Abstract
In Archaea, the importance of the proteasome system for basic biological processes is only poorly understood. Proteasomes were partially purified from Halobacterium by native gradient density ultracentrifugation. The peptidase activity profiles showed that the 20S proteasome accumulation is altered depending on the physiological state of the cells. The amount of active 20S particles increases in Halobacterium cells as a response to thermal and low salt stresses. In the same conditions, Northern experiments showed a positive transcriptional regulation of the alpha and beta proteasome subunits as well as of the two proteasome regulatory ATPases, PANA and PANB. Co-immunoprecipitation experiments demonstrated the existence of a physical interaction between the two Proteasome Activating Nucleotidase (PAN) proteins in cell extracts. Thus, a direct regulation occurs on the PAN–proteasome components to adjust the protein degradation activity to growth and environmental constraints. These results also indicate that, in extreme halophiles, proteasome mediated proteolysis is an important aspect of low salt stress response. The tri-peptide vinyl sulfone inhibitor NLVS was used in cell cultures to study the in vivo function of proteasome in Halobacterium. The chemical inhibition of proteasomes was measured in the cellular extracts. It has no effect on cell growth and mortality under normal growth conditions as well as under heat shock conditions. These results suggest that the PAN activators or other proteases compensate for loss of proteasome activity in stress conditions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Protein degradation is a controlled process that eliminates the damaged or the misfolded protein that could not be rescued by the chaperone system in order to prevent their intracellular aggregation (Sherman and Goldberg 2001). Protein hydrolysis therefore represents an important aspect of the environmental stress response as well as of the protein quality control (Goldberg 2003). A selective proteolysis also occurs in order to provide an irreversible way of inactivating, at the appropriate times, the regulatory molecules involved in cell cycle progression or in metabolic switches (Coux et al. 1996). In eukaryotes, the ATP-dependent ubiquitin/26S proteasome system is the main proteolytic machinery (Lee and Goldberg 1996). The 26S proteasome can be separated in two subparticles: the 20S self-compartmentalized peptidase core and the 19S dependent regulatory particle (RP) (Zwickl and Baumeister 1999). Six regulatory subunits (Rpt) form the base of the RP (Patel and Latterich 1998; Rubin et al. 1998). These proteins belong to a large family of proteins known as the AAA ATPases (Atpases Associated with a variety of cellular Activities) (Patel and Latterich 1998). They recognize specific proteins as targets and function as disassembly molecular machines (Sauer et al. 2004). Archaea possess a simplified version of the eukaryotic proteasome made of only one or two types of alpha subunits and one catalytic beta subunit (Maupin-Furlow et al. 2000). The energy dependent regulatory complex consists in a single type of Rpt-like protein that was named Proteasome Activating Nucleotidase (PAN) (Zwickl et al. 1999). The recombinant PAN complexes were found to stimulate the proteasome dependent hydrolysis of proteins (Wilson et al. 2000). In this process, the binding of the substrate activates ATP hydrolysis and promotes protein unfolding and opening of the axial gate of the peptidase 20S core (Benaroudj et al. 2003; Navon and Goldberg 2001). Considering its simplicity, the archaeal PAN-20S system may serve as a paradigm to study the basic role of the proteasome in cell physiology.
The cellular stress response is a reaction to any form of macromolecular damage independent of the underlying cause aimed to re-establish cellular homeostasis. In this response, chaperones are induced to mediate global protein stabilisation, to prevent or control aggregation and to eventually assist protein refolding (Hartl 1996). It is generally acknowledged that intra cellular proteolysis is involved in the stress response in order to remove molecular debris and to break down the misfolded proteins (Goldberg 2003). However, the biological significance of the PAN–proteasome system in archaeal cell physiology and environmental/extremophilic adaptation remains poorly documented. Halobacterium represents an archaeon model to address this question: it displays a high resistance to multiple stressors, including temperature, radiation and desiccation (Kottemann et al. 2005; Shukla 2006). As an extreme halophilic organism, Halobacterium accumulates multimolar salt concentration in the cytosol, and most of its proteins present an obligatory salt adaptation for their solubility and stability (Madern et al. 2000; Oren 2008). These organisms thrive in salterns where they often have to face changes in salt concentrations. Therefore, low salt conditions represent a unique natural stress for these types of cells, which is reflected by the synthesis of various stress response proteins (Bidle 2003; Franzetti et al. 2001; Kuo et al. 1997; Mojica et al. 1997). In addition, Halobacterium possesses 2 PAN proteins that strongly differ in their N- and C-terminal regions (Chamieh et al. 2008). In Rpt homologues, the C-terminal region is believed to be responsible for binding to the alpha rings 20S proteasome alpha subunits, while the N-ter region is responsible for substrate recognition (Verma et al. 2000). For these reasons, it was proposed that the two PANs from halophilic Archaea could play a non-redundant role in cell physiology.
The objective of the present work was to specify the role of the archaeal proteasome system in stress and growth response and to assess the respective role of the two PAN proteins as proteolytic regulators. For this purpose, we have measured the 20S proteasome intracellular activity and the transcriptional expression of the PAN–proteasome subunits during growth and exposition to heat and salt shocks. Since the capability of the two PAN proteins to interact in vivo remains unknown, co-immunoprecipitation experiments were also performed to assess the in vivo interaction between the two different PAN subunits. We also examined the effect of chemical proteasome inhibition on cell growth and viability under stress conditions.
