Abstract
The prokaryotic formate metabolism is considerably diversified. Prokaryotes use formate in the C1 metabolism, but also evolved to exploit the low reduction potential of formate to derive energy, by coupling its oxidation to the reduction of numerous electron acceptors. To fulfil these varied physiological roles, different types of formate dehydrogenase (FDH) enzymes have evolved to catalyse the reversible 2-electron oxidation of formate to carbon dioxide. This review will highlight our present knowledge about the diverse physiological roles of FDH in prokaryotes, their modular structural organisation and active site structures and the mechanistic strategies followed to accomplish the formate oxidation. In addition, the ability of FDH to catalyse the reverse reaction of carbon dioxide reduction, a potentially relevant reaction for carbon dioxide sequestration, will also be addressed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction and scope
This minireview focuses on prokaryotic formate dehydrogenase (FDH) enzymes, which catalyse the reversible 2-electron oxidation of formate to carbon dioxide. FDHs are a group of heterogeneous proteins, harbouring diverse or no redox centres and displaying different subunit compositions and quaternary structures. FDHs are involved in a multiplicity of pathways, in both biosynthetic and energy metabolism, where they participate in fermentation and respiratory chain pathways. Herein, we will describe some of the physiological roles of formate in prokaryotic metabolism, including also a very brief picture of formate metabolism in humans and plants. Subsequently, the basis for the FDH classification as metal-dependent and metal-independent enzymes will be described and a brief account on the molybdo- and tungstoenzymes’ families will be given. The article will be focussed, then, on describing the molybdenum- and tungsten-dependent FDH modular structural organisation and their active site structures (the enzymatic “machinery”). At this point, Table 1 aims to be a “roadmap” of the minireview, providing a summary of the structural characteristics and cellular roles of some of the FDHs herein described. Finally, the molybdenum- and tungsten-dependent FDH reaction mechanism to oxidise formate will be discussed and the catalytic properties towards the reverse reaction of carbon dioxide reduction of some FDHs will be reviewed.
Living with formate
Formate is the conjugated base of formic acid (methanoic acid, HCOOH). With a pK a of 3.77, it is present in cells as the formate anion. It is the simplest carboxylic acid and, as such, it is broadly used in C1 metabolism, in prokaryotes and eukaryotes. In addition, the low redox potential of formate [Eº′ (CO2/HCOO− (pH 7, formate 1 molal activity, CO2 (g) 1 atm)] = −0.43 V [1]), enables prokaryotes to also use it to derive energy, by coupling its oxidation (Eq. 1) to the reduction of several terminal electron acceptors.
Formate is found in all forms of life, from bacteria to man. In humans, it has long been known that formate is involved in C1 metabolism, where it participates in the tetrahydrofolate-mediated metabolism of nucleic acids and serine [2–4]. Nevertheless, mammals do not possess FDH enzymes and formate oxidation to carbon dioxide (for its elimination) is catalysed by the combined actions of 10-formyltetrahydrofolate synthetase and 10-formyltetrahydrofolate dehydrogenase or by catalase [4–6].
In contrast, plants possess FDH (NAD+-dependent, metal-independent enzymes). Actually, FDH accounts for 9 % of all mitochondrial proteins of potato non-photosynthesising tissues [7]. In higher plants, formate, formed via ferredoxin-dependent carbon dioxide fixation or as a side product of photorespiration and some other pathways, is a precursor of carbon-containing compounds, being also involved in serine metabolism, as in mammals [8, 9]. Recently, several studies showed that the FDH expression is highly increased under unfavourable conditions, such as drought, deficiency of both light and iron, hypoxia, presence of pathogenic microorganisms or chemical agents [9–13]. Although the formate sources in plants under stress remain a matter of discussion [10], those studies suggest the involvement of FDH in cellular stress responses, further emphasising the key role of FDH in higher plants.
The prokaryotic formate metabolism is considerably more diversified. As in higher organisms, prokaryotes can use formate for biosynthetic purposes, as a carbon source, inserting the methyl residue directly or in tetrahydrofolate-mediated reactions (formation of methyl purines, serine, methionine, thymine), or using it in biosynthetic redox reactions (ribonucleotides reduction to deoxyribonucleotides) [14–16]. However, prokaryotes can use formate also for the energy metabolism, in fermentation and respiratory chain pathways [14, 17–23]. As such, formate, formed in the pyruvate:formate lyase reaction, is an end product of many bacterial fermentations, such as the mixed-acid fermentations of enterobacteria. Formate is also an intermediate of the energy metabolism of acetogenic bacteria, where carbon dioxide is reduced to acetic acid. In addition, formate is also the substrate (electron donor) for a variety of inducible respiratory chains that take advantage of the very low reduction potential of formate to couple its oxidation to the reduction of different terminal electron acceptors, including nitrate, sulfate, polysulfide, fumarate, carbon dioxide, iron (Fe3+), arsenate, or even dioxygen.
In the following paragraphs, the main prokaryotic pathways designed to derive energy from formate/FDH will be outlined (some included in the Table 1).
