Abstract
A Gram-positive, motile, endospore-forming and rod-shaped halophilic bacterial strain MSS-155 (KCTC 3788 and KCCM 41687) was isolated from a marine solar saltern of the Yellow Sea in Korea and was subjected to a polyphasic taxonomic study. This organism grew at temperature of 10.0–42.0°C with an optimum of 35°C. Strain MSS-155 grew optimally in the presence of 10% NaCl and did not grow in the absence of NaCl. The cell wall peptidoglycan type of strain MSS-155 was A4β based on l-Orn-d-Asp. Strain MSS-155 was also characterized chemotaxonomically by having menaquinone-7 (MK-7) as the predominant isoprenoid quinone and anteiso-C15:0 as the major fatty acid. The DNA G+C content was 44.0 mol%. Phylogenetic analysis based on 16S rDNA sequences showed that strain MSS-155 falls within the radiation of the cluster comprising Halobacillus species. Levels of 16S rDNA sequence similarity between strain MSS-155 and the type strains of four Halobacillus species were in the range 97.6–98.8%. Strain MSS-155 exhibited levels of DNA-DNA relatedness of 6.2–11.2% to the type strains of Halobacillus species described previously. On the basis of phenotypic properties, phylogeny, and genomic data, strain MSS-155 should be placed in the genus Halobacillus as a member of a novel species, for which we propose the name Halobacillus locisalis sp. nov.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hypersaline environments are found worldwide and include a variety of habitats such as salt flats, evaporation ponds, natural inland salt lakes, soda lakes, subsurface salt formations, deep-sea hypersaline basins, and others (Vreeland et al. 1998; Rothschild and Mancinelli 2001; Sass et al. 2001; Litchfield and Gillevet 2002; Oren 2002). Studies on the microbiology of hypersaline environments have shown that halophilic members of the domain Archaea are dominant in these environments, whereas those of the domain Bacteria are minor components (Oren 1994; Rothschild and Mancinelli 2001; Ochsenreiter et al. 2002). However, recent studies based on phylogenetic techniques and fluorescence in situ hybridization have shown that members of the domain Bacteria play an important role in the microbial diversity and community of hypersaline environments (Antón et al. 1999, 2000). The halophilic bacteria inhabit a wide range of sources that are much less restricted than those of the halophilic archaea (Rodriguez-Valera 1986; Ventosa et al. 1998). The moderately halophilic bacteria constitute a heterogeneous group of microorganisms that belong to different genera (Ventosa et al. 1998), and the number of new species with salt-loving physiological properties has continuously increased.
Recently, in the course of screening useful microorganisms present in hypersaline environments, we isolated some bacterial strains from a marine solar saltern of the Yellow Sea in Korea and also characterized them taxonomically. Preliminary studies on the microbiology of this region have revealed the presence of a variety of bacterial strains. Of these isolates, one Gram-variable, halophilic, and rod-shaped bacterial strain (MSS-155) attracted our attention and was subjected to further taxonomic studies. Strain MSS-155 was considered to be a Halobacillus-like strain from the result of 16S rDNA sequence analysis. The genus Halobacillus was proposed by Spring et al. (1996) for two species, H. litoralis and H. trueperi, and Sporosarcina halophila (Claus et al. 1983) was transferred to Halobacillus as H. halophilus. The genus Halobacillus is clearly differentiated from other related genera by a cell wall peptidoglycan type based on l-Orn-d-Asp (Spring et al. 1996; Shida et al. 1997; Yoon et al. 2001). The aim of the present research was to determine the exact taxonomic status of strain MSS-155 by a polyphasic characterization, including phenotypic properties, detailed phylogenetic analysis based on 16S rDNA sequence, and genotypic relatedness.
Materials and methods
Bacterial strains and cultural conditions
Strain MSS-155 was isolated from a marine solar saltern located in Baekryung Island of the Yellow Sea in Korea. Isolation was performed aerobically on marine agar 2216 (MA) (Difco) at 30°C by the dilution plating technique. Strain MSS-155 was deposited in the Korean Collection for Type Cultures as KCTC 3788 and in the Korean Culture Center of Microorganisms as KCCM 41687. Halobacillus halophilus KCTC 3685T, Halobacillus litoralis KCTC 3687T, and Halobacillus trueperi KCTC 3686T were used as reference strains in this study. The type strain of Halobacillus salinus (strain HSL-3T = KCCM 41590T = JCM 11546T), which has recently been described as a new species of the genus Halobacillus (Yoon et al. 2003), was also used as reference strain in this study.