Experimental methods
Halobacterium cells cultivation, stress and proteasome inhibition experiments
Halobacterium salinarum R1 cells were grown in mediums containing 4.2 M NaCl, at 37°C with agitation as described before (Oesterhelt and Stoeckenius 1974). Cells were maintained as continuous cultures between OD600 0.3 and 0.8. For stress response experiments, mid-log exponential phase (OD620 = 0,6) cells were transferred to a 55°C incubator or diluted in salt-free medium to 2.5 M NaCl final. For anti-proteasome drug treatment, 30 μM of tri-peptide vinyl sulfone inhibitor carboxybenzyl-leucyl-leucyl-leucine vinyl sulfone (NLVS) purchased from Affinity were added to the cultivation medium. Aliquots were taken at different times. The LIVE/DEAD BacLight bacterial viability kit from Invitrogen was used to assess the rate of mortality in the different physiological conditions (Leuko et al. 2004). For RNA analysis the cells were centrifuged at 9000g for 5 min at 4°C. The pellets were frozen in liquid nitrogen and kept at −80°C until use.
Sucrose gradient experiments and peptidase activity measurements
For native sucrose gradient fractionation, 30 ml of cells suspension were collected at different times during growth, after heat and salt stress exposure or after cell treatments with the NLVS anti-proteasome drug. The cellular extracts were prepared and the proteins were processed by density-gradient sedimentation as described in Chamieh et al. (2008). The 20S proteasome activities present in the different cellular extracts and sucrose gradient fractions were measured by using the substrate Suc-LLVY-AMC as described (Chamieh et al. 2008). Protein concentrations were determined with the Bradford protein assay (Bio-Rad). These values were used to normalize the measured activities. To evaluate the specificity of the endopeptidase activities that were detected in the sucrose gradient fractions, parallel assays were performed in which the protein extracts were preincubated at 37°C for 30 min in the presence of 10 μM of the NLVS anti-proteasome drug prior to sucrose gradient ultracentrifugation.
Immunoprecipitations experiments
For native co-immunoprecipitations experiments, 1 gram of freshly pelleted Halobacterium cells were resuspended in 10 ml of a buffer containing 2.5 M KCl, 50 mM Tris–HCl pH 7.5; 1% triton and 0.04% of “Complete™” anti-proteases cocktail from GE Healthcare. The cells were lysed and homogenized by flushing several times through a needle connected to a syringe. Cell debris was eliminated by centrifugation at 13000 rpm for 10 min. Five micrograms of specific rabbit PANA or PANB antibodies were added to 1 ml of Halobacterium protein extract. The antibodies were previously purified on a peptide affinity column as described (Chamieh et al. 2008). The mixtures were incubated on a rolling shaker for 1 h at 37°C. The reactions were then incubated with 100 μl of Protein A-Sepharose beads suspension (GE Healthcare) to precipitate the antibody–antigen complexes for an additional hour at 37°C. The beads were then washed three times with a washing buffer containing 3 M KCL, 20 mM Tris–HCl pH 8.3 and 0,1% Triton-X100. The precipitated proteins were released by the addition of an SDS sample buffer (50 mM Tris–HCl pH 6, 8, 100 mM dithiothreitol, 2% SDS, 0.1% bromophenol blue, 10% glycerol) followed by boiling for 5 min. The immunoprecipitates were subjected to 12% SDS-polyacrylamide gels, and the resolved proteins were transferred to nitrocellulose membranes (Hybond-P, GE Healthcare). The blots were probed with anti-PAN A or PAN B. These antibodies were raised against synthetic peptides and are specific for each type of PAN proteins (Chamieh et al. 2008). The immunoprecipitates were also probed with different antibodies to assess the specificity of the detected PANA–PANB physical interaction. For this purpose we used anti-malate dehydrogenase (MalDH). All polysera were used at a 1:10000 dilution. Immunoreactives bands were visualized by chemiluminescence according to the supplier’s protocol (ECL detection kit; GE Healthcare). Details on antibodies source, suppliers and preparation are described in Oesterhelt and Stoeckenius (1974).
Northern blot analysis
Riboprobes directed against the proteasome alpha and beta subunits, the two PAN regulatory ATPases and the thermosome alpha subunit were synthesized. For probe construction, the following oligonucleotide sets were used for PCR amplifications: for the proteasome alpha subunit 5′-GCGTCGAGAAACTCCACAAG-3′ and 5′-CTTCCACTCGTTGGACGTG-3′; for the proteasome beta subunit 5′-AACGTCCAGAAAGTCGAGGA-3′ and 5′-TACGTGTCGGACATCGAACT-3′ for PANA 5′-AAGATCTTCCGCATCCACAC-3′ and 5′-TCGGAGTCCTGATCGAGTTT-3′; for PANB 5′-GAGTGGGGTGTTGTTGCAC-3′ and 5′-AGAGCAACTGCATCATCGTG-3′: for the thermosome alpha subunit 5′-TTCAGGAGACCGAGATCGAC-3′ and 5′-GATGTCGTTGACGTTCGAGA-3′. In each case, a polymerase chain reaction (PCR) product was generated using the Phusion® High-Fidelity DNA Polymerase and genomic Halobacterium DNA as a template. The PCR DNA products (300nt on average) were then directly cloned into PCR2.1-TOPO vector purchased from Invitrogen. The plasmids were sequenced to determine probe orientation on the plasmid and to transcribe anti-sense mRNA from T7 or SP6 promoters. The transcription reactions were carried out using the maxiscript kit purchased from Ambion according to the manufacturer’s instructions. For riboprobe labelling, 0.5 μl of a Bio11-UTP 10 mM solution (Ambion) was added to the reaction mixture.