-
(a)
Escherichia coli, for example, can differentially express three FDHs. E. coli can grow aerobically and anaerobically; the anaerobic growth can be sustained by anaerobic respiration in the presence of nitrate as electron acceptor or by mixed-acid fermentation of carbohydrates associated with the formate-hydrogen lyase system. Each condition/pathway requires the synthesis of a specific FDH.
-
(1)
Cytoplasmatic FDH of the formate-hydrogen lyase system, denominated formate dehydrogenase H (FDH-H) [24]. The E. coli formate-hydrogen lyase is a membrane-bound system involved in formate oxidation and molecular hydrogen formation under fermentative growth conditions [25, 26]. The system comprises two enzymes, a cytoplasmatic molybdenum-containing FDH and a membrane-bound, cytoplasmically oriented nickel/iron-containing hydrogenase (hydrogenase 3 or 4, depending on growth conditions [25]), and, probably, also small electron-transfer proteins. FDH oxidises formate to carbon dioxide and the resulting reducing equivalents are transferred, probably, through a ferredoxin-like protein, to the hydrogenase that reduces protons to molecular hydrogen (Eq. 1 → 2). The molecular hydrogen, thus formed in the cytoplasm, could be recycled (oxidised to protons) by the periplasmically oriented hydrogenases 1 and 2 and the electrons introduced in the membrane quinone pool [27–29]; this would suggest a hypothetical proton-translocating function for this system, which must wait for experimental confirmation.
-
(2)
Membrane-bound, periplasmically oriented FDH of the anaerobic nitrate–formate respiratory pathway, denominated formate dehydrogenase N (FDH-N). Under anaerobic conditions and in the presence of nitrate, E. coli co-expresses the FDH-N with the respiratory membrane-bound cytoplasmically oriented nitrate reductase NarGHI (both molybdenum-dependent) to form a supermolecular formate:nitrate oxidoreductase system [30–33]. Through this system, the periplasmatic FDH-N-catalysed formate oxidation is coupled to the cytoplasmatic nitrate reduction (Eq. 1→3), via the membrane menaquinone pool that mediates the electron transfer between the two enzymes. In this way, the FDH-mediated proton generation in the periplasm is coupled to the proton consumption in the cytoplasm (translocation of protons from cytoplasm to periplasm) and a proton motive force is generated [21, 32, 34–36].
-
(3)
In addition, under aerobiosis and in the presence of nitrate, E. coli is also able to express (although at low levels) a second membrane-bound periplasmically oriented FDH, denominated formate dehydrogenase O (FDH-O). This FDH is co-expressed with the nitrate reductase NarZWV and both enzymes participate in a nitrate–formate respiratory pathway that is suggested to operate in a similar way to the anaerobic system described above [37–39]. However, contrary to the FDH-N/NarGHI system that is expressed only in anaerobiosis, this system is also expressed in the presence of dioxygen, because it is not under the regulation of the FNR and ArcA (which control the genes for cellular anaerobic and aerobic functions, respectively) [27, 39, 40]. This differentiated regulation allows the bacterium to have some nitrate reductase activity under aerobic conditions, which facilitates the rapid adaptation during the transition from aerobic to anaerobic growth.
$$2{\text{H}}^{ + } + \, 2{\text{e}}^{-} \to {\text{H}}_{2}$$(2)$${\text{NO}}_{3}^{ - } + \, 2{\text{H}}^{ + } + \, 2{\text{e}}^{-} \to {\text{NO}}_{2}^{ - } + {\text{ H}}_{2} {\text{O}}$$(3)
-
(1)
-
(b)
Wolinella succinogenes constitutes a similar example, as it also expresses two membrane-bound periplasmically oriented FDH, but, in this case, the FDHs are suggested to be involved in the periplasmatic dissimilatory nitrite reduction to ammonium (Eq. 1→4) catalysed by cytochrome c-containing nitrite reductase [41–47]. In W. succinogenes, are the electrons provided by FDH to the membrane quinone pool that contributes to the proton motive force generation [46].
$${\text{NO}}_{ 2}^{ - } + {\text{ 8H}}^{ + } + {\text{ 6e}}^{ - } \to {\text{NH}}_{ 4}^{ + } + {\text{ 2H}}_{ 2} {\text{O}}$$(4) -
(c)
In Desulfovibrio species, periplasmatic FDHs are suggested to provide electrons not only to dissimilatory sulfate reduction (organisms grown on sulfate as energy source) [48–53], but also to dissimilatory nitrate reduction to ammonium (in those species able to grow on nitrate, D. desulfuricans and D. vulgaris) [33, 43], as well as to other alternative energy source pathways [54]. D. desulfuricans, in particular, in the presence of nitrate expresses a complex enzymatic system containing three different molybdenum-containing enzymes, a periplasmatic nitrate reductase, an aldehyde oxidoreductase and a FDH [44]. D. vulgaris, on the other hand, is also able to produce FDH-dependent formate when grown on molecular hydrogen and carbon dioxide, which allows the bacterium to reversibly store reducing power [53]. The electrons provided by Desulfovibrio FDH are transferred to a network of periplasmatic c-type cytochromes, which, in turn, are used to feed (reduce) several alternative respiratory pathways [55]. Nevertheless, how these systems generate a proton motive force remains a matter of debate [56–58].