In most cases for morphological and physiological characterization, strain MSS-155 was cultivated on MA and in marine broth 2216 (MB) (Difco) supplemented with approximately 8.1% (w/v) NaCl at 30°C. Cell biomass of strain MSS-155 for the analyses of the cell wall and menaquinones and for DNA extraction was produced in MB supplemented with approximately 8.1% NaCl at 30°C. Strain MSS-155 was cultivated on a gyratory shaker at 150 rpm; the broth cultures were checked for purity by microscopic examination before being harvested by centrifugation. For fatty acid methyl ester (FAME) analysis, cell mass of strain MSS-155 and the reference strains were obtained from agar plates after cultivation for 4 days at 30°C on MA supplemented with approximately 8.1% NaCl.
Morphological and physiological characterization
Cell morphology was examined by light microscopy and transmission electron microscopy (TEM). Flagellum type was examined by TEM using cells from exponentially growing cultures. The cells were negatively stained with 1% (w/v) phosphotungstic acid, and after air-drying, the grids were examined by using a model CM-20 transmission electron microscope (Philips, Eindhoven, The Netherlands). The Gram reaction was determined using bioMérieux Gram Strain kit (bioMérieux, Marcy-l'Etoile, France) according to the manufacturer's instructions. Catalase activity was determined by bubble production in a 3% (v/v) H2O2 solution. Oxidase activity was determined by oxidation of 1% p-aminodimethylaniline oxalate. Urease activity was determined as described by Cowan and Steel (1965) with addition of 10% NaCl. Hydrolysis of casein and starch was determined as described by Cowan and Steel (1965). Hydrolysis of aesculin, gelatin, Tween 20, Tween 40, Tween 60, and Tween 80 and nitrate reduction were determined as described by Lanyi (1987) with a modification in that modified artificial seawater (MASW) was used. The MASW contained (in l–1 distilled water) NaCl, 100 g; KCl, 0.64 g; MgCl2·6H2O, 4.53 g; MgSO4·7H2O, 5.94 g; CaCl2·2H2O, 1.3 g (Levring 1946). Hydrolysis of hypoxanthine, tyrosine, and xanthine was performed on MA supplemented with 8.1% (w/v) NaCl using substrate concentrations described previously (Cowan and Steel 1965). Acid production from carbohydrates was determined as described by Leifson (1963). Growth under anaerobic conditions was determined after incubation in an anaerobic chamber with anaerobically prepared MA supplemented with 8.1% (w/v) NaCl. Growth at various NaCl concentrations was investigated in MB. Growth at various temperatures and pH was measured at on MA supplemented with approximately 8.1% (w/v) NaCl.
Chemosystematic characterization
The presence or absence of diaminopimelic acid in the cell wall peptidoglycan was determined by the method described by Komagata and Suzuki (1987). Preparation of cell wall and determination of peptidoglycan structure were carried out by the methods described by Schleifer and Kandler (1972) with the modification that thin-layer chromatography (TLC) on cellulose sheets (Merck, Darmstadt, Germany) was used instead of paper chromatography. Menaquinones were analyzed as described previously (Komagata and Suzuki 1987) using reverse-phase high-performance liquid chromatography (HPLC). For quantitative analysis of cellular fatty acid compositions, a loop of cell mass was harvested and FAMEs were prepared and identified following the instructions of the Microbial Identification System (Microbial ID Inc., Newark, Del., USA).
DNA studies
Chromosomal DNA was isolated and purified according to a method described previously (Yoon et al. 1996), with the exception that ribonuclease T1 was used together with ribonuclease A. The G+C content was determined by the method of Tamaoka and Komagata (1984). DNA was hydrolyzed and the resultant nucleotides were analyzed by reverse-phase HPLC. DNA-DNA hybridization was performed fluorometrically by the method of Ezaki et al. (1989) using photobiotin-labeled DNA probes and microdilution wells. Hybridization was performed with five replications for each sample. Of the values obtained, the highest and lowest values in each sample were excluded and the remaining three values were used for the calculation of similarity values. DNA-DNA relatedness values were expressed as the mean of three values.