Total RNAs from Halobacterium cells were extracted from cell pellets by using the RNAwiz reagent (Ambion) according to the manufacturer’s instructions. RNA pellets were then resuspended in DEPC treated water and formaldehyde loading dye (Ambion) according to the manufacturer’s instructions. After heating at 65°C for 15 min, RNA samples were then subjected to electrophoresis through 1% formaldehyde agarose gel. Equal loading (between 5 and 10 μg) was normalized based on quantification of the fluorescence of ribosomal RNAs in ethidium bromide-stained gels. The fractionated RNAs were transferred to nylon membranes (Ambion-Brightstar-Plus) by capillary blotting in RNase free 20× SSC overnight. The blots were immobilized by UV cross-linking (Stratalinker 1800, Stratagene) and hybridized at 68°C in UltraHyb buffer (Ambion) containing the labelled riboprobes. The membranes were then washed in high stringency conditions for 2 × 5 min in 2× SSC, 0.1% SDS at 68°C and low stringency washes 2 × 15 min in 0.1 SSC, 0.1% SDS at 68°C. The signal was then detected by chemiluminescence by using the Bright Star Biodetect kit from Ambion according to the manufacturer’s instructions. The Northern blot experiments were repeated at least three times by using RNA samples from independent experiments.
Results
Activity levels of 20S particles fluctuate during the growth of Halobacterium cells
In order to assess the importance of the 20S proteasome function in the growth of Halobacterium, we measured chymotrypsin-like endopeptidase activities in cell lysates. The proteasome catalytic activity depends on the assembly of the alpha and beta subunits and, in vivo, the subunits oligomerisation rate represents an important level to regulate the proteasome function (Witt et al. 2006). Therefore, the intracellular 20S activity cannot be simply extrapolated from the total amount of proteasome alpha and beta proteins detected in crude cell extracts. To overcome this problem, the 720 kDa 20S particles were partially purified from Halobacterium extracts on calibrated sucrose gradients and the corresponding endopeptidase activity was measured as described previously by using the fluorogenic substrate Suc-LLVY-AMC (Chamieh et al. 2008). In these experiments NLVS, a proteasome specific drug, was used to make sure that no other proteasome-like hydrolysing activities were present in the high molecular weight protein fractions or induced depending upon growth. The same control was performed in the stress experiments. The peptidases activities were normalized based on the total protein contents in the extracts that were applied to the gradients. In our experimental conditions, Halobacterium cells showed a generation time of about 10 h. A typical growth curve is presented in Fig. 1a. To assess for growth-dependent proteasome activity modifications, we measured the 20S hydrolytic activity as a function of the cell cultures ODs. The result obtained from four independent experiments is presented in Fig. 1a. The maximum 20S activity was found during the early exponential phase. A marked drop in the peptidase activity was always observed during mid-exponentially growth phase, i.e. between OD 0.6 and 1.4. These results showed that the proteasome activity is regulated as a function of growth in a halophilic Archaea. Later (between OD 1.4 and 1.8), the level of proteasome activity was found to vary from one experiment to the other. This is reflected by large standard errors. This is probably due to different lag times, from one culture to the other, before entering into the stationary phase.
Modifications of the 20S proteasome activity and PAN–proteasome subunits genes expression levels during Halobacterium growth. a Halobacterium growth curve (circles). The sampling times used for total RNAs extractions and the Northerns experiments presented on b are indicated by arrows. The 20S peptidase activities (right Y axis) are plotted against growth time (triangles). In this experiment, post-membrane protein extracts were prepared from cells at different growth stages. Equal amounts of proteins were fractionated by sucrose gradient ultracentrifugation as described in experimental procedures. The endopeptidase activities were measured in the proteasome fractions by using the fluorogenic peptide substrate Suc-L-L-V-Y-AMC. B. Northern blot experiments: total RNAs were extracted from cells at different growth stages. Ten micrograms of RNA were loaded on denaturing agarose gels. The 23S and 16S rRNAs were visualized by ethidium bromide (BET) staining to control equal loading. After transfer on nitrocellulose membranes (Ambion), the transcripts encoding for proteasome subunit alpha (Proteα), proteasome subunit beta (Proteβ), proteasome activating subunit A (PANA) and proteasome activating subunit B (PANB) were visualized by using Bio11-UTP labeled anti-sense riboprobes. Results are representative of a minimum of three independent experiments
Growth phase dependent regulation of the 20S subunits genes expression
A transcript level analysis was carried out in order to assess whether the observed effect of growth on the 20S proteasome activity is due to changes in the alpha and beta subunits genes expression patterns. Total RNAs were extracted from cellular aliquots taken at different growth stages (Fig. 1a). To control for transfer artifacts and non-specific effects the blots were stained with ethidium bromide. The Northern blot experiments revealed that the two proteasomal subunits mRNAs resulted from the synthesis of monocistronic transcripts (Fig. 1b). The kinetic analysis of the catalytic beta subunits mRNAs accumulations showed clear depletions of the transcript levels while the cells are progressing through the exponential phase. In the experiment presented in Fig. 1b, a regain in transcriptional activity was detected in the later stationary phase. The alpha subunits transcript level was also found to fluctuate during growth, but with lesser amplitudes than the beta subunit transcript. Interestingly, the transcriptional alterations of the two 20S subunits genes during the exponential phase are consistent with the 20S endopeptidase activity patterns that we reported above (Fig. 1a) thus suggesting a direct control of 20S activity/accumulation by transcriptional regulation.
Pan A and Pan B are co-regulated during Halobacterium growth
It has been proposed that the PAN ATPases subunits stimulate the breakdown of targeted full proteins due to their unfoldases activities and their ability to promote the proteasome gate opening (Benaroudj et al. 2003). The expression of PANA and PANB, the two proteasome activating ATPases, was therefore studied in order to determine if their regulation could represent a way to control the rate of protein destruction during cell growth. In Halobacterium, the presence of two PAN-like proteins with different N- and C-terminal regions also raised the question of their respective roles in cell physiology. In particular, it was interesting to know whether they could display different expression profiles during cell growth. As for the 20S subunits, the PANs mRNA abundance was analyzed by Northern blot (Fig. 1b). The positions of the detected PAN signals corresponded to monocistronic transcripts. The results showed that two PAN transcripts are co-regulated during growth. The Pan transcripts accumulated to significantly higher levels during the early exponential phase and when cells entered the stationary phase. This finding indicates an involvement of the PAN proteins in the regulation of specific cellular process during archaeal cell growth.