-
(d)
FDHs are also involved in methane oxidation pathways. In methylotrophic bacteria, a NAD-dependent FDH (NAD-FDH) is employed as a terminal enzyme in the oxidative pathways that make use of C1 compounds as sole sources of carbon and energy and, in obligate methanotrophs, NAD-FDH is also involved in NADH regeneration (Eq. 5→8) [59, 60]. In this way, FDHs are involved in the generation of reducing equivalents. Formate can also be used in other aerobic pathways. For example, the facultative chemoautotrophic, hydrogen-oxidising, Ralstonia eutropha can grow aerobically using formate as an alternative energy source [61, 62]. R. eutropha expresses two FDHs, a membrane-bound enzyme involved in a respiratory chain [61] and a cytoplasmatic NAD-FDH that couples the formate oxidation to the reduction of NAD+ [63].
$${\text{CH}}_{4} + {\text{ NADH }} + {\text{ O}}_{2} + {\text{ H}}^{ + } \longrightarrow {\text{CH}}_{3} {\text{OH}} + {\text{ NAD}}^{ + } + {\text{ H}}_{2} {\text{O}}$$(5)$${\text{CH}}_{3} {\text{OH}} + {\text{ Q}} \longrightarrow {\text{HCOH}} + {\text{ QH}}_{2}$$(6)$${\text{HCOH}} + {\text{ NAD}}^{ + } + {\text{H}}_{2} {\text{O}} \longrightarrow {\text{HCOO}}^{ - } + {\text{ NADH }} + \, 2{\text{H}}^{ + }$$(7) (8)
(8) -
(e)
In turn, methanogenic organisms use a coenzyme F420-dependent FDH to oxidise formate; the reduced F420 is subsequently used in two steps of the pathway of carbon dioxide reduction to methane [18, 64, 65]. In these organisms, F420-dependent FDH and F420-reducing hydrogenase form a formate-hydrogen lyase system. In addition, a protein system was identified in the hydrogenotrophic methanogen Methanococcus maripaludis that comprises heterodisulfide reductase, formylmethanofuran dehydrogenase, F420-nonreducing hydrogenase and formate dehydrogenase [66, 67]. Formate can donate electrons to the heterodisulfide reductase via FDH (as can dihydrogen via F420-nonreducing hydrogenase). Thus, formate can be used as electron donor for methanogenesis, through heterodisulfide reductase, when hydrogen is limited.
-
(f)
Acetogens use FDH in a carbon dioxide fixation metabolic pathway that forms acetate. In these anaerobic organisms, the first step of the energy metabolism is the FDH-catalysed carbon dioxide reduction to formate, using often dihydrogen as ultimate physiological reductant [68–71]. This is the case, e.g. of Clostridium carboxidivorans that can grow autotrophically with carbon dioxide and dihydrogen as carbon and energy sources, using a NAD-FDH [72–75]. The Acetobacterium woodii constitutes another example [76]. This acetogenic bacterium was recently described to hold a hydrogen-dependent carbon dioxide reductase complex that directly uses dihydrogen for the reduction of carbon dioxide to formate, without the intervention of NAD(P)H or an external electron-transfer protein. A. woodii, as well as other acetogens, lives close to the thermodynamic limit of life, depending on several thermodynamically unfavourable reactions, and the direct use of dihydrogen for carbon dioxide reduction would be beneficial for the organism [76, 77].
-
(g)
In syntrophic cultures of acetogenic bacteria and hydrogenotrophic methanogenic archaea [78, 79], the dihydrogen and formate produced by acetogens are consumed by the methanogens [80–84]. This interspecies dihydrogen/formate transfer is crucial for acetogens, that depend on methanogens to scavenge those metabolites, and for methanogens that use them in their own metabolism, in the conversion of biological polymers to methane and carbon dioxide [85, 86]. Syntrophobacter fumaroxidans, e.g. can grow on propionate, oxidising it to acetate and using the reducing equivalents to reduce protons to dihydrogen or carbon dioxide to formate. During syntrophic growth with Methanospirillum hungatei, S. fumaroxidans optimises its metabolism by transferring formate and dihydrogen to the syntrophic partner that acts as a formate and dihydrogen scavenger [87, 88]. In this way, formate acts as a mediator of reducing equivalents between the two organisms, and both depend on FDH to reduce carbon dioxide (to yield formate) and oxidise formate.
-
(h)
Formate oxidation can also be coupled to the dissimilatory reduction of Fe3+ or Mn4+ in, e.g. Geobacter or Shewanella [89, 90] or to the dissimilatory arsenate reduction in Desulfitobacterium [91].
