Kolosov PM, Sokolov AS, Shlyapnikov MG, Kryukov VM, Granovsky IE (2008) Phage T4 SegB protein is a homing endonuclease required for the preferred inheritance of T4 tRNA gene region occurring in co-infection with a related phage. Nucleic Acids Res 36:2094–2105
Canchaya C, Proux C, Fournous G, Bruttin A, Brüssow H (2003) Prophage genomics. Microbiol Mol Biol Rev 67:238–276
Cardarelli L, Lam R, Tuite A, Baker LA, Sadowski PD, Radford DR, Rubinstein JL, Battaile KP, Chirgadze N, Maxwell KL, Davidson AR (2010) The crystal structure of bacteriophage HK97 gp6: defining a large family of head-tail connector proteins. J Mol Biol 395:754–768
Cary SC, McDonald IR, Barrett JE, Cowan DA (2010) On the rocks: the microbiology of Antarctic Dry Valley soils. Nat Rev Microbiol 8:129–138
Casjens SR (2005) Comparative genomics and evolution of the tailed-bacteriophages. Curr Opin Microbiol 8:451–458
Casjens SR (2008) Diversity among the tailed-bacteriophages that infect the Enterobacteriaceae. Res Microbiol 159:340–348
Darling AE, Mau B, Perna NT (2010) progressiveMauve: multiple genome alignment with gene gain, loss, and rearrangement. PLoS One 5:e11147
Doran PT, Priscu JC, Lyons WB, Walsh JE, Fountain AG, McKnight DM, Moorhead DL, Virginia RA, Wall DH, Clow GD, Fritsen CH, McKay CP, Parsons AN (2002) Antarctic climate cooling and terrestrial ecosystem response. Nature 415:517–520
Fouts DE (2006) Phage_Finder: automated identification and classification of prophage regions in complete bacterial genome sequences. Nucleic Acids Res 34:5839–5851
Hansen MC, Tolker-Neilson T, Givskov M, Molin S (1998) Biased 16S rDNA PCR amplification caused by interference from DNA flanking the template region. FEMS Microbiol Ecol 26:141–149
Horowitz NH, Cameron RE, Hubbard JS (1972) Microbiology of the dry valleys of Antarctica. Antarct Sci 176:242–245
Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L et al (2009) InterPro: the integrative protein signature database. Nucleic Acids Res 37:D211–D215
Karzai AW, Roche ED, Sauer RT (2000) The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue. Nat Struct Biol 7:449–455
Katsura I, Hendrix RW (1984) Length determination in bacteriophage lambda tails. Cell 39:691–698
Krogh A, Larsson B, von Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580
Kropinski AM (2000) Sequence of the genome of the temperate, serotype-converting, Pseudomonas aeruginosa bacteriophage D3. J Bacteriol 182:6066–6074
Laslett D, Canback B (2004) ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res 32:11–16
Laslett D, Canback B, Andersson S (2002) BRUCE: a program for the detection of transfer-messenger RNA genes in nucleotide sequences. Nucleic Acids Res 30:3449–3453
Lisle JT, Priscu JC (2004) The occurrence of lysogenic bacteria and microbial aggregates in the lakes of the McMurdo Dry Valleys, Antarctica. Microb Ecol 47:427–439
Liu B, Zhang X (2008) Deep-sea thermophilic Geobacillus bacteriophage GVE2 transcriptional profile and proteomic characterization of virions. Appl Microbiol Biotechnol 80:697–707
Liu B, Wu S, Song Q, Zhang X, Xie L (2006) Two novel bacteriophages of thermophilic bacteris isolated from deep-sea hydrothermal fields. Curr Microbiol 53:163–166
López-Bueno A, Tamames J, Velázquez D, Moya A, Quesada A, Alcamí A (2009) High diversity of the viral community from an Antarctic lake. Science 326:858–861
Marcaida MJ, Munõz IG, Blanco FJ, Prieto J, Montoya G (2010) Homing endonucleases: from basics to therapeutic applications. Cell Mol Life Sci 67:727–748
Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M et al (2009) CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res 37:D205–D210
Maruyama A, Honda D, Yamamoto H, Kitamura K, Higashihara T (2000) Phylogenetic analysis of psychrophilic bacteria isolated from the Japan Trench, including a description of the deep-sea species Psychrobacter pacificensis sp. nov. Int J Syst Evol Microbiol 50:835–846
Paul JH (2008) Prophages in marine bacteria: dangerous molecular time bombs or the key to survival in the seas? ISME J 2:579–589
Pointing SB, Chan Y, Lacap DC, Lau MC, Jurgens JA, Farrell RL (2009) Highly specialized microbial diversity in hyper-arid polar desert. Proc Natl Acad Sci USA 106:19964–19969
Rao VB, Feiss M (2008) The bacteriophage DNA packaging motor. Annu Rev Genet 42:647–681
Reysenbach A-L, Pace NR (1995) Thermophiles. In: Robb FT, Place AR (eds) Archaea: A laboratory manual. Cold Spring Harbour Laboratory Press, New York, pp 101–107
Romanenko LA, Lysenko AM, Rohde M, Mikhailov VV, Stackebrandt E (2004) Psychrobacter maritimus sp. nov. and Psychrobacter arenosus sp. nov., isolated from coastal sea ice and sediments of the Sea of Japan. Int J Syst Evol Microbiol 54:1741–1745
Ronning CM, Losada L, Brinkac L, Inman J, Ulrich RL, Schell M, Nierman WC, Deshazer D (2010) Genetic and phenotypic diversity in Burkholderia: contributions by prophage and phage-like elements. BMC Microbiol 10:202
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York
Säwström C, Pearce I, Davidson AT, Rosén P, Laybourn-Parry J (2008) Influence of environmental conditions, bacterial activity and viability on the viral component in 10 Antarctic lakes. FEMS Microbiol Ecol 63:12–22
Siebert PD, Chenchik A, Kellogg DE, Lukyanov KA, Lukyanov SA (1995) An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res 25:1087–1088
Smith JJ, Ah Tow L, Stafford W, Cary C, Cowan DA (2006) Bacterial diversity in three different Antarctic cold desert mineral soils. Microbiol Ecol 51:413–421
Vincent WF (1988) Microbial ecosystems of Antarctica. Cambridge University Press, Cambridge
Wang I-N, Smith DL, Young R (2000) Holins: the protein clocks of bacteriophage infections. Ann Rev Microbiol 54:799–825
Wei D, Zhang X (2010) Proteomic analysis of interactions between a deep-sea thermophilic bacteriophage and its host at high temperature. J Virol 84:2365–2373
Williamson KE, Radosevich M, Smith DW, Wommack KE (2007) Incidence of lysogeny within temperate and extreme soil environments. Environ Microbiol 9:2563–2574
Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, Brinkman FSL (2010) PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615
Acknowledgments
The authors wish to thank Antarctica New Zealand, the National Research Foundation of South Africa and the Claude Leon Foundation for financial support.
Conflict of interest
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by L. Huang.
Rights and permissions
About this article
Cite this article
Meiring, T.L., Marla Tuffin, I., Cary, C. et al. Genome sequence of temperate bacteriophage Psymv2 from Antarctic Dry Valley soil isolate Psychrobacter sp. MV2. Extremophiles 16, 715–726 (2012). https://doi.org/10.1007/s00792-012-0467-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-012-0467-7