Abstract
Two 16S rRNA gene clone libraries (KF and KS) were constructed using two soil samples (K7s and K8s) collected near Kafni Glacier, Himalayas. The two libraries yielded a total of 648 clones. Phyla Actinobacteria, Bacteroidetes, Chloroflexi Firmicutes, Proteobacteria, Spirochaetae, Tenericutes and Verrucomicrobia were common to the two libraries. Phyla Acidobacteria, Chlamydiae and Nitrospirae were present only in KF library, whereas Lentisphaerae and TM7 were detected only in KS. In the two libraries, clones belonging to phyla Bacteroidetes and Proteobacteria were the most predominant. Principal component analysis (PCA) revealed that KF and KS were different and arsenic content influenced the differences in the percentage of OTUs. PCA indicated that high water content in the K8s sample results in high total bacterial count. PCA also indicated that bacterial diversity of KF and KS was similar to soils from the Pindari Glacier, Himalayas; Samoylov Island, Siberia; Schrimacher Oasis, Antarctica and Siberian tundra. The eleven bacterial strains isolated from the above two soil samples were phylogenetically related to six different genera. All the isolates were psychro-, halo- and alkalitolerant. Amylase, lipase and urease activities were detected in the majority of the strains. Long chain, saturated, unsaturated and branched fatty acids were predominant in the psychrotolerant bacteria.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Investigating microbial populations with respect to various habitats is of enormous importance in establishing the role of microorganisms in these ecosystems. Further, the advent of molecular approaches such as cloning of the 16S rRNA genes and sequencing (Pace et al. 1986; Woese 1987) DGGE and T-RFLP have indeed enhanced our understanding of the dynamics of microbial populations in various habitats (Wani et al. 2006; Pradhan et al. 2010; Shivaji et al. 2011a, b) and have provided compelling evidence for the existence of many novel types of microorganisms in the environment (Giovannoni et al. 1990; Ward et al. 1990; Hugenholtz et al. 1998a; Shivaji et al. 2005, 2011a, b; Prabagaran et al. 2007) including entirely new prokaryotic lineages (Torsvik et al. 1996; Head et al. 1998; Hugenholtz et al. 1998b).
Bacterial diversity of various cold habitats (water, soil, ice, etc.) from cold regions such as Arctic, Antarctica and other temperate regions based on the rRNA approach is limited (Torsvik et al. 1996; Brambilla et al. 2001; Shivaji et al. 2005, 2011a). Studies on glaciers are even more scarce (Christner et al. 2001, 2003; Zhang et al. 2002; Miteva et al. 2004; Miteva and Brenchley 2005; Skidmore et al. 2005; Shivaji et al. 2005, 2011b; Bhatia et al. 2006; Cheng and Foght 2007; Prabagaran et al. 2007; Nemergut et al. 2007; Pradhan et al. 2010). The few studies of bacterial diversity in glaciers have clearly revealed that the diversity of glacial water, sediment and basal ice are distinct from microorganisms in the supraglacial (top of the glacier) and proglacial (in front of the glacier) environments, thus implying that each of the habitats needs to be analysed separately.
In this study, both culture-independent and culture-dependent approaches were used to establish the bacterial diversity of soil samples collected near the Kafni Glacier located in the Himalayan Mountain ranges, India. In addition, the culturable bacteria were screened for cold-active enzymes and fatty acid profiles. The primary objectives of the study were to establish the bacterial diversity of a Himalayan glacier, to compare the diversity with other Himalayan, alpine and polar glaciers and other cold habitats and to biopropspect for cold-active enzymes.
Materials and methods
Sampling site and sample collection
The Kafni Glacier (30°13′05″ N 80°03′20″ E) is located in the Kumoan Himalayas of Uttaranchal, a state in India, at an altitude of about 3,853 m above sea level. Two soil samples (K7s and K8s) were collected near the glacier (Supplementary Fig. S1) in the month of September 2005, at altitudes of 3,300 and 3,500 m, respectively. Ice-free soil samples (K7s and K8s) 2 and 1 km away from the glacial snout were collected in sterile polythene bags. The surface soil (about 1 cm) was removed with a sterile spatula and using another sterile spatula the soil was collected. The soil samples were transported to the laboratory under sterile conditions and stored at −20°C until use. It was not possible to reach the snout of the glacier.
Soil analysis
Soil samples were thawed, dried, ground and allowed to pass through a 2-mm sieve. Soil pH (Rhoades 1982), water content (Blakemore et al. 1987) and the chemical characteristics (Jackson 1967) were determined using standard methods. The total element content in the soil was determined after acid digestion. Micronutrients/elements (Li, Be, Al, V, Cr, Mn, Fe, Co, Ni, Zn, Ga, As, Se, Sr, Cd, In, Cs and U) were determined using an atomic absorption spectrophotometer (Perkin Elmer Aanalyst 100 model). Each sample was analysed in triplicate and the mean values were calculated.
Bacterial count
BacLight™ Bacterial Viability Kit (Invitrogen) was used for the determination of the total bacterial count (TBC) in the sediment samples as per the instructions given in the kit. Bacteria were counted in a Petroff-Hausser Counter using a fluorescent microscope (Axioplan 2, Zeiss).
Isolation and characterisation of bacterial strains
Bacterial colonies were isolated on Luria-Bertani (LB) agar plates [tryptone (1.0% w/v), yeast extract (0.5% w/v), NaCl (1.0% w/v) and agar (2.0% w/v)] and 1/10 of LB was then incubated at 4°C for 15 days (Reddy et al. 2008a). For this purpose, 1 g of the soil sample was suspended in 0.9% (w/v) NaCl solution and shaken for 2 h at 15°C. Subsequently, 100 μl of the water sample was plated on LB agar plates, colony counts recorded, and different morphotypes purified and maintained on LB agar medium (Shivaji et al. 1989).
The growth of the bacterial strains at different temperatures, pH and various salt concentrations was checked using ABM [peptone (3% w/v), yeast extract (2% w/v) and agar (2% w/v)] plates (Shivaji et al. 1992, 2011b). Extracellular enzymatic activities of amylase (Priest 1977), lipase (Booth 1978), protease (Priest 1977) and urease were checked by streaking the cultures on ABM agar plates supplemented with 0.2% soluble starch, 1% Tween-60 along with 0.01% CaCl2, 0.3% casein and 2% urea (filter sterilised), respectively, and incubating the plates at 4 and 18°C for 5–10 days. For fatty acid analysis, all the isolates were grown on trypticase soy agar plates at 18°C for 2 days. Fatty acid methyl esters were prepared and analysed as previously described (Shivaji et al. 2007). DNA was isolated from all the cultures and the 16S rRNA gene was amplified and sequenced, and phylogenetic analysis was carried out as described earlier (Reddy et al. 2000, 2009; Srinivas et al. 2009; Shivaji et al. 2011b).
Extraction of total DNA from soil and PCR amplification of the 16S rRNA gene
Total DNA was isolated from the soil samples essentially according to the methods described earlier (Shivaji et al. 2004; Tsai and Olson 1991). Primers 16S3 (5′-TCC TAC GGG AGG CAG CAG-3′) and 16S4 (5′-GGC GGT GTG TAC AAG GCC C-3′) corresponding to positions 339–356 and 1,402–1,384, respectively, of the Escherichia coli 16S rRNA gene (Lane 1991) were used to amplify about 1.0-kb fragment of the total 1.5-kb 16S rRNA gene. Amplification was done as described earlier (Reddy et al. 2000; Shivaji et al. 2004, 2011a). The PCR product was purified with the Quiaquick PCR purification kit (Qiagen Inc.) according to the instructions provided.