Formate dehydrogenases
Classes of formate dehydrogenases
FDH can be divided into two major classes, based on their metal content/structure and consequent catalytic strategies. One class, the metal-independent FDH class, comprises the NAD+-dependent FDH belonging to the D-specific dehydrogenases of the 2-oxyacid family [92–94]. These enzymes are more widespread, being found in aerobic bacteria, yeasts, fungi and plants. Because these enzymes have no redox cofactors or metal ions, the formate oxidation to carbon dioxide has been suggested to involve the direct hydride transfer from formate to NAD+ (Eq. 9). The protein role would be to position formate and NAD+ in proximity to each other and making NAD+ acquiring a bipolar form during the reaction transition state; this conformational change would increase the NAD+ electrophilicity and facilitate the rate-limiting hydride transfer [95–102]. (Because this review is focused on the molybdenum- and tungsten-containing FDH, these metal-independent enzymes will not be further discussed.)
The other class, the metal-containing FDH class, comprises only prokaryotic FDH belonging to the molybdenum and tungsten-containing enzymes’ families and will be herein discussed in detail (unless where explicitly indicated, the abbreviation “FDH” will henceforth only refer to metal-containing enzymes).
This class of FDH is composed by complex proteins, that hold different redox cofactors, and whose active site harbours one molybdenum or tungsten atom that mediates the formate oxidation. Accordingly, metal-containing FDH can be sub-divided as molybdenum-containing FDH (Mo-FDH) and tungsten-containing FDH (W-FDH). In contrast to the first class, in metal-containing FDH, the transfer of protons and electrons (Eq. 1) is mediated by the active site (i.e. there is no direct proton/electrons transfer between formate and the physiological electron acceptor). Noteworthy, this class also comprises NAD+-dependent FDH (NAD-FDH). However, contrary to the metal-independent enzymes, these FDHs have a molybdenum or tungsten atom in their active site, where formate oxidation takes place and which mediates the proton/electrons transfer, and these enzymes use NAD+ only as the terminal electron acceptor (co-substrate).
Molybdenum and tungsten-containing enzymes’ families
Molybdenum is essential to most organisms, from bacteria to man, being part of the active site of enzymes that catalyse important redox reactions of the metabolism of carbon, nitrogen, and sulfur, many of which constitute critical steps in the global biogeochemical cycles of those elements [103–108]. Tungsten, possibly because of its different and limited bioavailability [109], is, by far, less used by living organisms, being found in anaerobic prokaryotes, most of which are thermophiles [110–113].
With the exception of [MoFe7S9] cofactor of nitrogenase (see both Hu and Ribbe and Bjornsson, Neese, Schrock, Einsle and DeBeer contributions in this JBIC issue), Mo–S–Cu cofactor of carbon monoxide dehydrogenaseFootnote 1 (see Hille et al. contribution in this issue) and a few other heteronuclear centres, whose physiological function is not yet fully understood [114–117], molybdenum is found in the active site in a mononuclear form, hereafter designated only as molybdenum centre. In these centres, one molybdenum atom is coordinated by the cis-dithiolene group of one or two pyranopterin cofactor molecules (Fig. 1) and by oxygen, sulfur, or selenium atoms, in a diversity of arrangements that determinates the classification of molybdoenzymes into three big families [103], xanthine oxidase, sulfite oxidase and dimethylsulfoxide reductase (DMSOR) families (Fig. 1). The DMSOR family is the larger and more diverse family, comprising prokaryotic enzymes of different functions and structures (subunit composition and nature and number of redox cofactors), such as DMSOR itself, Mo-FDH, dissimilatory and assimilatory nitrate reductases, among many others. The DMSOR family enzymes (in oxidised form) hold a trigonal prismatic L2MoXY core, where L stands for the pyranopterin cofactor and X and Y represent terminal =O, –OH, =S, and –SH groups and/or oxygen, sulfur or selenium atoms from cysteine, selenocysteine, serine or aspartate residue side chains (Fig. 1) [118].
Active site structures of the molybdenum- and tungsten-containing enzymes. a Structure of the pyranopterin cofactor. The cofactor is a pyranopterin-dithiolate moiety, which forms a five-membered ene-1,2-dithiolate chelate ring with the molybdenum atom; in eukaryotes, the cofactor is found in the simplest monophosphate form (R is an hydrogen atom), while in prokaryotes it is found esterificated with several nucleotides (R can be one cytidine monophosphate, guanosine monophosphate, or adenosine monophosphate). b Structures of the molybdenum centres of the three families of molybdoenzymes and of the tungsten centres of tungstoenzymes; for simplicity, only the dithiolate moiety of the pyranopterin cofactor is represented. The images were produced with Accelrys Draw 4.0 (Accelrys Software Inc.)
Tungsten is found in the active site of the enzymes also in a mononuclear form (herein denominated tungsten centre), where it is coordinated by the cis-dithiolene group of two molecules of the same pyranopterin cofactor found on molybdoenzymes (Fig. 1) [103–108]. The tungsten coordination sphere is completed with oxygen and/or sulfur atoms from terminal groups or from amino acid residue side chains, in the same trigonal prismatic geometry found in DMSOR family members. The tungstoenzymes can be grouped in a single family that comprises all the tungsten-pyranopterin-containing enzymes, including the W-FDH and aldehyde:ferredoxin oxidoreductases [104, 110, 119]. Some authors classify the aldehyde:ferredoxin oxidoreductases separately, in a distinct family, and include the other tungsten-containing enzymes (also the W-FDH) in the DMSOR family. We suggest that a systematic organisation, based on the metal/cofactor structure, should be followed and all tungsten-pyranopterin-containing enzymes should be grouped together. This tungstoenzymes’ family can be, subsequently, sub-divided to account for differences between members, as is presently done with the families of molybdoenzymes. When the aim is to highlight the structural/functional similarities between tungstoenzymes and DMSOR family enzymes, as it is the case with the homologous W-FDH and Mo-FDH, both families could be gathered under a unique super-family denominated “molybdenum/tungsten-bis pyranopterin guanosine dinucleotide-containing enzymes”, with the acronym Mo/W-bis PGD, as suggested by others (Fig. 1) [26].