16S rDNA sequencing and phylogenetic analysis
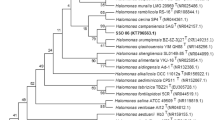
16S rDNA was amplified by PCR as described previously (Yoon et al. 1998) using two universal primers; 9F (5′-GAGTTTGATCCTGGCTCAG-3′) and 1542R (5′-AGAAAGGAGGTGATCCAGCC-3′). The PCR product was purified with a QIAquick PCR purification kit (QIAGEN GmbH, Hilden, Germany). The purified 16S rDNA was sequenced using an ABI PRISM BigDye Terminator cycle sequencing ready reaction kit (Applied Biosystems, Foster City, Calif., USA) as recommended by the manufacturer. The sequencing reaction mixtures were electrophoresed automatically using an Applied Biosystems model 377 automatic DNA sequencer. Alignment of sequences was carried out with CLUSTAL W software (Thompson et al. 1994). Gaps at the 5′ and 3′ ends of the alignment were omitted from further analysis. Phylogenetic trees were inferred by using three tree-making algorithms, i.e., the neighbor-joining (Saitou and Nei 1987), maximum-likelihood (Felsenstein 1981), and maximum-parsimony (Kluge and Farris 1969) methods contained within the PHYLIP package (Felsenstein 1993). Evolutionary distance matrices for the neighbor-joining method were calculated with the algorithm of Jukes and Cantor (1969) with the program DNADIST. The stability of relationships was assessed by a bootstrap analysis based on 1,000 resamplings of the neighbor-joining dataset by using the programs SEQBOOT, DNADIST, NEIGHBOR, and CONSENSE of the PHYLIP package. The designations and 16S rDNA sequence accession numbers of the reference strains used in the phylogenetic analysis are shown in Fig. 1.
Neighbor-joining tree based on 16S rDNA sequences showing the phylogenetic positions of strain MSS-155, Halobacillus species, and the representatives of some other related taxa. Scale bar represents 0.01 substitutions per nucleotide position. Bootstrap values (expressed as percentages of 1,000 replications) greater than 50% are shown at the branch points
Nucleotide sequence accession number
The GenBank accession number for the 16S rDNA sequence of strain MSS-155 is AY190534.
Results and discussion
Phenotypic characteristics
Strain MSS-155 was strictly aerobic and Gram-positive but changed to Gram-variable as cultures aged. Its cells were straight rods measuring approximately 0.8–1.0 μm wide and 1.5–4.0 μm long after cultivation for 3 days under optimum conditions. Strain MSS-155 was motile by means of a single polar flagellum. Ellipsoidal endospores were observed at central or subterminal positions in swollen sporangia. Colonies of strain MSS-155 were smooth, circular to slightly irregular in shape, slightly raised, light orange-yellow in color, and 2–3 mm in diameter after 3 days' culture.
Strain MSS-155 grew at temperature range of 10.0–42.0°C, with an optimum temperature at 30–35°C. The optimal pH for growth was 7.0–8.0; growth was observed at pH 5.0 and pH 9.5 but not at pH 4.5 or pH 10.0. Strain MSS-155 required NaCl for growth. It grew optimally in the presence of 10% NaCl and grew in the presence of 23% NaCl but not in the presence of more than 24% NaCl.
Strain MSS-155 was positive for catalase and oxidase but negative for urease activity and nitrate reduction. Aesculin, starch, Tween 40, and Tween 60 were hydrolyzed, and Tween 20 and Tween 80 were weakly hydrolyzed. No hydrolysis of casein, gelatin, hypoxanthine, tyrosine, and xanthine was observed. Acid was produced from d-cellobiose, d-fructose, d-glucose, d-melezitose, d-ribose, sucrose, and d-trehalose. The phenotypic properties of strain MSS-155 and Halobacillus species are summarized in Table 1.
Chemosystematic characteristics
Strain MSS-155 did not contain any diaminopimelic acid as the diagnostic diamino acid in the cell wall peptidoglycan. From the result of the cell wall analysis, strain MSS-155 had the peptidoglycan type of A4β, based on l-Orn-d-Asp, as described by Schleifer and Kandler (1972). This cell wall peptidoglycan type is a key marker differentiating strain MSS-155 and the genus Halobacillus from other aerobic or facultative anaerobic, endospore-forming, rod-shaped genera. The genus Halobacillus has the peptidoglycan type based on l-Orn-d-Asp (Spring et al. 1996), whereas other related genera contain meso-diaminopimelic acid or l-lysine at position 3 of the cell wall peptidoglycan (Shida et al. 1997; Wainø et al. 1999; Yoon et al. 2001). The genus Filobacillus, which has recently been described, contains l-ornithine at position 3 of the cell wall peptidoglycan, but it has the peptidoglycan type based on l-Orn-d-Glu (Schlesner et al. 2001). The predominant menaquinone found in strain MSS-155 was an unsaturated menaquinone with seven isoprene units (MK-7). Strain MSS-155 had a cellular fatty acid profile containing large amounts of branched fatty acids (Table 2). The major fatty acid detected in strain MSS-155 was anteiso-C15:0; significant amounts of iso-C16:0, anteiso-C17:0 and iso-C14:0 were also present (Table 2). The DNA G+C content of strain MSS-155 was 44.0 mol%, which is in the range for known Halobacillus species (Table 1). These chemosystematic data analyzed in strain MSS-155 were shown to be most similar to those of Halobacillus species described previously (Spring et al. 1996).