Cloning and library construction of soil 16S rRNA gene sequences
The purified PCR product was cloned into pMOS Blue Blunt End vector system (Amersham Biosciences) following the instructions of the manual. Transformants were selected on an LB agar plate containing 20 μg/ml of X-gal and 60 μg/ml of ampicillin and incubated at 37°C overnight. Clones were maintained on LB agar plates containing X-gal and ampicillin.
PCR amplification, sequencing of the 16S rRNA clone libraries and phylogenetic analysis
The 16S rRNA gene was amplified from the transformants by colony PCR using the vector-targeted M13 forward (5′-GTA AAA CGA CGG CCA GT-3′) and M13 reverse (5′-GGA AAC AGC TAT GAC CAT G-3′) primers, respectively, and sequenced using the primers M13 forward, M13 reverse, pD (5′-CAG CAG CCG CGG TAA TAC-3′) and pF* (5′-ACG AGC TGA CGA CAG CCA TG-3′) (Pradhan et al. 2010). Vector and chimeric sequences were eliminated using Gene Tool version 2 (www.biotools.com). Sequences were then subjected to BLAST to identify the nearest taxa and aligned with sequences belonging to the nearest taxa (downloaded from NCBI database http://www.ncbi.nlm.nih.gov/) using CLUSTAL X. Phylogenetic trees were constructed using neighbour joining and maximum likelihood methods (Pradhan et al. 2010; Shivaji et al. 2011b). Bootstrap analysis, based on 1,000 replicate data sets, was performed to assess stability among the clades.
Statistical analyses of the cloned libraries
To compare the bacterial diversity within the two samples, 16S rRNA gene sequences of of the clones from the two libraries showing ≥97% sequence similarity were grouped into the same OTU (phylotype). Shannon–Wiener Diversity Index (http://www.changbioscience.com/genetics/shannon.html) was used to calculate Shannon Index (H′), evenness (Lefauconnier et al. 1994) and the Simpson’s index (D) (Magurran 1996). Rarefaction analysis was done using the site Online Calculation (http://biome.sdsu.edu/fastgroup/cal_tools.htm). Coverage of 16S rRNA gene clone libraries was calculated as described previously (Good 1954). Rarefaction curves were generated to compare the relative diversity and coverage of each library.
The two 16S rRNA gene clone libraries from this study (KF and KS) were compared using a JAVA-based software Comm Cluster (Hur and Chun 2004) with three different cutoff values (97, 90 and 80%) and an ordination analysis was performed using principle component analysis (PCA). PCA was performed using the SPSS statistical computing package (version 16.0; SPSS Inc.) to group or separate samples based on the biogeochemical parameters (altitude, total bacterial count, water content, pH value and micronutrients/elements such as Li, Be, Al, V, Cr, Mn, Fe, Co, Ni, Zn, Ga, As, Se, Sr, Cd, In, Cs and U) and the percentages of OTUs in each sample (higher taxa ≥1% in the libraries were considered). Phylotypes (showing ≥97% similarity in 16S rRNA gene sequences) from KF and KS libraries were also compared with 16S rRNA gene clone libraries of 14 other glacier samples from different regions of the world [RKS1 and RKS6, soil samples from Roopkund Glacier Lake, India (Pradhan et al. 2010); RKS7, soil sample from Roopkund Glacier, India (Pradhan et al. 2010); P1S, P4S and P8S, soil samples, Pindari Glacier, India (Shivaji et al. 2011b); TPG, Tibetan Plateau Glacier (Liu et al. 2009), MLME, moraine Lakes, Mount Everest (Liu et al. 2006); GMWME, glacial meltwater, Mount Everest (Liu et al. 2006); JEGC, John Evans Glacier, Canada (Cheng and Foght 2007); BGA, Bench Glacier, Alaska (Skidmore et al. 2005); SOA, soil sample, Schirmacher Oasis, Antarctica (Shivaji et al. 2004); ST, soil sample, Siberian tundra; SIS, Samoylov Island, Siberia (Wagner et al. 2009)]. PCA was performed on the number of phylotypes in each of the libraries.
Nucleotide sequence accession numbers
All the sequences of the 16S rRNA gene clone libraries were deposited in GenBank with accession numbers EF421990 to EF421997; EF421999 to EF422011; EF422013 to EF422015; EF422018 to EF422022; EF422026 to EF422033; EF422035; EF422036; EF422038 to EF422044; EF422046; EF422047; EF422049 to EF422063; EF422065; EF422066; EF434184 to EF434196; EF434198 to EF434202; EF434204 to EF434221; EF434223 to EF434225; EF434227 to EF434262; EF445432 to EF445444; EF445446 to EF445457; EF445459 to EF445478; EF445480 to EF445493; EF445495 to EF445503; EF445507 to EF445515; EF445517 to EF445523; EF445525 to EF445531; EF445533 to EF445535; EF445537 to EF445544; and EU809512 to EU809923.
Results
Bacterial abundance and the physicochemical characteristics of the soil samples collected near Kafni Glacier
In the two soil samples (K7s and K8s) collected near the Kafni Glacier at an altitude of 3,300 and 3,500 m, respectively, it was observed that the pH, water content, soil texture and elemental chemical characteristics were different (Table 1). The total bacterial count in K7s and K8s was also different (6.25 ± 0.25 and 30.71 ± 0.23 × 108 bacteria g−1 of soil respectively) (Table 1).
Culture-independent bacterial diversity from soil samples collected near the Kafni Glacier
16S rRNA gene clone libraries
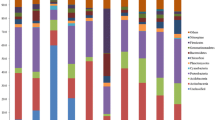
The soil samples (K7s and K8s) yielded about 50 and 60 μg DNA g−1 of soil. About 200 ng of the DNA from K7s and K8s was used for the construction of two 16S rRNA gene libraries KF and KS, respectively. The number of clones in KF and KS were 237 and 411 clones, respectively, with an insert size of approximately 1 kb. BLAST sequence similarity analysis of the two clone libraries indicated that clones in KF and KS were affiliated to 11 and 10 phyla, respectively. Clones affiliated to eight phyla Actinobacteria, Bacteroidetes, Chloroflexi, Firmicutes, Proteobacteria, Spirochaetae, Tenericutes and Verrucomicrobia were common to both libraries. Phyla Acidobacteria, Chlamydiae and Nitrospirae were present only in the KF library and clones belonging to the phylum Lentisphaerae and candidate phylum TM7 were present only in the KS library (Fig. 1; Supplementary Table 1). Based on the BLAST analysis, the 237 clones from KF belong to 106 bacterial taxa and the 411 clones from KS belong to 93 bacterial taxa (Supplementary Table 1). In the two libraries (KF and KS), clones belonging to phyla Bacteroidetes (23.2 and 34.8%) and Proteobacteria (51.5 and 51.3%) were the most predominant (representing >10%) of the total clones (Supplementary Table 1).