In general, the enzymes of all families catalyse the transfer of an oxygen atom from water to the product or from the substrate to water, in reactions that imply a net exchange of two electrons and in which the molybdenum or tungsten cycles between Mo6+ and Mo4+ or W6+ and W4+ [103–105, 120–125]. It is based on this catalytic feature that these enzymes are commonly, but inaccurately, referred to as oxotransferases. One important exception is precisely the FDH-catalysed formate oxidation to carbon dioxide, which does not involve any oxygen atom transfer (Eq. 1). In particular, regarding the Mo/W-bis PGD super-family enzymes, they catalyse diverse reactions, including (1) proper transfer of oxygen atom (e.g. nitrate reduction by nitrate reductases), (2) cleavage of C–H bonds (e.g. formate oxidation by Mo-FDH and W-FDH), (3) transfer of sulfur atom (e.g. inorganic sulfur reduction to sulfide by polysulfide reductase), (4) simultaneous oxidation and reduction (e.g. reductive dehydroxylation and concomitant oxidative hydroxylation by pyrogallol:phloroglucinol hydroxyltransferase) and (5) even hydration reactions (e.g. hydration of acetylene to acetaldehyde (a non-redox reaction) by acetylene hydratase) [36, 118, 126, 127]. The electrons derived from, or necessary to carry out, these reactions are intramolecularly transferred to the electron acceptor, or from the electron donor, through different redox centres, such as iron–sulfur centres (Fe/S), haems and flavins.
Molybdenum and tungsten-containing formate dehydrogenases: enzymatic machineries
Prokaryotic FDHs are involved in different biochemical pathways (as described above) and each pathway requires a specific enzymatic “machinery” to accomplish the respective function. There are membrane-bound enzymes that must be “anchored” to the membrane and interact with membrane-associated electron acceptors; but there are also cytoplasmatic and periplasmatic enzymes that, instead, need an appropriate “interface” to use cytochromes, ferredoxins, NAD or coenzyme F420 (an obligate two-electron acceptor, flavin derivative) as electron acceptors. As a result, FDHs are a group of heterogeneous proteins, displaying diverse redox centres, subunit compositions and quaternary structures.
The FDH enzymatic “machineries” will be described here using some representative enzymes as models, giving particular emphasis to the enzymes whose 3D structure is known, but also describing, as much as possible, each of different “types” of enzyme. A summary is provided in Table 1.
E. coli formate dehydrogenase H (Mo-FDH)
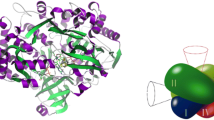
E. coli FDH-H (product of the fdhF gene) is a monomeric molybdenum-containing enzyme (≈80 kDa), folded into four domains, that contains one [4Fe–4S] centre and one molybdenum centre (Fig. 2) [128–131]. The molybdenum centre is the active site, where formate is oxidised, and the Fe/S centre is responsible for the subsequent intramolecular electron transfer to the physiological acceptor, probably a ferredoxin protein. The Fe/S centre is bound by the N-terminal domain, just below the protein surface and adjacent to the tetrahydropterin-like Q (proximal) pyranopterin of the molybdenum centre. The molybdenum centre, bound mainly through the other three domains, holds the molybdenum atom coordinated by the four sulfur atoms of two pyranopterin guanosine dinucleotide cofactor (PGD) molecules, characteristic of these Mo/W-bis PGD enzymes.
E coli formate dehydrogenase H. a Three-dimensional structure view. b Arrangement of the two redox centres shown in the same orientation (but not same scale) as in (a). c Molybdenum catalytic centre of oxidised enzyme as described in [131]. d Molybdenum catalytic centre of oxidised enzyme complexed with the inhibitor nitrite as described in [131]. e Molybdenum catalytic centre of reduced enzyme as described in [131]. f Molybdenum catalytic centre of reduced enzyme as described in [132]. The structures shown are based on the PDB files 1FDI (a, b, d), 1FDO (c), 1AA6 (e) and 2IV2 (f). α helices and β sheets are shown in red and cyan, respectively (a); pyranopterin cofactor c–f is represented in dark red. The images were produced with Accelrys DS Visualizer, Accelrys Software Inc.