Phylogenetic analysis
An almost complete 16S rDNA sequence of strain MSS-155was directly determined after PCR amplification. The 16S rDNA sequence of strain MSS-155 determined in this study comprised 1,522 nucleotides, representing approximately 96% of the Escherichia coli 16S rRNA sequence. This sequence was subjected to similarity searches with a public database (GenBank) to infer a possible phylogenetic classification of strain MSS-155. The result revealed that MSS-155 is a member of the genus Halobacillus. This became clear from the phylogenetic analysis and nucleotide sequence similarity values. In the phylogenetic tree based on 16S rDNA sequences and a neighbor joining algorithm, strain MSS-155 fell within the radiation of the cluster comprising three valid Halobacillus species and Halobacillus salinus, which has been accepted for publication as a new species of the genus Halobacillus (Fig. 1). Similar tree topology was also found in the tree generated with a maximum-parsimony algorithm (data not shown). Strain MSS-155 exhibited levels of 16S rDNA similarity of 97.6%, 98.8%, 98.8%, and 98.3% with the type strains of H. halophilus, H. litoralis, H. trueperi, and H. salinus, respectively. Levels of 16S rDNA similarity between strain MSS-155 and other members used in the phylogenetic analysis were less than 95% (Fig. 1).
DNA-DNA relatedness
DNA-DNA hybridization was performed to determine the genotypic relatedness between strain MSS-155 and the type strains of all Halobacillus species. Strain MSS-155 exhibited levels of DNA-DNA relatedness of 6.2%, 8.3%, 8.9%, and 11.2% to H. halophilus KCTC 3685T, H. litoralis KCTC 3687T, H. trueperi KCTC 3686T, and H. salinus HSL-3T, respectively. There are widely accepted criteria for delineating species in current bacteriology, stating that strains with a level of DNA relatedness less than 70% or with greater than 3% difference in 16S rDNA similarity are considered as being different species (Wayne et al. 1987; Stackebrandt and Goebel 1994).
Strain MSS-155 exhibited the closest phylogenetic affiliation to Halobacillus species from 16S rDNA sequence comparison (Fig. 1). The chemotaxonomic data obtained from strain MSS-155 was the most similar to that of members of the genus Halobacillus (Spring et al. 1996). Therefore, both phylogenetic and chemotaxonomic results clearly indicate that strain MSS-155 belongs to the genus Halobacillus. Strain MSS-155 is similar to Halobacillus species in its morphological and most of its physiological characteristics (Table 1). However, there are some minor differences between strain MSS-155 and Halobacillus species, including tolerance of NaCl, temperature, and pH for growth, their ability to hydrolyze some substrates, and acid production from carbohydrates (Table 1). The levels of DNA-DNA relatedness, together with some differential phenotypic properties and phylogenetic distinctiveness, confirm that strain MSS-155 is separate from Halobacillus species described previously (Wayne et al. 1987). Therefore, on the basis of phenotypic, chemotaxonomic, and phylogenetic data and genomic distinctiveness, strain MSS-155 should be placed in the genus Halobacillus as a novel species, for which we propose the name Halobacillus locisalis sp. nov. The properties of the new species are summarized below.
Description of Halobacillus locisalis sp. nov.