Comparison of the bacterial diversity in different cold habitats as determined by 16S rRNA gene clone libraries. K7s and K8s, soil samples, Kafni Glacier, India (present study); RKS1, RKS6 and RKS7 Roopkund Glacier; P1S, P4S and P8S, Pindari Glacier; TPG, Tibetan Plateau Glacier; MLME, moraine lakes, Mount Everest; GMWME, glacial meltwater, Mount Everest; JEGC, John Evans Glacier, Canada; BGA, Bench Glacier, Alaska; SOA, Schirmacher Oasis, Antarctica and ST, Siberian tundra; SIS, Samoylov Island, Siberia. Pro, Proteobacteria, Aci, Acidobacteria; Act, Actinobacteria; CFB, Cytophaga–Flavobacterium–Bacteroides; Chf, Chloroflexi; Cya, Cyanobacteria; De-Th, Deinococcus and Themomicrobia; Elu, Elusimicrobia; Fir, Firmicutes; Gem, Gemmatimonadetes; Len, Lentosphaerae; Nit, Nitrospirae; OP11, candidate divisions OP11; Pla, Planctomycetes; Spi, Spirochaetae; Ten, Tenericutes; TM7, TM7_s TM7a (candidate phylum); Unc, unculturables; and Ver, Verrucomicrobia. Values represent the percentage of clones
Proteobacteria
Clones affiliated to all classes of the phylum Proteobacteria namely Alpha-, Beta-, Gamma-, Delta- and Epsilonproteobaceria were present in libraries KF and KS (122 and 211 clones) (Supplementary Table 1). Phylogenetic analysis indicated that 108 out of the 122 clones in KF and 161 out of the 211 clones in KS that were affiliated to the phylum Proteobacteria clustered with their nearest phylogenetic neighbour (Figs. 2a, 3a) (Supplementary Table 1). Some of the clones (13 and 46 clones in KF and KS, respectively) did not form a cluster with the nearest phylogenetic neighbour, but formed a cluster within the same class of phylum Proteobacteria. In contrast, clones KF-225, KS-106, KS-365, KS-504 and KS-202 did not cluster within the same class (Figs. 2a, 3a).
Neighbour joining phylogenetic tree of 16S rRNA gene clones from KF library, showing the phylogenetic relationship of clones affiliated to Proteobacteria (a) and clones affiliated to phyla Acidobacteria, Actinobacteria, Bacteroidetes, Chlamydiae, Chloroflexi, Firmicutes, Nitrospirae, Spirochaetae, Tenericutes and Verrucomicrobia (b). Aquifex pyrophilus Kol5aT was taken as an out-group. Numbers at the nodes are bootstrap values. The bar represents 0.05 substitutions per alignment position in a and b
Neighbour joining phylogenetic tree of 16S rRNA gene clones from KS library, showing the phylogenetic relationship of clones affiliated to Proteobacteria (a) and to Actinobacteria, Bacteroidetes, Chloroflexi, Firmicutes, Lentosphaerae, Spirochaetae, Tenericutes, TM7_s TM7a (candidate phylum) and Verrucomicrobia (b). Aquifex pyrophilus Kol5aT was taken as an out-group. Numbers at nodes are bootstrap values. The bar represents 0.02 and 0.05 substitutions per alignment position in a and b, respectively
Bacteroidetes
Clones affiliated to three classes of the phylum Bacteroidetes, namely Bacteroidia, Flavobacteria and Sphingobacteria were present in both the libraries (KF and KS) (Supplementary Table 1). The majority of the clones, 51 out of the 55 clones in KF and 126 out of the 143 clones, clustered with their nearest phylogenetic neighbour (Figs. 2b, 3b). A few of the clones (KS-177 and KS-228) clustered outside of the phylum (Fig. 3b).
Firmicutes
Clones affiliated to four classes of the phylum Firmicutes, namely Bacilli, Clostridia, Erysipelotrichia and Negativicutes, were present in KF, whereas clones related to the class Negativicutes were absent in the KS library (Supplementary Table 1). All clones which belonged to the phylum Firmicutes clustered with their nearest phylogenetic neighbour in the KF library (Fig. 2b), but only 17 out of the 26 clones clustered with their nearest phylogenetic neighbour in the KS library (Fig. 3b). Clones KS-437, KS-159, KS-327, KS-420, KS-470 and KS-483 clustered with the phylum Bacteroidetes instead of in Firmicutes (Fig. 3b) clade 5.
Actinobacteria
Clones affiliated to Actinobacteria in KF and KS (12 and 4 clones) (Supplementary Table 1) clustered with the related sequences (Figs. 2b, 3b). One clone KS-78 appeared to be phylogenetically affiliated to Cryobacterium roopkundense RuGl7T (which was earlier isolated by us from the same habitat (Reddy et al. 2010) (Fig. 3b).
Verrucomicrobia
Clones affiliated to two classes of the phylum Verrucomicrobia, namely Opitutae and Verrucomicrobiae, were present in KF, but in KS clones related to class Opitutae were absent (Supplementary Table 1). KF-288 clustered with the phylum Chlamydiae (Fig. 2b).
Nitrospirae and Spirochaetae
Clones affiliated to Nitrospirae were present in only the KF library (9 clones), whereas those affiliated to Spirochaetae were present in both KF and KS (8 and 6 clones) (Supplementary Table 1). All the clones of Nitrospirae and Spirochaetae clustered with the nearest phylogenetic neighbours (Figs. 2b, 3b).
Acidobacteria, Chlamydiae, Chloroflexi, Lentisphaerae, Tenericutes and candidate phylum TM7_s TM7a
Clones affiliated to the phyla Chloroflexi and Tenericutes were present in both KF and KS. Clones affiliated to the phyla Acidobacteria and Chlamydiae were specific only to KF; those affiliated to Lentisphaerae and candidate phylum TM7_s TM7a were specific only to the KS library and appeared at a very low frequency (Supplementary Table 1). Phylogenetic analysis indicated that clones affiliated to the above phyla clustered with their respective phylogenetic neighbours (Figs. 2b, 3b), except KS-330 which was clustered within the phylum Bacteroidetes (Fig. 3b).
Statistical analysis
The 16S rRNA gene clones from the 2 soil samples (K7s and K8s) based on 16S rRNA gene sequence similarity criteria of ≥97% could be categorised into 125 and 238 phylotypes (Supplementary Fig. S2; Table 2). The common phylotypes in KF and KS libraries are shown in Supplementary Table 1. Species richness at 97% cutoff was 125 and 238, and the diversity coverage 47.3 and 42.1% for KF and KS libraries, respectively. Reducing the cutoff from 97 to 90% and from 97 to 80% reduced the species richness to 12 and 11 and increased the coverage to 94.9 and 97.3%, respectively (Table 2). The rarefaction curves indicated that at 80% cutoff, both the curves in K7s and K8s soil samples plateaued, while only K7s soil sample plateaued when 90% cutoff was considered (Supplementary Fig. S2). The rarefaction curve analysis implied that these were likely to be minimal estimates of diversity. These observations are supported by bacterial diversity parameters, such as Shannon–Wiener Diversity Index, Shannon entropy, Simpson’s Index, coverage, Chao1 and evenness (Table 2). Further based on principal component analysis, K7s and K8s clustered distantly at 97% cutoff value, but came closer at both 90 and 80% cutoff values (Fig. 4a, b, c).