Stress regulation of the 20S peptidase activity
Intracellular proteolysis is believed to be especially important for emergency reactions such as general stress response, both to control the degradation of stress regulatory factors and to eliminate the stress-damaged proteins (Smith et al. 2006). For studying the regulation of the proteasome system during environmental stress response, mid-exponential phase Halobacterium cells were exposed to different stress conditions. Beside temperature, low salt was also considered as a major stressor for Halobacterium cells. These halophilic cells grow optimally above 4 M salt and accumulate multimolar salt concentrations in their cytosol to counterbalance the osmotic pressure (Ginzburg et al. 1970). Due to a peculiar solvation mechanism, the proteins from extreme halophilic Archaea depend on high salt concentrations to remain soluble and stable, in vitro (Madern et al. 2000). Because of this adaptive mechanism, a decrease in the external salt concentration is expected to elicit cytosolic stress conditions. In fact, several studies in halophilic Archaea have reported that changing salinities in the environment greatly influence the expression of a define set of gene and the accumulation of specific proteins, including members of the Hsp family (Bidle 2003; Mojica et al. 1997). To investigate whether the components of the proteasome system also participate in the heat and/or salt shock response, we determined the optimal stress conditions for Halobacterium. To do so, we measured by Northern blot the time course induction of the thermosome alpha subunit (TF55) a bona fide heat shock transcript (Figs. 2b, 3b). The cellular growth and the mortality rates were also measured to assess that, under the chosen conditions, the physiological states of the cells still allow cellular stress responses to take place (data not shown). We selected the conditions that combined the maintenance of cell growth and viability with optimal TF55 induction. For thermal stress, optimal conditions were obtained when the cells were grown at 37°C and then shifted to 55°C for 8 h. For the low salt stress, the optimal condition was obtained when the cultivation media NaCl concentration was diluted from 4.2 to 2.5 M.
The intracellular 20S proteasome activity and the four genes encoding for PAN–proteasome subunits are induced as a response to thermal stress in Halobacterium. a 20S intracellular peptidase activities. Samples were removed at different times after a shift of the culture from 37C to 55°C. Equal amounts of proteins were fractionated on the sucrose gradient. The 20S peptidase activity was examined as described in experimental procedures. b Northern blot analysis of proteasomeα, proteasomeβ, PANA and PANB transcripts after a shift in temperature from 37°C to 55°C. RNA was isolated at the indicated times. Transcripts were identified using labelled riboprobes specific to each gene
A decrease in the external salt concentration result in an accumulation of 20S proteasome particles and in an induction of the gene encoding for PAN and proteasome subunits. a 20S intracellular peptidase activities. Samples were removed at different times after a shift of the culture from 4.2 to 2.5 M NaCl in the cultivation medium. The protein extracts were fractionated on sucrose gradient and examined for proteasome activity as described in experimental procedures. b Northern blot analysis of proteasomeα, proteasomeβ, PanA and PanB transcripts after a hyposaline shock from 4.2 to 2.5 M NaCl in the cultivation medium. RNA was isolated at the indicated times. Transcripts were identified using labelled riboprobes specific to each gene
The 20S proteasome activity was measured as described above to investigate whether the 20S proteasome accumulation undergoes changes when the Halobacterium cells where placed into heat or salt stress conditions. Compared to the non-shocked control experiment, we found that a weak stimulation (about 0.5-fold) of the intracellular 20S peptidase activity occurred progressively until 8 h after exposure to heat shock (Fig. 2a). To determine if this response is specific to heat stresses or is more generally associated to general environmental stresses acting on the proteome fate, we studied the effect of salt stress on the intracellular 20S catalytic activity. In these conditions, the 20S activity was found to increase rapidly by a factor of 0.8 in the first 4 h after the salt dilution shock (Fig. 3a). These changes in specific activity indicated that there are more active 20S particles in cells grown under stress conditions and suggested that, in Archaea, changes in the 20S particle accumulation could participate in the cellular environmental stress response.
Transcriptional stimulation of the PAN and proteasomes subunits by heat and low salt stresses
Northern blot analyses were carried out in order to determine if the stimulations of the 20S activities that we observed under stress conditions were due to a gene regulation process. As mentioned before, the induction of the thermosome alpha subunit transcript was used as a positive control for heat and salt shock responses. The transcription of the heat shock TF55 subunit was found to be strongly induced by the temperature and salt dilution stress. The transcript level exhibited a rapid and transient induction pattern, typical of many heat shock proteins (Figs. 2b, 3b). In the same mRNA preparations, we found that the transcripts encoding for the two proteasome subunits gradually increased when the cells were subjected to both heat and salt stress (Figs. 2b, 3b). These results showed that, in Halobacterium, the proteasome subunits genes are stress regulated. As expected the transcriptional induction patterns of the catalytic beta subunit precede the induction in 20S activity patterns that we measured after stress exposure (Figs. 2a, 3a). These results suggest that, under stress conditions, the 20S cellular activity is importantly controlled by the transcription rate of the catalytic beta subunit.