Halobacillus locisalis (lo.ci.sa'lis L. n. locus place, locality; L. gen. n. salis of salt; N.L. gen. n. locisalis from a place of salt)
Cells are straight rods measuring 0.8–1.0 μm in width and 1.5–4.0 μm in length after 3 days' cultivation at 35°C on MA. Strictly aerobic, and Gram-positive but Gram-variable in old cultures. Motile by means of a single polar flagellum. Central or subterminal ellipsoidal endospores are observed in swollen sporangia. Colonies are smooth, circular to slightly irregular in shape, slightly raised, light orange-yellow in color, and 2–3 mm in diameter after cultivation of 3 days. Optimal growth temperature is 30–35°C. Temperature range for growth is 10–42°C. Optimal growth pH is 7–8; growth occurs at pH 5 and pH 9.5 but not at pH 4.5 or pH 10. Optimal growth occurs in the presence of 10% NaCl. No growth occurs in the absence of NaCl. Growth occurs in the presence of 23.0% NaCl but not in the presence of more than 24% NaCl. Catalase and oxidase-positive. Urease-negative. Aesculin, starch, Tween 40, and Tween 60 are hydrolyzed. Tween 20 and Tween 80 are weakly hydrolyzed. Casein, gelatin, hypoxanthine, tyrosine, and xanthine are not hydrolyzed. Nitrate is not reduced to nitrite. Acid is produced from the following sugars: d-cellobiose, d-fructose, d-glucose, d-melezitose, d-ribose, sucrose, and d-trehalose. Acid is not produced from the following sugars: adonitol, l-arabinose, d-galactose, d-mannitol, d-mannose, melibiose, myo-inositol, lactose, maltose, d-raffinose, l-rhamnose, d-sorbitol, stachyose, and d-xylose. The predominant menaquinone is MK-7. The major fatty acid is anteiso-C15:0. The DNA G+C content is 44 mol% (determined by HPLC). Isolated from a marine solar saltern located in Baekryung Island of the Yellow Sea in Korea. Strain MSS-155 has been deposited in the Korean Collection for Type Cultures as KCTC 3788T and in the Korean Culture Center of Microorganisms as KCCM 41687T.
References
Antón J, Llobet-Brossa E, Rodríguez-Valera F, Amann R (1999) Fluorescence in situ hybridization analysis of the prokaryotic community in habiting crystallizer ponds. Environ Microbiol 1:517–523
Antón J, Rosselló-Mora R, Rodríguez-Valera F, Amann R (2000) Extremely halophilic Bacteria in crystallizer ponds from solar salterns. Appl Environ Microbiol 66:3052–3057
Claus D, Fahmy F, Rolf HJ, Tosunoglu N (1983) Sporosarcina halophila sp. nov., an obligate, slightly halophilic bacterium from salt marsh soils. Syst Appl Microbiol 4:496–506
Cowan ST, Steel KJ (1965) Manual for the identification of medical bacteria. Cambridge University Press, London
Ezaki T, Hashimoto Y, Yabuuchi E (1989) Fluorometric deoxyribonucleic acid- deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int J Syst Bacteriol 39:224–229
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1993) PHYLIP: phylogenetic inference package (version 3.5). University of Washington, Seattle, Washington
Jukes TH, Cantor CR (1969) Evolution of protein molecules. In: Munro HN (ed) Mammalian protein metabolism,vol 3. Academic Press, New York, pp 21–132
Kluge AG, Farris FS (1969) Quantitative phyletics and the evolution of anurans. Syst Zool 18:1–32
Komagata K, Suzuki K (1987) Lipids and cell-wall analysis in bacterial systematics. Methods Microbiol 19:161–203
Lanyi B (1987) Classical and rapid identification methods for medically important bacteria. Methods Microbiol 19:1–67
Leifson E (1963) Determination of carbohydrate metabolism of marine bacteria. J Bacteriol 85:1183–1184
Levring T (1946) Some culture experiments with Ulva and artificial seawater. Kungl Fysiografiska Sällsk Lund Förhandlingar 16:45–56
Litchfield CD, Gillevet PM (2002) Microbial diversity and complexity in hypersaline environments: A preliminary assessment. J Ind Microbiol Biotechnol 28:48–55
Ochsenreiter T, Pfeifer F, Schleper C (2002) Diversity of Archaea in hypersaline environments characterized by molecular-phylogenetic and cultivation studies. Extremophiles 6:267–274
Oren A (1994) The ecology of the extremely halophilic archaea. FEMS Microbiol Rev 13:415–440
Oren A (2002) Diversity of halophilic microorganisms: Environments, phylogeny, physiology, and applications. J Ind Microbiol Biotechnol 28:56–63