Ordination diagrams based on principal component analysis using frequency tables obtained from the two 16S rRNA gene clone libraries (KF and KS) defined at different cutoff similarity values of 97% (a), 90% (b) and 80% (c) using Comm Cluster Software. PC 1, principal component analysis factor 1; PC 2, principal component analysis factor 2
The result of PCA based on percentage of a specific OTU (Phyla with ≥1% clones were considered) in the two libraries (KF and KS) is shown in Fig. 5a, and it appears that the principal component factors 1 and 2 (PC1-72.419 and PC2-27.581%) explain 100% of the total variances. The phyla Acidobacteria, Chlamydiae and Nitrospirae, which were present only in the KF library, clustered together, whereas phyla Lentisphaerae and candidate phylum TM7, which were present only in the KS library, clustered together. The phyla Chloroflexi, Actinobacteria, Spirochaetae and Tenericutes, which were dominant in the KF library, clusterd together close to the right side horizontal line of PCA plot. The other phyla Firmicutes, Proteobacteria, Verrucomicrobia and Bacteroidetes, which were nearly equal in both the libraries or higher in the KS library, clustered together away from the other phyla.
a Principal component analysis based on percentage of specific OTUs from two 16S rRNA gene clone libraries (PC1, 72.419% and PC2, 27.581%). b PCA plot based on biogeochemical properties (altitude, total bacterial count, pH value and micronutrients Li, Be, Al, V, Cr, Mn, Fe, Co, Ni, Cu, Zn, Ga, As, Se, Rb, Sr, Cd, In, Cs, Pb, Bi and U) of two soil samples (PC1, 89.464% and PC2, 10.535%)
The result of PCA based on the biogeochemical properties is shown in Fig. 5b and the principal component factors 1 and 2 (PC1, 89.464 and PC2, 10.535%) explained 100% of the total variances. The PCA plot indicated that arsenic and water content which are higher in the K8s soil sample grouped closely (Fig. 5b; Table 1). All other parameters which are numerically high in K7s, such as Ga, In, Cd, Zn, V, Co, Ni, Cr, Al, Cs, Be, Li, Se, Mn, Sr, U, altitude and pH, clustered together (Fig. 5b). Thus, it may imply that As and water content are the key parameters that influence the observed differences in the percentage of specific OTUs in the two 16S rRNA gene clone libraries. The high bacterial content in K8s may also be influenced by water content (Fig. 5b). Thus, PCA does discriminate the two soil samples.
The result of PCA based on phylotypes with 97% cutoff value of 16 clone libraries, 2 from the present study (KF and KS libraries) and 14 16S rRNA gene clone libraries from other glaciers, is shown in Fig. 6. The 16 libraries grouped into 5 clusters represented by cluster 1 (KF and KS from the Kafni Glacier; P1s and P8s from Pindari Glacier; SOA from Schirmacher Oasis, Antarctica; ST from Siberian tundra and and SIS from Samoylov Island, Siberia), cluster 2 (P4s sample from Pindari Glacier; TPG from Tibetan Plateau Glacier, Tibet and MLME from moraine lakes of Mount Everest) cluster 3 (BGA from Bench Glacier, Alaska; GMWME from glacial meltwater of Mount Everest and JEGC from John Evans Glacier, Canada) cluster 4 (RUGL1 and RUGL6 from Roopkund Glacier, Himalayas, India) and cluster 5 (RUPGL7 also from Roopkund Glacier which clustered between the RUGL1, RUGL6 and KF and KS libraries). Cluster 2 also appeared to be close to cluster 1 and 3.
Principal component analysis based on phylotypes with 97% cutoff value from sixteen 16S rRNA gene clone libraries KF and KS, Kafni Glacier, India; P1s, P4s and P8s, Pindari Glacier, India; RUGL1 and RUGL6, Roopkund Glacier Lake, India; RUPGL7, Roopkund Glacier, India; TPG, Tibetan Plateau Glaciers; MLME, moraine lakes, Mount Everest; GMWME, glacial meltwater, Mount Everest; JEGC, John Evans Glacier, Canada; BGA, Bench Glacier, Alaska; SOA, Schirmacher Oasis, Antarctica; ST, Siberian tundra; and SIS, Samoylov Island, Siberia
Characterisation of the bacterial strains isolated from the soil samples collected near the Kafni Glacier
A total of 11 bacterial strains were isolated from the 2 soil samples (Table 3). The 11 isolates were psychro-, halo- and alkalitolerant (Table 3). Amylase, lipase and urease activities were detected in the majority of the strains (7, 9 and 11, respectively) at 4 and 18°C (Table 3), but protease activity was not detected at 4 and 18°C.
Fatty acid profiles of the isolated strains from soil samples collected near the Kafni Glacier
The fatty acid composition in the 11 isolates was different and not even a single fatty acid was common to all the isolates. Some fatty acids such as iso-C10:0, C12:0, C14:0, iso-C15:0, anteiso-C15:0, anteiso-C15:1, iso-C16:0, C16:0, anteiso-C17:1, C18:0, C18:1 ω9c and summed feature 3 were present in the majority of the isolates (>6 out of 11 isolates) (Supplementary Table 2). The results indicate that in the psychrotolerent bacteria, saturated, iso-, anteiso-, hydroxyl- and unsaturated fatty acids are predominant and together constitute a significant proportion of the total fatty acid composition. The composition of iso-fatty acids ranged from 0 to 29.0%, antesio-fatty acids from 0 to 66.2%, hydroxyl-fatty acids from 0 to 6.8%, saturated fatty acids from 0 to 19.8% and unsaturated fatty acids from 0 to 50.0% (Supplementary Table 2). Out of the 11 strains, only one strain (K7SC-13) contained 1 different polyunsaturated fatty acid (C18:3 ω6, 9,12c) with 0.2% (Supplementary Table 2).
Identification of bacterial strains isolated from soil samples collected near the Kafni Glacier
Taxonomic analysis of all the 11 strains of different morphotypes, indicated that 4 strains were gram-negative and 7 were gram-positive. BLAST sequence similarity search based on 16S rRNA gene sequence indicated that 2 strains belonged to the genus Acinetobacter, 3 strains each to the genera Bacillus and Viridibacillus, and 1 strain each to the genera Lysinibacillus, Pseudomonas and Psychrobacter, respectively (Table 3). Phylogenetic analysis confirmed the affiliation of all the 11 isolates with their nearest phylogenetic neighbour (Supplementary Fig. S3; Table 3).
Discussion
In the present study, bacterial diversity was studied using two soil samples K7s and K8s collected from the viccinity of the Kafni Glacier using viable isolates and 16S rRNA gene clones from two libraries (KF and KS) constructed using the soil DNA.
The majority of the clone sequences, 92% in the KF and 81.5% in the KS library, could be assigned to the nearest phylogenetic neighbour. A few of the clones exhibited <97% similarity with already known species at the 16S rRNA gene level and did not cluster with the nearest phylogenetic neighbour (for example KF-225, KS-106, KS-365, KS-504 and KS-202). Such bacteria need to be isolated and characterised.
In the present study, clones belonging to phyla Actinobacteria, Bacteroidetes, Chloroflexi, Firmicutes, Proteobacteria, Spirochaetae, Tenericutes and Verrucomicrobia were common to KF and KS (Fig. 1; Supplementary Table 1). Other clones belonging to the phyla Acidobacteria, Chlamydiae, Lentisphaerae, Nitrospirae, and candidate phylum TM7s TM7a were present only in one of the libraries (0.2–3.7%; Supplementary Table 1). In fact, except for the phyla affiliated to Bacteroidetes and Proteobacteria, all the other phyla common to both the libraries were present in low frequency (<10%). These observations are in agreement with earlier studies, which had also indicated that clones affiliated to Verrucomicrobia (Cheng and Foght 2007; Liu et al. 2006, 2009; Pradhan et al. 2010; Wagner et al. 2009; Zhou et al. 1997), Chlamydiae (Shivaji et al. 2004), Chloroflexi (Costello and Schmidt 2006; Li et al. 2008; Liu et al. 2006, 2008, 2009; Lysnes et al. 2004; Pradhan et al. 2010; Reed et al. 2006; Shivaji et al. 2004; Wagner et al. 2009), Nitrospirae (Pradhan et al. 2010; Shivaji et al. 2004) and candidate phylum TM7 s TM7a (Liu et al. 2009; Pradhan et al. 2010; Shivaji et al. 2011b) are present in other cold habitats (Fig. 1) at a low frequency. Phyla Lentisphaerae and Tenericutes are represented only by a single clone, and reports on the presence of these bacteria in cold habitats is lacking. This study also differs with the above studies in the absence of Cyanobacteria (alpine tundra wet meadow soil, alpine dry meadows soil, unvegetated deglaciated soil and periglacial soil), Deinococcus (unvegetated deglaciated soil), Elusimicrobia (glacial lake soil), Gemmatimonadetes (glacial lake soil and periglacial soil), Planctomycetes (glacial lake soil, periglacial soil, alpine tundra wet meadow soil and alpine dry meadows soil), Themomicrobia (permafrost-affected soil) and candidate divisions like OS-K and OP10 (alpine tundra wet meadow soil).