The expression of the genes encoding for PANA and PANB, the two proteasome activating ATPases, was also studied in order to ascertain if these proteins may represent global regulators of protein destruction under physico-chemical stress conditions. PanA and PanB transcripts were both detected under normal growth conditions as monocistronic RNA fragments with apparent lengths of 1070 and 1105 bases, respectively. Replicate experiments demonstrated that the PanA and PanB transcripts abundance consistently increase in response to high temperature and salt dilution. Under heat stress conditions both PAN transcripts increased gradually up to 8 h (Fig. 2b). The low salt regime resulted in a dramatic, but transient induction of the two transcripts. The same transient induction was observed for the thermosome subunit. Interestingly, the PanA induction peak was found to be delayed compared to the one from PanB and the thermosome (Fig. 3b). This study showed that the PAN proteasome activators are global stress response proteins that are tightly regulated at the transcript level.
The low salt induction of PAN transcripts was followed by a sharp down regulation. A similar transient induction pattern was also observed for the TF55 alpha subunit, indicating that a salinity mediated signalling process exists for PAN proteins and the TF55 chaperonin system. In all salt stress experiments, the PanB gene was induced before PanA. In some experiments the induction of PanB was even already detectable after 15 min, while the PANA gene induction occurred only after 1 h. These observations suggested that the two regulatory factors are not co-regulated under salt stress.
Oligomerisation state and physical interactions of PANA and PANB in Halobacterium cells
We have previously shown that, in Halobacterium cellular extracts, the PAN ATPases formed labile complexes with the 20S particle (Chamieh et al. 2008). Since the PAN proteins expression is growth and stress regulated, we investigated whether their association rates could also be modified depending on the growth stages or the stress conditions. The cellular extracts were fractionated on sucrose gradients and the PAN proteins were immunodetected as described (Chamieh et al. 2008). We tested cell extracts from mid-log and stationary phases as well as heat-shocked and salt-shocked cells. In all experiments, the sedimentation profiles of the PAN proteins in the gradients were found to be identical compared to unstressed control experiments (data not shown). From these negative results one can conclude that the formation of stable PAN–proteasome complexes do not occur depending on the physiological conditions, in order to increase the proteolytic activity under stress conditions.
In previous experiments we showed that the PAN A and PAN B proteins displayed a different ability to interact physically with the 20S proteasome complex (Chamieh et al. 2008). In this paper we showed that, apart from under low salt conditions, the PAN proteins are co-regulated. These findings raised questions about the respective role of these proteins and their capacity to form heteroligomers. Co-immunoprecipitation experiments were used to answer this question. For these experiments, the protein and salt concentrations were chosen in order to allow antigen–antibody interactions while maintaining the salt dependent solubility of halophilic proteins. The presence of PAN A or PAN B in the immunoprecipitates was analyzed by Western blot (Fig. 4). Control experiments were performed with different protein specific antibodies to make sure that the detected PAN proteins were specifically pulled-down by the immunoprecipitation process and not by non-specific interaction with the Sepharose beads because of artefactual aggregation. The results presented in Fig. 4 showed that PAN B co-immunoprecipitated with PAN A. In the reverse experiment in which PAN B immunoprecipitates were probed with PAN A antibodies, a weak PAN A signal was detected. These data indicated that PAN A and PAN B can interact together in the Halobacterium cellular extract.
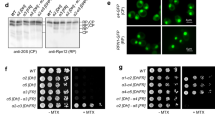
Physical interactions between PANA and PANB proteasome regulatory ATPases can be detected by co-immunoprecipitation. Halobacterium protein extracts were incubated with anti-PANA or anti-PANB antibodies. The immunocomplexes were precipitated by using protein A Sepharose beads, boiled and separated by reducing 12% SDS-PAGE. Control experiments in which the proteins extracts were incubated with the protein A Sepharose beads in the absence of antibodies were also performed (C). The precipitated proteins were transferred on nitrocellulose membranes and analyzed by anti-PANA (middle) or anti-PANB (right) Western blots. A control experiment in which the immunoprecipitates are probed with anti-MalDH is presented on the left panel
Proteasome deficient Halobacterium cells can maintain cell growth under normal and stress conditions
Experiments done with T. acidophilum have demonstrated that covalent modification of the proteasome beta subunit actives sites by the NLVS peptidomimetic drug did not prevent cell growth under normal conditions, but causes growth arrest in heat shocked culture (Ruepp et al. 1998). These data indicated that the proteasome could play a role in the survival of T. acidophilum under stress. We performed similar experiments with Halobacterium cells. Exponential cultures of Halobacterium were incubated with 30 μM of NLVS inhibitor for different times. After washing the cells in a cultivation medium without inhibitor, the intracellular efficiency of the drug was evaluated by measuring the residual 20S proteasome activity in sucrose gradients fractionated lysates, as described in “Experimental methods”. Compared to the untreated samples, about 96 to 92% of the 20S chymotryptic activity was lost during the first 8 h of drug treatment, as shown of Fig. 5a. Note that in these experiments we loaded an equal amount of proteins on the sucrose gradient. The same results were obtained by normalizing the loadings on a cell number basis. After 24 h, the proteasome activity was still reduced by about 85%. Cultivations in the presence of increased amount of inhibitors did not lead to a more complete inhibition of the 20S proteasome activity. In these proteasome deficient conditions, we found that the cells doubling time was not significantly affected. However, the treated cells were found to have a slightly lower OD when entering the stationary phase compared to the untreated control. No significant differences were detected when we studied, at different incubation times, the cell morphologies and mortality rates by using the LIVE/DEAD BacLight bacterial viability assay (data not shown). Thus, these experiments revealed that, in Halobacterium, an important loss in proteasome activities do not hamper dramatically the cellular metabolism under normal conditions. To test if the proteasome activity is important for thermal stress resistance, untreated and treated cells were shifted to 55°C. Under heat stress the untreated Halobacterium cells showed a significant decrease in growth rate during the first 20 h and then enter the stationary phase. Signs of cellular mortality, cell shape modifications and bleaching began to be significant after 48 h at 55°C. In the presence of NLVS, the cells behaved exactly as the untreated ones. These data showed that, in Halobacterium, a high level of 20S proteasome activity is not essential for the cells to survive for prolonged periods in thermal stress conditions.