The molybdenum coordination sphere, in the oxidised state, was initially suggested to be completed by a conserved essential selenocysteine residue, SeCys140, and a hydroxyl group, in a trigonal prismatic coordination geometry (Fig. 2c) [131]. After FDH-H reduction with formate, the molybdenum centre was described to loose the sixth ligand and acquire an approximate square pyramidal geometry, with the bis PGD sulfur atoms as the four equatorial ligands and the SeCys140 selenium atom as the axial ligand (Fig. 2e) [131]. However, a subsequent reinterpretation [132], revealed that the sixth ligand would be a sulfur and not an oxygen atom. Moreover, although the global original structure was corroborated, it was observed that the loop containing the SeCys140 was in a different conformation and, most important, shifted away (9 Å) from the molybdenum centre in the formate-reduced FDH. Accordingly, while this residue would be coordinated to the molybdenum in the oxidised state, it would be no longer bound after enzyme reduction. As a result, in the reduced state, the molybdenum centre is presently being thought as a square pyramid, with the bis PGD sulfur atoms as the four equatorial ligands and a terminal sulfur atom as the axial ligand (Fig. 2f) [132]. Notably, the recent identification of a sulfurtransferase, which would insert the sulfur atom into the FDH molybdenum centre [133], supports that all FDH could have a terminal sulfur atom. The active site comprises, in addition to the molybdenum centre, arginine (Arg333) and histidine (His141) residues that are strictly conserved (Fig. 2c–f). A deep crevice constitutes the substrate-binding pocket; at its bottom, the Arg333 provides both a positive charge and a critical hydrogen bond for orienting and binding the substrate at the active site.
E. coli formate dehydrogenase N (Mo-FDH)
E. coli FDH-N (product of the fdnGHI operon) is a complex trimer of trimers, (αβγ)3 (≈510 kDa) that harbours two b haems, five [4Fe–4S] centres and one molybdenum centre (Fig. 3) [32, 134]. Notably, the overall organisation of the αβγ unit is similar to the one of respiratory nitrate reductase NarGHI,Footnote 2 although the two enzymes have the catalytic subunits on opposite sides of membrane (NarGHI is a cytoplasmically oriented and FDH-N periplasmically oriented enzyme [31, 32, 138–140]). The FDH-N αβγ unit is constituted by (Fig. 3a–c) [32]: (1) a periplasmatic formate-oxidising α subunit (product of fdnG gene, ≈115 kDa), that holds one molybdenum centre and one [4Fe–4S] centre (labelled FS0); this subunit is folded into five domains and, notably, the four domains involved in the Fe/S and molybdenum centres binding are structurally similar to the FDH-H monomer; (2) a periplasmatic electron-transfer β subunit (product of fdnH, ≈35 kDa) that harbours four [4Fe–4S] centres (labelled FS1 to FS4); (3) and a membrane-bound menaquinone-reducing γ subunit (product of fdnI, ≈20 kDa) that holds two b-type bis-histidinyl-coordinated haems (named b P and b C, for haem closer to the periplasm side and haem closer to the cytoplasm side, respectively). The trimer of αβγ units, (αβγ)3, is very tightly packed and a cardiolipin molecule is maintained at the trimer interface, thus suggesting that this complex arrangement is physiologically meaningful.
E. coli formate dehydrogenase N. a, b Three-dimensional structure view of the αβγ unit coloured according to structural motifs (α helices and β sheets are shown in red and cyan, respectively) and subunits. (α, green, β, violet, γ, dark blue). c Arrangement of the redox centres shown in the same orientation (but not same scale) as in (a) and (b) (see text for details). d Molybdenum catalytic centre (pyranopterin cofactor is represented in dark red). The structures shown are based on the PDB file 1KQF [32]. The images were produced with Accelrys DS Visualizer, Accelrys Software Inc.
As expected the molybdenum centre is the active site where formate is oxidised. The Fe/S centres and haems form a “wire” that facilitates the fast and effective electron transfer from the active site to the physiological electron acceptor, menaquinone, against the membrane potential and across a ≈90 Å distance (from the α to the γ subunit). In detail, the electrons transferred to the molybdenum centre during formate oxidation are, subsequently, transferred to the β subunit though the α subunit FS0. In the β subunit, the electrons flow through FS1 → FS4 → FS2 → FS3 and are eventually transferred to the b P haem in the γ subunit and, then, across the membrane to b C haem. Finally, menaquinone binds to the b C haem histidine ligand, Hisγ169, from which it can directly accept electrons.
The active site holds the molybdenum atom coordinated by the four sulfur atoms of two PGD molecules and one selenium atom from the SeCysα196. A sixth ligand, initially modelled as a hydroxyl group [32], but presently believed to be a sulfur atom (as described above for FDH-H [133] ), completes the molybdenum coordination sphere, in trigonal prismatic geometry (Fig. 3d). In addition, the active site harbours also the conserved arginine (Argα446) and histidine (Hisα197) residues. The FDH-N Hisα197 was found to be in an orientation, with its Nδ1 pointing towards the substrate-binding site, that supports its direct involvement in the abstraction of the formate Cα proton, during catalysis [32]. Overall, the FDH-N active site is suggested to be rather similar to the FDH-H one.