The current data supports the earlier data that Proteobacteria, Cytophaga–Flavobacterium–Bacteroides (CFB) and high G + C gram-positive bacteria are common in most cold habitats (Cheng and Foght 2007; Steven et al. 2007; Shivaji et al. 2011a, b; Srinivas et al. 2009; Reddy et al. 2009). In both the libraries, the most predominant bacteria observed were gram-negative belonging to phyla Proteobacteria (51.5 and 51.3%) and Bacteroidetes (23.2 and 34.8%). The frequency of gram-positive bacteria belonging to the phyla Actinobacteria (5.1 and 1.0%) and Firmicutes (6.7 and 6.3%) was significantly low compared with gram-negative bacterial diversity in these samples. These results are in agreement with the earlier studies from similar habitats such as from Roopkund Glacier Lake, India (Pradhan et al. 2010); Pindari Glacier, India (Shivaji et al. 2011b); Tibetan Plateau Glacier (Liu et al. 2009), moraine lakes and glacial meltwaters of Mount Everest (Liu et al. 2006); John Evans Glacier, Canada (Cheng and Foght 2007); Bench Glacier, Alaska (Skidmore et al. 2005); Schirmacher Oasis, Antarctica (Shivaji et al. 2004); Siberian tundra and Samoylov Island, Siberia (Wagner et al. 2009), which had also indicated that gram-negative bacteria predominated over gram-positive bacteria.
The diversity of bacteria in the two soil samples near the Kafni Glacier are comparable with that reported (Figs. 1, 6) for the ice core from the Muztag Ata Glacier (Xiang et al. 2005), solid ice, firn and snow of the Kuytun 51 Glacier (Xiang et al. 2009), soil from the western Himalayas (Gangwar et al. 2009) and Puruogangri ice (Zhang et al. 2006, 2008) with respect to the presence of Actinobacteria, Proteobacteria and Acidobacteria. Xiang et al. (2004) and Liu et al. (2006, 2009) reported only Proteobacteria and Cytophaga–Flavobacterium–Bacteroides from the Malan ice core, Tibetan Plateau and glacial meltwater from the Mount Everest, which are cold deserts. In Roopkund glacial soil, the predominant phyla were Actinobacteria and Firmicutes (Pradhan et al. 2010) and in the Pindari glacial soil the predominant phyla were Acidobacteria, Actinobacteria, Firmicutes and Proteobacteria (Shivaji et al. 2011b). Similarly, Skidmore et al. (2005) reported only four phyla Acidobacteria, Cytophaga–Flavobacterium–Bacteroides, Proteobacteria and Spirochaetes from the Bench Glacier, Alaska. These studies indicate that though the number of dominant phyla is limited, the phyla are not identical.
Comparison of the biodiversity of the Kafni Glacier soil samples with those of two other geographically close glaciers, namely Pindari and Roopkund glaciers, at species level indicated that out of 174 species or taxa (including candidate taxa and uncultured clones belonging to different taxa) from the present study, only 16 were common to Pindari Glacier samples (Arenimonas oryziterrae, Candidatus Solibacter usitatus, Cryobacterium roopkundense, Desulfacinum hydrothermale, Devosia insulae, Geoalkalibacter ferrihydriticus, Georgfuchsia toluolica, Haliangium tepidum, Ilumatobacter fluminis, Lysobacter ginsengisoli, Methylotenera mobilis, Nitrospira moscoviensis, Propionibacterium acnes, Pseudolabrys taiwanensis, Steroidobacter denitrificans and Sulfuricella denitrificans), 28 taxa were common to Roopkund Glacier samples (Alkaliflexus imshenetskii, Anaeromyxobacter dehalogenans, Azospira oryzae, Bellilinea caldifistulae, Cryobacterium roopkundense, Cytophaga fermentans, Dechloromonas hortensis, Devosia neptuniae, Geoalkalibacter ferrihydriticus, Geobacter psychrophilus, Haliangium tepidum, Haloferula helveola, Ilumatobacter fluminis, Longilinea arvoryzae, Nitrospira moscoviensis, Owenweeksia hongkongensis, Prolixibacter bellariivorans, Propionivibrio limicola, Pseudolabrys taiwanensis, Rhodobacter ovatus, Rhodoferax antarcticus, Sphingomonas jaspsi, Steroidobacter denitrificans, Syntrophus gentianae, Thiohalomonas denitrificans, Variovorax boronicumulans, Verrucomicrobium spinosum and Zoogloea caeni) and only seven taxa were common to all three samples (Cryobacterium roopkundense, Geoalkalibacter ferrihydriticus, Haliangium tepidum, Ilumatobacter fluminis, Nitrospira moscoviensis, Pseudolabrys taiwanensis and Steroidobacter denitrificans) representing 9.2, 16.1 and 4%, respectively (Pradhan et al. 2010; Shivaji et al. 2011b). Thus, this implies that bacterial diversity associated with glaciers or in the vicinity of glaciers is likely to be influenced not only by temperature and water content, but also by other physicochemical characteristics of the soil.
The observed difference in the bacterial diversity in the two libraries KF and KS (Supplementary Table 1) may reflect the inherent changes observed in the physicochemical characteristics of the two samples. PCA analysis indicated that As (7.3 times more in K8s sample) was the key parameter that influenced the observed diversity (Fig. 5b), thus confirming an earlier finding that increased As content resulted in low diversity (Shivaji et al. 2011b). Clones related to Sulfurospirillum arsenophilum, Clostridium, Rhodoferax and Bacillus, which are supposed to utilise arsenate, were also detected in the present libraries. Further, in addition to As, factors such as pH (Eichorst et al. 2007), water content (Treves et al. 2003) and texture of soil are also known to influence microbial community composition and diversity (Zhang et al. 2008). The texture was very similar. Therefore, the high bacterial count in the K8s soil sample (30.71 × 108) compared to the K7s soil sample (6.25 × 108) may be attributed to the high water content in K8s. In fact, the total bacterial counts from the Kafni Glacier were higher than in most of the glaciers worldwide (Liu et al. 2009; Miteva et al. 2004; Pradhan et al. 2010; Shivaji et al. 2011b; Skidmore et al. 2005; Xiang et al. 2006, 2009; Zhang et al. 2007). This could be attributed to the fact that the Kafni Glacier is located near human settlements and influenced by anthropogenic activities (Zhang et al. 2007).