Effects of proteasome inhibition under normal growth conditions as well as under conditions of heat shock induced stress. a Assessment of the Halobacterium 20S proteasome inhibition by the proteasome specific inhibitor NLVS. After 8 h of cell treatment with 30 μM of NLVS, total proteins were extracted and equal amounts were separated on 5–25% sucrose gradients. The 20S peptidase activity was measured in the different fractions as described in the experimental procedures. Empty circles untreated sample, black triangles treated sample, black circles correspond to a negative control in which 5 μM of NLVS was added to the protein extract prior to gradient fractionation. b Growth curves of Halobacterium cultures in the presence (30 μM) and absence of the proteasome inhibitor NLVS, respectively. Empty circles and squares correspond to untreated cells grown at 37 and 55°C, respectively. Black circles and squares correspond to treated cells grown at 37 and 55°C, respectively. Results are representative of a minimum of three independent experiments
Discussion
The present work aimed to provide information on the role of the proteasome system in the basic cell physiology of a prokaryotic cell, with a focus on stress response and halophilic adaptation. We studied the regulation of the eukaryotic-like PAN–proteasome system from Halobacterium, a halophilic archaeon that comprises only two types of subunits forming the peptidase 20S core and two ATPases, PANA and PANB, forming the proteasome activating particle that stimulates the breakdown of large proteins in an energy dependent manner.
To specify the importance of proteasome regulation during the growth of Archaea, we kinetically measured the 20S peptidase activity and the gene expression of the PAN–proteasome subunits in Halobacterium. A maximum in 20S endopeptidase activities and transcripts accumulation was measured in cell extracts at the beginning of the exponential phase. During exponential growth, we observed a drastic reduction of both the activity and the mRNAs accumulation followed by a regain in the late stationary phase. A similar coincidental transcriptional regulation was also observed for the two PAN activating subunits. In Halobacterium NRC1 a transcriptomic study revealed an decrease in the beta subunit transcripts during exponentially growth phase while increased levels were detected in H. volcanii at the stationary phase (Facciotti et al. 2007; Reuter et al. 2004). Taken together these observations suggest that, a transcriptional regulation acting on all the components of the PAN–proteasome system, is important to control the intracellular proteolytic activity as a function of the growth stage. The observed increased in proteolytical activity combined with the accumulation of PAN–proteasome transcripts could be necessary to eliminate the defective ribosomal translation products when cells are actively translating de novo proteins and to eliminate unwanted proteins for metabolic and regulatory purposes during stationary phases.
The regulation of the archaeal proteasome system in response to physical stresses remains poorly studied. Previous transcriptomic studies showed that, in hyperthermophilic Archaea, the amount of one of the proteasome beta subunits transcripts was increased in heat stressed cells (β1) while the alpha subunit was down regulated (Shockley et al. 2003). In stressed Halobacterium cells, we measured a significant increase in the proteasome hydrolyzing activity as well as a strong transcriptional accumulation of the alpha, beta proteasome subunits and of the two PAN subunits. Our observations suggest that, in Archaea, the coordinated transcriptional regulation of the PAN–proteasome subunits play an important role in cellular environmental stress response and extremophilic adaptation to compensate for the deleterious effect of temperature and salt variations on protein stability. This hypothesis is supported by several studies: (1) in T. acidophilum, a partial drug inhibition of the 20S peptidase activity significantly reduced the ability of cells to overcome heat shock (Ruepp et al. 1998), (2) in Haloferax volcanii, the proteomic analysis of an isogenic panA mutant strain revealed an up-regulation of stress response proteins (Kirkland et al. 2008) and (3) the purified 20S proteasome complexes from T. acidophilum and H. marismortui remain stable under thermal and salt stress conditions and can break down partially unfolded proteins (Akopian et al. 1997; Franzetti et al. 2002).
Halobacterium cells accumulate salt in their cytosol and their proteins depend on multi molar salt concentration to remain soluble and stable (Madern et al. 2000). Therefore, in Halobacterium, a decreased salt concentration in the environment represents a severe threat for intracellular protein folding. Several studies have reported that, in halophiles, changing salinities in the environment greatly influence the expression of members of the heat shock protein family (Franzetti et al. 2001; Kuo et al. 1997; Mojica et al. 1997). In a previous study we showed that the halophilic proteasome is more stable under salt stress conditions than most halophilic enzymes and can efficiently degrade the halophilic malate dehydrogenase while the enzyme is exposed to low salt denaturation (Franzetti et al. 2002). In this work, we report an important accumulation of active 20S proteasome particles and a transcriptional induction of the 20S and PAN subunits genes under low salt conditions. These observations suggest that the PAN–proteasome system also plays an important role in halophilic adaptation by helping the cells to get rid of misfolded proteins under low salt conditions.
In yeast, mutagenesis studies have revealed an unexpected diversity of non-redundant functions among the 6 different Rpt proteasomal ATPases (Rubin et al. 1998). In all AAA-ATPases, the N-terminal domain is essential for substrate recognition. The coiled coil motifs present in the N-terminal regions from AAA ATPases have been proposed to be involved in the interaction with substrates proteins (Martin et al. 2004). In Halobacterium the presence of two PAN proteins with different N-terminal regions might be indicative of a different role in cell physiology due to different substrate specificities. Our Northern blot studies showed that, in Halobacterium, the two PAN subunits do not display different expression profiles during cell growth and stress response. These data are in favour of a common role of PANA and PANB in cell physiology. The co-immunoprecipitations experiments presented in this paper indicated that the two PAN proteins interact with each other within Halobacterium cell extracts. This finding suggests that the two PAN subunits form hetero-oligomeric rings complexes within cells, which is also in good agreement with the assumption that PANA and PANB are not specialized in different cellular function. This conclusion is consistent with a report covering the essentiality of the proteasome components in Haloferax volcanii that showed that the two PAN proteins have to be knocked down to give a lethal phenotype, implying that they are involved in similar functions (Zhou et al. 2008). However it remains possible that, depending on the subunit composition of the oligomers the PAN complexes display different substrate affinity, which could result in a finer tuning in the protein degradation profiles.