E. coli formate dehydrogenase O (Mo-FDH)
The E. coli FDH-O (product of the fdoGHI operon) 3D structure is not known, but this enzyme is believed to be similar to the “anaerobic” homologous FDH-N. FDH-O is also a membrane-bound heterotrimer constituted by two periplasmatic subunits (αβ, 107, 34 kDa), associated with a third integral membrane subunit (γ, 22 kDa) [38, 141]. Genome analysis revealed a remarkable 75 % sequence identity in the α subunit, including the conserved active site residues (selenocysteine, histidine and arginine); 76 % sequence identity in the β subunit, where the iron–sulfur binding region signature was found to be 100 % identical; and a more modest 45 % sequence identity in the γ subunit [142].
D. gigas formate dehydrogenase (W-FDH)
D. gigas FDH (product of fdhA and fdhB genes) is a periplasmatic, heterodimeric (αβ; ≈135 kDa) tungsten-containing enzyme that harbours four [4Fe–4S] centres and one tungsten centre (Fig. 4) [143–145]. The tungsten centre is the active site, as anticipated, and the Fe/S centres are responsible for the subsequent intramolecular electron transfer to the physiological acceptor, periplasmatic c-type cytochromes. The β subunit (≈24 kDa), located opposite from the cleft leading to the active site, is wrapped by the N-terminal residues of the α subunit, which contributes to the stability of the dimer (Fig. 4a). This subunit is folded into two domains, one containing one Fe/S and the other two Fe/S centres (total of three). Interestingly, a similar arrangement is found the FDH-N β subunit, although in FDH-N both domains contain two Fe/S centres each (total of four). The α subunit (≈110 kDa), folded into four domains, is homologous to the E. coli FDH-H and contains one Fe/S centre (bound by the N-terminal domain) and the tungsten centre (bound mainly through the other three domains).
D. gigas formate dehydrogenase. a, b Three-dimensional structure view of the heterodimeric enzyme (αβ) coloured according to structural motifs (α helices and β sheets are shown in red and cyan, respectively) and subunits (α, green, β, violet); the Cysα817 and Cysα844 are represented by grey spheres (see text for details). c Arrangement of the redox centres shown in the same orientation (but not same scale) as in a and b. d Tungsten catalytic centre (pyranopterin cofactor is represented in dark red). The structures shown are based on the PDB file 1H0H [145]. The images were produced with Accelrys DS Visualizer, Accelrys Software Inc.
The tungsten centre, buried at the interior of the protein, holds the tungsten atom coordinated by the four sulfur atoms of two PGD molecules and one selenium atom of SeCysα158 and by one sulfur atom, in distorted octahedral geometry (Fig. 4d). The conserved histidine and arginine residues (Hisα159 and Argα407) complete the active site pocket. The active site is accessible through a positively charged tunnel, with the arginine residue at its bottom, as in FDH-H. Carbon dioxide release may be facilitated through a hydrophobic channel.
Noteworthy, D. gigas FDH displays one disulfide bridge, Cysα817–Cysα844 (Fig. 4a, b). This disulfide bridge is located close to one of the PGD molecules, on the surface of the protein (thus, accessible to reducers/oxidisers). Its reduction would allow wider opening and movement of the formate entry cleft and would alter the position of residues 818–848. The segment 818–843 is absent in FDH-H, but topologically, those two cysteine residues would correspond to the FDH-H Asp567 and Leu569. Because the D. gigas FDH was described to be activated by 2-mercaptoethanol, it was suggested that it is this disulfide bridge reduction that is needed to activate the enzyme [145]. Furthermore, it was hypothesised that the disulfide bridge could have evolved as a regulatory strategy to tune the FDH activity under mildly oxidised environments, in contrast to FDH-H that is permanently inactivated by oxygen.
D. desulfuricans formate dehydrogenase (Mo-FDH)
D. desulfuricans FDH (product of fdhABC3 operon) is also a periplasmatic enzyme, but its composition is quite different from the D. gigas protein. D. desulfuricans FDH is a heterotrimeric (αβγ; ≈135 kDa) molybdenum-containing enzyme, containing four c haems, two [4Fe–4S] centres and one molybdenum centre [44, 146]. The α subunit (≈90 kDa) harbours the molybdenum centre and one Fe/S centre, while the β (≈30 kDa) and γ (≈15 kDa) subunits hold one Fe/S centres and four c haems, respectively.
D. alaskensis formate dehydrogenases (Mo/W-FDH, W-FDH)
Genome analysis of the D. alaskensis bacterium revealed the presence of three FDHs [147]. Two of them were characterised and one was found to be a W-FDH (product of W-fdh genes), while the second was shown to incorporate either molybdenum or tungsten (Mo/W-FDH; product of Mo/W-fdh genes), thus being the first FDH known to be active with both metals [147, 148]. The Mo/W-FDH enzyme, in spite of incorporating tungsten, should preferentially harbour molybdenum, as is suggested by the slight upregulation of the Mo/W-fdh genes, when the bacterium is grown under molybdenum supplementation, and strong downregulation under tungsten supplementation [147]. The W-FDH is a dimer of heterodimers [(αβ)2; ≈110 and 30 kDa], while the Mo/W-FDH was purified as an heterodimer (αβ; ≈110 and 30 kDa); both enzymes have the characteristic PGD cofactor and several Fe/S centres [147, 148].