Comparison of the bacterial diversity of the Kafni Glacier soil with that of other nonpolar cold habitats such as alpine tundra wet meadow soil (Costello and Schmidt 2006; Zhou et al. 1997), surface snow (Segawa et al. 2005), alpine dry meadows soil (Lipson and Schmidt 2004), unvegetated deglaciated soil (Nemergut et al. 2007), periglacial soil (Schmidt et al. 2009) and permafrost-affected soil (Wagner et al. 2009) indicated that Betaproteobacteria, Bacteroidetes, Chloroflexi, Spirochaetes Verrucomicrobia, Actinobacteria and Acidobacteria were present in most of these habitats, though one or more of these taxa were absent in some of the habitats as in the permafrost-affected soil (Wagner et al. 2009). Further clones belonging to Proteobacteria, Acidobacteria, Actinobacteria and Cytophaga–Flavobacterium–Bacteroides have also been reported from: the Tibetan Plateau Glacier, China; Mount Everest, Nepal; John Evans Glacier, Canada; Bench Glacier, Alaska; Schirmacher Oasis soil, Antarctica and Siberian tundra soil samples (Fig. 1). The occurrence of identical phyla in geographically diverse cold environments is indicative of the ability of microorganisms to adapt to similar strategies to survive freezing and remain active at low temperatures (Abyzov et al. 1998; Priscu and Christner 2003). The bacterial diversity of K7s and K8s was very different compared to polar habitats. Bacterial taxa such as Arthrobacter, Bacillus, Cryobacterium, Hymenobacter, Janthinobacterium, Micrococcus, Myxococcus, Planococcus, Pseudomonas, Psychrobacter and Sphingobacterium (Alam et al. 2003; Bowman et al. 1996; Shivaji et al. 1992, 2004; Reddy et al. 2009) were predominant in polar habitats and this variation in diversity could be attributed to the pristine environment and severe climatic conditions in the polar regions.
The 11 bacterial strains isolated from the soil samples were psychro-, halo- and alkalitolerant. The majority of the strains resembled the nearest phylogenetic neighbour in these characteristics. For instance, strains K7Sc-3b, K7Sc-2, K7Sc-4, K8Sc-3, K7Sc-8a, K7Sc-11a and K7Sc-13 were similar to their nearest phylogenetic neighbours Lysinibacillus parviboronicapiens BAM-582T (Miwa et al. 2009), Viridibacillus arvi LMG 22165T (Albert et al. 2007), Acinetobacter lwoffii (Reddy et al. 2009) and Psychrobacter nivimaris (Heuchert et al. 2004; Srinivas et al. 2009). The isolates belonging to the genera Bacillus, Lysinibacillus and Psychrobacter were not observed in the clone library analysis. A few strains such as those affiliated to Bacillus simplex and Pseudomonas lutea have been reported to be mesophilic, and psychrotolerant strains have not been reported. Noticeably, neither enteric nor pathogenic bacteria were detected in this study, though enteric bacteria were isolated from glacial ice on the Ellesmere Island (Dancer et al. 1997) and surface snow of the Tateyama Mountains in Japan (Segawa et al. 2005). In addition when the culturable bacterial diversity of the Kafni Glacier soil samples was compared with that of the Pindari Glacier, only one taxa Viridibacillus arvi was common to both (Shivaji et al. 2011b). As of now, only one new species of Bacillus has been described from a Himalayan Glacier (Reddy et al. 2008b).
In cold habitats, psychrophilic bacteria release hydrolytic enzymes, which can degrade complex biomolecules into smaller and easily accessible compounds. Strain K7Sc-3a was similar to its phylogenetic neighbour Pseudomonas lutea with respect to amylase activity. Strains K7Sc-7a, K7Sc-9b, K8Sc-2, K7Sc-2 and K8Sc-3 were similar to their phylogenetic neighbours Bacillus simplex and Viridibacillus arvi with respect to amylase activity, but differed from the same strain with respect to protease and urease activities. Surprisingly, none of the strains produced extracellular proteases.
The synthesis of unsaturated fatty acids is very vital for survival at low temperature (Nishida and Murata 1996). In accordance with previous studies, medium chain fatty acids with 10–12 carbons, unsaturated fatty acids and branched fatty acids (including the iso- and anteiso-fatty acids) that maintain the membrane in a fluid state and which are common in psychrophilic bacteria (Chintalapati et al. 2004; Shivaji et al. 2007) were also present in the bacterial isolates from the Kafni Glacier soil samples.
Conclusions/novel findings
-
1.
Bacterial diversity of two soil samples (K7s and K8s) collected near the Kafni Glacier based on 16S rRNA gene sequence analysis led to the identification of clones affiliated to Actinobacteria (5.1 and 1.0%), Bacteroidetes (23.2 and 34.8%), Chloroflexi (1.7 and 0.2%), Firmicutes (6.7 and 6.3%), Proteobacteria (51.5 and 51.3%), Spirochaetae (3.4 and 1.5%), Tenericutes (0.4 and 0.2%) and Verrucomicrobia (3.4 and 4.1%) as the common bacterial phyla. Clones affiliated to the phyla Lentisphaerae and Tenericutes have so far only been reported from mesophilic habitats.
-
2.
A total of 106 and 93 bacterial taxa were identified from K7s and K8s soil samples, respectively. Arsenic content is probably the key parameter that influences the heterogeneity observed in the bacterial diversity in K8s.
-
3.
Clones related to bacterial taxa that have a role in arsenic cycle, such as Sulfurospirillum arsenophilum, Clostridium and Rhodoferax, and strains related to the genus Bacillus were identified from both the libraries. Clones related to Sulfurospirillum arsenophilum were predominant in the KS library (42) compared to the KF library (3).
-
4.
Comparison of the bacterial diversity of the Kafni Glacier with the diversity in other Himalayan cold habitats, other non-polar high-altitude habitats and various polar habitats indicated that it was different in terms of distribution and abundance of the taxa.
-
5.
PCA indicated that bacterial diversity of KF and KS libraries were closely affiliated to the diversity in the Pindari (P8s), SIS (Samoylov Island, Siberia), SOA (Schirmacher Oasis, Antarctica) and ST (Siberian tundra) libraries.
-
6.
All the bacterial isolates were alkali-, halo- and psychrotolerant. Cold-active enzymes and fatty acids isolated from bacterial strains of the Kafni Glacier might offer novel opportunities for biotechnological exploitation.