In Haloferax volcanii the knock out mutant of the proteasome catalytic beta subunits is lethal (Zhou et al. 2008). Therefore, partial inhibition by specific chemical inhibitors represents a tool for investigating the contribution of the Halobacterium proteasome to different cellular functions, including growth and stress response. In this study, we used peptide vinyl sulfone, a specific proteasome inhibitor that was shown to irreversibly modify the β subunits in eukaryotes and in Archaea (Glas et al. 1998; Ruepp et al. 1998). In our experiments, the growth rate of Halobacterium cells was not significantly affected despite a dramatic loss in 20S peptidase activity. This implies that a high level of active 20S particles is not required for growth. The absence of significant modifications in the growth rate and viability of heat-shocked Halobacterium cells that have been treated with the anti-proteasome drug is more surprising. As indicated by the strong induction of the thermosome subunit, the conditions that we used should provoke a deleterious accumulation of misfolded proteins in the cytosol that represents substrates for the proteases. With a strong impairment of the proteasomal enzyme activity, one should have expected a drastic effect on the Halobacterium ability to overcome the stress conditions. An explanation would be that, in Halobacterium, other cytosolic or membrane associated proteases such as Lon can operate on the same substrates as the 20S proteasome and may compensate for losses in the proteasome function. In fact, in eukaryotes, increased peptidase activities were measured in the cytosol of proteasome deficient cells (Glas et al. 1998). This result is also consistent with the observation that, in Haloferax volcanii, the degradation rate of a GFP reporter proteasome substrate was only slightly reduced in the presence of the proteasome inhibitor clastro lactacystine (Reuter and Maupin-Furlow 2004). In Thermoplasma acidophilum, similar stress experiments, also using the NLVS inhibitor, have shown severe effects on cellular growth (Ruepp et al. 1998). Since halophiles have evolved to withstand extreme and multiples stress experienced in the hypersalines environments such as temperature, salt, radiation, heavy metals, they may have developed a higher tolerance limit towards stress compared to other Archaea. In Halobacterium, the strong up-regulation of the PAN regulatory unfoldases under stress might reflect such an adaptation and suggest that improved chaperones and protease systems may compensate for losses in 20S peptidase activity.
References
Akopian TN, Kisselev AF, Goldberg AL (1997) Processive degradation of proteins and other catalytic properties of the proteasome from Thermoplasma acidophilum. J Biol Chem 272:1791–1798
Benaroudj N, Zwickl P, Seemuller E, Baumeister W, Goldberg AL (2003) ATP hydrolysis by the proteasome regulatory complex PAN serves multiple functions in protein degradation. Mol Cell 11:69–78
Bidle KA (2003) Differential expression of genes influenced by changing salinity using RNA arbitrarily primed PCR in the archaeal halophile Haloferax volcanii. Extremophiles 7:1–7
Chamieh H, Guetta D, Franzetti B (2008) The two PAN ATPases from Halobacterium display N-terminal heterogeneity and form labile complexes with the 20S proteasome. Biochem J 411:387–397
Coux O, Tanaka K, Goldberg AL (1996) Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem 65:801–847
Facciotti MT, Reiss DJ, Pan M, Kaur A, Vuthoori M, Bonneau R, Shannon P, Srivastava A, Donohoe SM, Hood LE, Baliga NS (2007) General transcription factor specified global gene regulation in archaea. Proc Natl Acad Sci USA 104:4630–4635
Franzetti B, Schoehn G, Ebel C, Gagnon J, Ruigrok RW, Zaccai G (2001) Characterization of a novel complex from halophilic archaebacteria, which displays chaperone-like activities in vitro. J Biol Chem 276:29906–29914
Franzetti B, Schoehn G, Garcia D, Ruigrok RW, Zaccai G (2002) Characterization of the proteasome from the extremely halophilic archaeon Haloarcula marismortui. Archaea 1:53–61
Ginzburg M, Sachs L, Ginzburg BZ (1970) Ion metabolism in a Halobacterium. I. Influence of age of culture on intracellular concentrations. J Gen Physiol 55:187–207
Glas R, Bogyo M, McMaster JS, Gaczynska M, Ploegh HL (1998) A proteolytic system that compensates for loss of proteasome function. Nature 392:618–622
Goldberg AL (2003) Protein degradation and protection against misfolded or damaged proteins. Nature 426:895–899
Hartl FU (1996) Molecular chaperones in cellular protein folding. Nature 381:571–579
Kirkland PA, Gil MA, Karadzic IM, Maupin-Furlow JA (2008) Genetic and proteomic analyses of a proteasome-activating nucleotidase A mutant of the haloarchaeon Haloferax volcanii. J Bacteriol 190:193–205
Kottemann M, Kish A, Iloanusi C, Bjork S, Diruggiero J (2005) Physiological responses of the halophilic archaeon Halobacterium sp. strain NRC1 to desiccation and gamma irradiation. Extremophiles 9:219–227
Kuo YP, Thompson DK, St. Jean A, Charlebois RL, Daniels CJ (1997) Characterization of two heat shock genes from Haloferax volcanii: a model system for transcription regulation in the Archaea. J Bacteriol 179:6318–6324