Rodriguez-Valera F (1986) The ecology and taxonomy of aerobic chemoorganotrophic halophilic eubacteria. FEMS Microbiol Rev 39:17–22
Rothschild LJ, Mancinelli RL (2001) Life in extreme environments. Nature 409:1092–1101
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sass AM, Sass H, Coolen MJL, Cypionka H, Overmann J (2001) Microbial communities in the chemocline of a hypersaline deep-sea basin (Urania basin, Mediterranean Sea). Appl Environ Microbiol 67:5392–5402
Schleifer KH, Kandler O (1972) Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev 36:407–477
Schlesner H, Lawson PA, Collins MD, Weiss N, Wehmeyer U, Völker H, Thomm M (2001) Filobacillus milensis gen. nov., sp. nov., a new halophilic spore-forming bacterium with Orn-d-Glu-type peptidoglycan. Int J Syst Evol Microbiol 51:425–431
Shida O, Takagi H, Kadowaki K, Nakamura LK, Komagata K (1997) Transfer of Bacillus alginolyticus, Bacillus chondroitinus, Bacillus curdlanolyticus, Bacillus glucanolyticus, Bacillus kobensis, and Bacillus thiaminolyticus to the genus Paenibacillus and emended description of the genus Paenibacillus. Int J Syst Bacteriol 47:289–298
Spring S, Ludwig W, Marquez MC, Ventosa A, Schleifer K-H (1996) Halobacillus gen. nov., with description of Halobacillus litoralis sp. nov. and Halobacillus truperi sp. nov., and transfer of Sporosarcina halophilia to Halobacillus halophilus comb. nov. Int J Syst Bacteriol 46:492–496
Stackebrandt E, Goebel BM (1994) Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44:846–849
Tamaoka J, Komagata K (1984) Determination of DNA base composition by reverse-phase high-performance liquid chromatography. FEMS Microbiol Lett 25:125–128
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Ventosa A, Nieto JJ, Oren A (1998) Biology of moderately halophilic aerobic bacteria. Microbiol Mol Biol Rev 62:504–544
Vreeland RH, Piselli Jr. AF, McDonnough S, Meyers SS (1998) Distribution and diversity of halophilic bacteria in a subsurface salt formation. Extremophiles 2:321–331
Wainø M, Tindall BJ, Schumann P, Ingvorsen K (1999) Gracilibacillus gen. nov., with description of Gracilibacillus halotolerans gen. nov., sp. nov.; transfer of Bacillus dipsosauri to Gracilibacillus dipsosauri comb. nov., and Bacillus salexigens to the genus Salibacillus gen. nov., as Salibacillus salexigens comb. nov. Int J Syst Bacteriol 49:821–831
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE, Stackebrandt E, Starr MP, Trüper HG (1987) Report of the Ad Hoc Committee on Reconciliation of Approaches to Bacterial Systematics. Int J Syst Bacteriol 37:463–464
Yoon J-H, Kim H, Kim S-B, Kim H-J, Kim WY, Lee ST, Goodfellow M, Park Y-H (1996) Identification of Saccharomonospora strains by the use of genomic DNA fragments and rRNA gene probes. Int J Syst Bacteriol 46:502–505
Yoon J-H, Lee ST, Park Y-H (1998) Inter- and intraspecific phylogenetic analysis of the genus Nocardioides and related taxa based on 16S rDNA sequences. Int J Syst Bacteriol 48:187–194
Yoon J-H, Weiss N, Lee K-C, Lee I-S, Kang KH, Park Y-H (2001) Jeotgalibacillus alimentarius gen. nov., sp. nov., a novel bacterium isolated from jeotgal with l-lysine in the cell wall, and reclassification of Bacillus marinus Rüger 1983 as Marinibacillus marinus gen. nov., comb. nov. Int J Syst Evol Microbiol 51:2087–2093
Yoon J-H, Kang KH, Park Y-H (2003) Halobacillus salinus sp. nov., isolated from a salt lake on the coast of the East Sea in Korea. Int J Syst Evol Microbiol 53:687–693
Acknowledgments
This work was supported by the 21C Frontier program of Microbial Genomics and Applications (grant MG02-0401-001-1-0-0), the NRL research program (grants M10104000294-01J000012800 and M10104000294-01J000012811) of the Ministry of Science and Technology (MOST) of the Republic of Korea, and by the research fund of Probionic Corporation of Korea.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by W.D. Grant
Rights and permissions
About this article
Cite this article
Yoon, JH., Kang, K.H., Oh, TK. et al. Halobacillus locisalis sp. nov., a halophilic bacterium isolated from a marine solar saltern of the Yellow Sea in Korea. Extremophiles 8, 23–28 (2004). https://doi.org/10.1007/s00792-003-0352-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-003-0352-5