References
Abyzov SS, Mitskevich IN, Poglazova MN (1998) Microflora of the deep glacier horizons of central Antarctica. Microbiol 67:66–73
Alam SI, Singh L, Dube S, Reddy GSN, Shivaji S (2003) Psychrophilic Planococcus matriensis sp. nov. from Antarctica. Syst Appl Microbiol 26:505–510
Albert RA, Archambault J, Lempa M, Hurst B, Richardson C, Gruenloh S, Duran M, Worliczek HL, Huber BE, Rosselló-Mora R, Schumann P, Busse HJ (2007) Proposal of Viridibacillus gen. nov. and reclassification of Bacillus arvi, Bacillus arenosi and Bacillus neidei as Viridibacillus arvi gen. nov., comb. nov., Viridibacillus arenosi comb. nov. and Viridibacillus neidei comb. nov. Int J Syst Evol Microbiol 57:2729–2737
Bhatia M, Sharp M, Foght J (2006) Distinct bacterial communities exist beneath a high Arctic polythermal glacier. Appl Environ Microbiol 72:5838–5845
Blakemore LC, Searle PL, Daly BK (1987) Methods for chemical analysis of soils. NZ Soil Bureau Scientific Report 80
Booth C (1978) Introduction to general methods. In: Booth C (ed) Methods microbiol. Academic Press, New York, pp 57–91
Bowman JP, Cavanagh J, Austin JJ, Sanderson K (1996) Novel Psychrobacter species from Antarctic ornithogenic soils. Int J Syst Bacteriol 46:841–848
Brambilla E, Hippe H, Hagelstein A, Tindall BJ, Stackebrandt E (2001) 16S rDNA diversity of cultured and uncultured prokaryotes of a mat sample from Lake Fryxell, McMurdo Dry Valleys, Antarctica. Extremophiles 5:23–33
Cheng SM, Foght JM (2007) Cultivation-independent and -dependent characterization of Bacteria resident beneath John Evans glacier. FEMS Microbiol Ecol 59:318–330
Chintalapati S, Kiran MD, Shivaji S (2004) Role of membrane lipid fatty acids in cold adaptation. Cell Mol Biol 50:631–642
Christner BC, Mosley-Thompson E, Thompson LG, Reeve JN (2001) Isolation of bacteria and 16S rDNAs from Lake Vostok accretion ice. Environ Microbiol 3:570–577
Christner BC, Mosley-Thompson E, Thompson LG, Reeve JN (2003) Bacterial recovery from ancient glacial ice. Environ Microbiol 5:433–436
Costello EK, Schmidt SK (2006) Microbial diversity in alpine tundra wet meadow soil: novel Chloroflexi from a cold, water-saturated environment. Environ Microbiol 8:1471–1486
Dancer SJ, Shears P, Platt DJ (1997) Isolation and characterization of coliforms from glacial ice and water in Canada’s High Arctic. J Appl Microbiol 82:597–609
Eichorst SA, Breznak JA, Schmidt TM (2007) Isolation and characterization of soil bacteria that define Terriglobus gen. nov. in the phylum Acidobacteria. Appl Environ Microbiol 73:2708–2717
Gangwar P, Alam SI, Bansod S, Singh L (2009) Bacterial diversity of soil samples from the western Himalayas, India. Can J Microbiol 55:564–577
Giovannoni SJ, Britschgi TB, Moyer CL, Field KG (1990) Genetic diversity in Sargasso Sea bacterioplankton. Nature 345:60–63
Good IJ (1954) The population frequencies of species and the estimation of population parameters. Biometrica 40:237–264
Hall LL, George SE, Kohan MJ, Styblo M, Thomas DJ (1997) In vitro methylation of inorganic arsenic in mouse intestinal cecum. Toxicol Appl Pharmacol 147:101–109
Head IM, Saunders JR, Pickup RW (1998) Microbial evolution diversity and ecology: a decade of ribosomal RNA analysis of uncultivated microorganisms. Microbial Ecol 35:1–21
Heuchert A, Glöckner FO, Amann R, Fischer U (2004) Psychrobacter nivimaris sp. nov., a heterotrophic bacterium attached to organic particles isolated from the South Atlantic (Antarctica). Syst Appl Microbiol 27:399–406
Hugenholtz P, Goebel BM, Pace NR (1998a) Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol 180:4765–4774
Hugenholtz P, Pitulle C, Hershberger KL, Pace NR (1998b) Novel division level bacterial diversity in a Yellowstone hot spring. J Bacteriol 180:366–376
Hur I, Chun J (2004) A method for comparing multiple bacterial community structures from 16S rDNA clone library sequences. J Microbiol 42:9–13
Jackson ML (1967) Soil chemical analysis. Prentice-Hall of India, New Delhi
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, New York, pp 115–147
Lefauconnier B, Hagen JO, Rudant JP (1994) Flow speed and calving rate of Kongsbreen glacier, Svalbard, using SPOT images. Pol Res 10:56–65
Li Y, Li F, Zhang X, Qin S, Zeng Z, Dang H, Qin Y (2008) Vertical distribution of bacterial and archaeal communities along discrete layers of a deep-sea cold sediment sample at the East Pacific Rise (approximately 13 degrees N). Extremophiles 12:573–585
Lipson DA, Schmidt SK (2004) Seasonal changes in an alpine soil bacterial community in the Colorado Rocky Mountains. Appl Environ Microbiol 70:2867–2879
Liu Y, Yao T, Jiao N, Kang S, Zeng Y, Huang S (2006) Microbial community structure in moraine lakes and glacial meltwaters, Mount Everest. FEMS Microbiol Lett 265:98–105
Liu Y, Yao T, Jiao N, Kang S, Xu B, Zeng Y, Huang S, Liu X (2009) Bacterial diversity in the snow over Tibetan Plateau glaciers. Extremophiles 13:411–423
Lysnes K, Thorseth IH, Steinsbu BO, Øvreås L, Torsvik T, Pedersen RB (2004) Microbial community diversity in seafloor basalt from the Arctic spreading ridges. FEMS Microbiol Ecol 50:213–230
Magurran AE (ed) (1996) Ecological diversity and its measurement. Chapman-Hall, London, pp 274–286
Miteva VI, Brenchley JE (2005) Detection and isolation of ultrasmall microorganisms from a 1, 20, 000-year-old Greenland glacier ice core. Appl Environ Microbiol 71:7806–7818
Miteva VI, Sheridan PP, Brenchley JE (2004) Phylogenetic and physiological diversity of microorganisms isolated from a deep Greenland glacier ice core. Appl Environ Microbiol 70:202–213
Miwa H, Ahmed I, Yokota A, Fujiwara T (2009) Lysinibacillus parviboronicapiens sp. nov., a low-boron-containing bacterium isolated from soil. Int J Syst Evol Microbiol 59:1427–1432
Nemergut DR, Anderson SP, Cleveland CC, Martin AP, Miller AE, Seimon A, Schmidt SK (2007) Microbial community succession in an unvegetated, recently deglaciated soil. Microbiol Ecol 53:110–122
Nishida I, Murata N (1996) Chilling sensitivity in plants and cyanobacteria: the crucial contribution of membrane lipids. Annu Rev Plant Physiol Plant Mol Biol 47:541–568
Pace NR, Stahl DA, Lane DJ, Olsen GJ (1986) The analysis of natural microbial populations by ribosomal RNA sequences. Adv Microb Ecol 9:1–55
Prabagaran SR, Manorama R, Delille D, Shivaji S (2007) Predominance of Roseobacter, Sulfitobacter, Glaciecola and Psychrobacter in seawater collected off Ushuaia, Argentina, Sub-Antarctica. FEMS Microbiol Ecol 59:342–355
Pradhan S, Srinivas TNR, Pindi PK, Kishore KH, Begum Z, Singh PK, Singh AK, Pratibha MS, Yasala AK, Reddy GS, Shivaji S (2010) Bacterial diversity from Roopkund Glacier, Himalayan Mountain ranges, India. Extremophiles 14:377–395
Priest FG (1977) Extracellular enzyme synthesis in the genus Bacillus. Bacteriol Rev 41:711–753
Priscu JC, Christner BC (2003) Microbial diversity and bioprocessing. In: Bull AT (ed) Earth’s icy biosphere. ASM press, Washington DC, pp 130–145
Reddy GSN, Aggarwal RK, Matsumotto GI, Shivaji S (2000) Arthrobacter flavus sp. nov., a psychrophilic bacterium isolate from a pond in McMurdo Dry Valley, Antarctica. Int J Syst Evol Microbiol 50:1553–1561
Reddy GSN, Prabagaran SR, Shivaji S (2008a) Leifsonia pindariensis sp. nov., isolated from the Pindari glacier of the Indian Himalayas, and emended description of the genus Leifsonia. Int J Syst Evol Microbiol 58:2229–2234