Lee DH, Goldberg AL (1996) Selective inhibitors of the proteasome-dependent and vacuolar pathways of protein degradation in Saccharomyces cerevisiae. J Biol Chem 271:27280–27284
Leuko S, Legat A, Fendrihan S, Stan-Lotter H (2004) Evaluation of the LIVE/DEAD BacLight kit for detection of extremophilic archaea and visualization of microorganisms in environmental hypersaline samples. Appl Environ Microbiol 70:6884–6886
Madern D, Ebel C, Zaccai G (2000) Halophilic adaptation of enzymes. Extremophiles 4:91–98
Martin J, Gruber M, Lupas AN (2004) Coiled coils meet the chaperone world. Trends Biochem Sci 29:455–458
Maupin-Furlow JA, Wilson HL, Kaczowka SJ, Ou MS (2000) Proteasomes in the archaea: from structure to function. Front Biosci 5:D837–D865
Mojica FJ, Cisneros E, Ferrer C, Rodriguez-Valera F, Juez G (1997) Osmotically induced response in representatives of halophilic prokaryotes: the bacterium Halomonas elongata and the archaeon Haloferax volcanii. J Bacteriol 179:5471–5481
Navon A, Goldberg AL (2001) Proteins are unfolded on the surface of the ATPase ring before transport into the proteasome. Mol Cell 8:1339–1349
Oesterhelt D, Stoeckenius W (1974) Isolation of the cell membrane of Halobacterium halobium and its fractionation into red and purple membrane. Methods Enzymol 31(Pt A):667–678
Oren A (2008) Microbial life at high salt concentrations: phylogenetic and metabolic diversity. Saline Syst 4:2
Patel S, Latterich M (1998) The AAA team: related ATPases with diverse functions. Trends Cell Biol 8:65–71
Reuter CJ, Maupin-Furlow JA (2004) Analysis of proteasome-dependent proteolysis in Haloferax volcanii cells, using short-lived green fluorescent proteins. Appl Environ Microbiol 70:7530–7538
Reuter CJ, Kaczowka SJ, Maupin-Furlow JA (2004) Differential regulation of the PanA and PanB proteasome-activating nucleotidase and 20S proteasomal proteins of the haloarchaeon Haloferax volcanii. J Bacteriol 186:7763–7772
Rubin DM, Glickman MH, Larsen CN, Dhruvakumar S, Finley D (1998) Active site mutants in the six regulatory particle ATPases reveal multiple roles for ATP in the proteasome. EMBO J 17:4909–4919
Ruepp A, Eckerskorn C, Bogyo M, Baumeister W (1998) Proteasome function is dispensable under normal but not under heat shock conditions in Thermoplasma acidophilum. FEBS Lett 425:87–90
Sauer RT, Bolon DN, Burton BM, Burton RE, Flynn JM, Grant RA, Hersch GL, Joshi SA, Kenniston JA, Levchenko I, Neher SB, Oakes ES, Siddiqui SM, Wah DA, Baker TA (2004) Sculpting the proteome with AAA(+) proteases and disassembly machines. Cell 119:9–18
Sherman MY, Goldberg AL (2001) Cellular defenses against unfolded proteins: a cell biologist thinks about neurodegenerative diseases. Neuron 29:15–32
Shockley KR, Ward DE, Chhabra SR, Conners SB, Montero CI, Kelly RM (2003) Heat shock response by the hyperthermophilic archaeon Pyrococcus furiosus. Appl Environ Microbiol 69:2365–2371
Shukla HD (2006) Proteomic analysis of acidic chaperones, and stress proteins in extreme halophile Halobacterium NRC-1: a comparative proteomic approach to study heat shock response. Proteome Sci 4:6
Smith DM, Benaroudj N, Goldberg A (2006) Proteasomes and their associated ATPases: a destructive combination. J Struct Biol 156:72–83
Verma R, Chen S, Feldman R, Schieltz D, Yates J, Dohmen J, Deshaies RJ (2000) Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol Biol Cell 11:3425–3439
Wilson HL, Ou MS, Aldrich HC, Maupin-Furlow J (2000) Biochemical and physical properties of the Methanococcus jannaschii 20S proteasome and PAN, a homolog of the ATPase (Rpt) subunits of the eucaryal 26S proteasome. J Bacteriol 182:1680–1692
Witt S, Kwon YD, Sharon M, Felderer K, Beuttler M, Robinson CV, Baumeister W, Jap BK (2006) Proteasome assembly triggers a switch required for active-site maturation. Structure 14:1179–1188
Zhou G, Kowalczyk D, Humbard MA, Rohatgi S, Maupin-Furlow JA (2008) Proteasomal components required for cell growth and stress responses in the haloarchaeon Haloferax volcanii. J Bacteriol 190:8096–8105
Zwickl P, Baumeister W (1999) AAA-ATPases at the crossroads of protein life and death. Nat Cell Biol 1:E97–E98
Zwickl P, Ng D, Woo KM, Klenk HP, Goldberg AL (1999) An archaebacterial ATPase, homologous to ATPases in the eukaryotic 26 S proteasome, activates protein breakdown by 20 S proteasomes. J Biol Chem 274:26008–26014
Acknowledgments
This work was funded by Grants form the French National Research Agency (ANR Project MacroTET; BLAN07-3_204002) and the CNRS interdisciplinary program Origins of Planets and Life (OPV). H.C. and V.M. were supported by PhD Grants from the French ministry for Research and Technology.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Albers.
Rights and permissions
About this article
Cite this article
Chamieh, H., Marty, V., Guetta, D. et al. Stress regulation of the PAN–proteasome system in the extreme halophilic archaeon Halobacterium . Extremophiles 16, 215–225 (2012). https://doi.org/10.1007/s00792-011-0421-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-011-0421-0