Reddy GSN, Uttam A, Shivaji S (2008b) Bacillus cecembensis sp. nov., isolated from the Pindari glacier of the Indian Himalayas. Int J Syst Evol Microbiol 58:2330–2335
Reddy PVV, Rao SSSN, Pratibha MS, Sailaja B, Kavya B, Manorama RR, Singh SM, Srinivas TNR, Shivaji S (2009) Bacterial diversity and bioprospecting for cold-active enzymes from culturable bacteria associated with sediment of melt water stream of Midtre Lov′enbreen glacier, an Arctic glacier. Res Microbiol 160:538–546
Reddy GSN, Pradhan S, Manorama R, Shivaji S (2010) Cryobacterium roopkundense sp. nov., a psychrophilic bacterium from a Himalayan glacier. Int J Syst Evol Microbiol 60:866–870
Reed AJ, Lutz RA, Vetriani C (2006) Vertical distribution and diversity of bacteria and archaea in sulfide and methane-rich cold seep sediments located at the base of the Florida Escarpment. Extremophiles 10:199–211
Rhoades JD (1982) Soluble salts. In: Page AL (ed) Methods of soil analysis. Part 2, 2nd edn. Agronomy monograph No. 9. American Society of Agronomy, Madison, pp 167–179
Schmidt SK, Nemergut DR, Miller AE, Freeman KR, King AJ, Seimon A (2009) Microbial activity and diversity during extreme freeze–thaw cycles in periglacial soils, 5400 m elevation, Cordillera Vilcanota, Perú. Extremophiles 13:807–816
Segawa T, Miyamoto K, Ushida K, Agata K, Okada N, Kohshima S (2005) Seasonal change in bacterial flora and biomass in mountain snow from the Tateyama Mountains, Japan, analyzed by 16S rRNA gene sequencing and real-time PCR. Appl Environ Microbiol 71:123–130
Shivaji S, Shyamala Rao N, Saisree L, Sheth V, Reddy GSN, Bhargava PM (1989) Isolation and identification of Pseudomonas species from Schirmacher Oasis, Antarctica. Appl Environ Microbiol 55:767–771
Shivaji S, Ray MK, Rao NS, Saisree L, Jagannadham MV, Kumar GS, Reddy GSN, Bhargava PM (1992) Sphingobacterium antarcticus sp. nov., a psychrotrophic bacterium from the soils of Schirmacher Oasis, Antarctica. Int J Syst Bacteriol 42:102–106
Shivaji S, Reddy GSN, Aduri RP, Kutty R, Ravenschlag K (2004) Bacterial diversity of a soil sample from Schirmacher Oasis, Antarctica. Cell Mol Biol 50:525–536
Shivaji S, Chaturvedi P, Reddy GS, Suresh K (2005) Pedobacter himalayensis sp. nov., from the Hamta glacier located in the Himalayan mountain ranges of India. Int J Syst Evol Microbiol 55:1083–1088
Shivaji S, Kiran MD, Chintalapati S (2007) Perception and transduction of low temperature in bacteria. In: Gerday C, Glansdorff N (eds) Physiology and biochemistry of extremophiles. ASM Press, Washington DC, pp 194–207
Shivaji S, Kumari K, Kishore KH, Pindi PK, Rao PS, Radha Srinivas TN, Asthana R, Ravindra R (2011a) Vertical distribution of bacteria in a lake sediment from Antarctica by culture-independent and culture-dependent approaches. Res Microbiol 162:191–203
Shivaji S, Pratibha MS, Sailaja B, Hara Kishore K, Singh AK, Begum Z, Anarasi U, Prabagaran SR, Reddy GSN, Srinivas TNR (2011b) Bacterial diversity of soil in the vicinity of Pindari glacier, Himalayan mountain ranges, India, using culturable bacteria and soil 16S rRNA gene clones. Extremophiles 15:1–22
Skidmore M, Anderson SP, Sharp M, Foght J, Lanoil BD (2005) Comparison of microbial community composition in two subglacial environments reveals a possible role for microbes in chemical weathering processes. Appl Environ Microbiol 71:6986–6997
Srinivas TNR, Nageswara Rao SSS, Vishnu Vardhan Reddy P, Pratibha MS, Sailaja B, Kavya B, Hara Kishore K, Begum Z, Singh SM, Shivaji S (2009) Bacterial diversity and bioprospecting for cold-active lipases, amylases and proteases, from culturable bacteria of Kongsfjorden and Ny-Ålesund, Svalbard, Arctic. Curr Microbiol 59:537–547
Steven B, Briggs G, McKay CP, Pollard WH, Greer CW, Whyte LG (2007) Characterization of the microbial diversity in a permafrost sample from the Canadian high Arctic using culture-dependent and culture-independent methods. FEMS Microbiol Ecol 59:513–523
Torsvik V, Sorheim R, Goksoyr J (1996) Total bacterial diversity in soil and sediment communities—a review. J Ind Microbiol 17:170–178
Treves DS, Xia B, Tiedje JM (2003) A two-species test of the hypothesis that spatial isolation influences microbial diversity in soil. Microbiol Ecol 45:20–28
Tsai YL, Olson BH (1991) Rapid method for direct extraction of DNA from soil and sediments. Appl Environ Microbiol 57:1070–1074
Wagner D, Kobabe S, Liebner S (2009) Bacterial community structure and carbon turnover in permafrost-affected soils of the Lena Delta, northeastern Siberia. Can J Microbiol 55:77–83
Wani AA, Surakasi VP, Siddharth J, Raghavan RG, Patole MS, Ranade D, Shouche YS (2006) Molecular analyses of microbial diversity associated with the Lonar soda lake in India: an impact crater in a basalt area. Res Microbiol 157:928–937
Ward DM, Weller R, Bateson MM (1990) 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature 345:63–65
Woese CR (1987) Bacterial evolution. Microbiol Rev 51:221–271
Xiang SR, Yao TD, An LZ, Xu BQ, Li Z, Wu GJ, Wang YQ, Ma S, Chen XR (2004) Bacterial diversity in Malan ice core from the Tibetan Plateau. Folia Microbiol 49:269–275
Xiang S, Yao T, An L, Xu B, Wang J (2005) 16S rRNA sequences and differences in bacteria isolated from the Muztag Ata glacier at increasing depths. Appl Environ Microbiol 71:4619–4627
Xiang SR, Yao TD, Wu GJ, Chen Y, Shang TC, Pu LL, An LZ (2006) Deposition properties of bacterial populations in the Muztag Ata ice core. Quat Sci 26:185–191
Xiang SR, Shang TC, Chen Y, Jing ZF, Yao T (2009) Dominant bacteria and biomass in the Kuytun 51 Glacier. Appl Environ Microbiol 75:7287–7290
Zhang XJ, Yao TD, Ma XJ, Wang NL (2002) Microorganisms in a high altitude glacier ice in Tibet. Folia Microbiol (Praha) 47:241–245
Zhang X, Tandong Y, Lizhe A, Lide T, Shijian X (2006) A study on the vertical profile of bacterial DNA structure in the Puruogangri (Tibetan Plateau) ice core using denaturing gradient gel electrophoresis. Anals Glaciol 43:160–166
Zhang S, Hou S, Ma X, Qin D, Chen T (2007) Culturable bacteria in Himalayan glacial ice in response to atmospheric circulation. Biogeoscience 4:1–9
Zhang X, Yao T, Tian L, Xu S, An L (2008) Phylogenetic and physiological diversity of bacteria isolated from Puruogangri ice core. Microbiol Ecol 55:476–488
Zhou J, Davey ME, Figueras JB, Rivkina E, Gilichinsky D, Tiedje JM (1997) Phylogenetic diversity of a bacterial community determined from Siberian tundra soil DNA. Microbiol 143:3913–3919
Acknowledgments
We would like to thank the Department of Biotechnology and Council of Scientific and Industrial Research, Government of India, for financial support to Dr. S. Shivaji. TNRS acknowledges the CSIR, Government of India, for the award of research associateship.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Oren.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Srinivas, T.N.R., Singh, S.M., Pradhan, S. et al. Comparison of bacterial diversity in proglacial soil from Kafni Glacier, Himalayan Mountain ranges, India, with the bacterial diversity of other glaciers in the world. Extremophiles 15, 673 (2011). https://doi.org/10.1007/s00792-011-0398-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00792-011-0398-8