D. vulgaris formate dehydrogenases (Mo/W-FDH, Mo-FDH)
D. vulgaris genome analysis also suggested the presence of three periplasmatic FDHs [149]. The characterised D. vulgaris FDH is a heterotrimeric molybdoenzyme (≈84, 27 and 14 kDa) homologous to the D. desulfuricans molybdoenzyme [43, 150]. Notably, D. vulgaris also expresses a dimeric FDH that can incorporate both molybdenum and tungsten [151].
In D. desulfuricans and D. vulgaris enzymes, formate oxidation occurs at the molybdenum/tungsten centre and the other redox centres are involved in the electron transfer to the physiological acceptor, mono-haemic c-type cytochromes [43, 44, 50, 152]. For none of these three Desulfovibrio enzymes is the 3D structure known.
Ralstonia eutropha NAD+-dependent formate dehydrogenase (Mo-FDH)
The R. eutropha NAD-FDH (product of the fdsGBACD operon) 3D structure is presently not known. This NAD-dependent enzyme is a cytoplasmatic heteromultimeric (αβγδ or αβγ) molybdenum-containing enzyme that harbours one FMN, seven Fe/S centres and one molybdenum centre [63, 153, 154]. As anticipated from the presence of molybdenum, the α subunit (≈105 kDa) has no similarity with the metal-independent NAD+-dependent FDH. Instead, it shows ≈55 % sequence similarity with the α (formate oxidising) subunits of NAD+-independent Mo-FDH and W-FDH, indicating that this subunit would catalyse the formate oxidation [154]. The chemical identification of PGD and inhibition by cyanide [63] further suggest that the molybdenum atom would be coordinated by the two characteristic PGD cofactor molecules and by a terminal sulfur atom. Although the possible conserved histidine and arginine residues (His379 and Arg579) were identified, the position corresponding to the conserved selenocysteine was occupied by a cysteine residue (Cys378) in R. eutropha, which was suggested to contribute to the air stability of this molybdenum centre [154]. The N-terminal segment of this subunit, absent in NAD+-independent FDH, exhibits considerable similarity with NuoG of the E. coli NADH:ubiquinone oxidoreductase, as well as to NAD(P)+-reducing hydrogenases and iron-containing hydrogenases, suggesting that it probably harbours one [2Fe–2S] and three [4Fe–4S] centres. The overall identified conserved cysteine and histidine residues suggest that the α subunit would contain a total of two [2Fe–2S] and three [4Fe–4S] centres, besides the molybdenum centre [154]. The β subunit (≈55 kDa) displayed no significant sequence similarity to other FDH subunits. However, it shows ≈40 % identity to the NAD+-reducing (α) subunit of hydrogenase and to the E. coli NuoF, both of which have NADH dehydrogenase activity and contain a NAD+ binding site, one FMN and one [4Fe–4S] centre [154]. This similarity suggests that the R. eutropha FDH β subunit is involved in NAD+ reduction and that it may contain a fourth [4Fe–4S] centre and one FMN. The γ-subunit (≈20 kDa) is closely related (≈30 % sequence identity) to E. coli NuoE, and was suggested to harbour a third [2Fe–2S] centre [154]. Concerning the δ subunit (≈15 kDa; product of FdsD), no significant similarity to any known protein was described. Since it is, apparently, devoid of redox cofactors, this subunit would not participate in the intramolecular electron transfer. Instead, it was suggested that it may play a role in maintaining the FDH quaternary structure [154], or, most probably, be involved in the FDH maturation.
Similar to the other FDH, the NAD-FDH molybdenum centre should be the active site responsible for formate oxidation, with the Fe/S centres forming a “wire” that transfers the electrons to FMN, and, ultimately to NAD+, probably through α → γ → β subunits [154] The β and γ subunits, together with the N-terminal extension of the α subunit, form a functional NADH dehydrogenase entity, parallel to the one of the respiratory chain complex I (NADH:ubiquinone oxidoreductase) or the NAD(P)-dependent hydrogenases [155]. Based on the sequence similarities described, Hille et al. proposed the 3D structural model for R. eutropha schematised in Fig. 5 [118, 156, 157].
A model for the structure of the R. eutropha FDH, as described by Hille et al. [171]. The model was obtained by superimposing of the FS0 [4Fe–4S] centre of E. coli FDH-H (PDB 1AA6) with the N7 [4Fe–4S] centre of the T. thermophilus NADH dehydrogenase Nqo3 subunit (PDB 3IAM), with the Nqo1 and Nqo2 subunits (which have strong homologies to FdsB and G, respectively) included in the model. See [171] for details. a Three-dimensional structure view of the model. b Arrangement of the redox centres in the model, with the approximate position of the additional R. eutropha Fe/S centre indicated by the orange ball. Red arrow indicates the direction of electron transfer in the R. eutropha FDH; blue arrow indicates the direction of electron transfer in NADH dehydrogenase. Reproduced with permission from reference 175. Copyright 2014 American Chemical Society
Other NAD+-dependent formate dehydrogenases (Mo-FDH and W-FDH)
The utilisation of molybdenum in a FDH dependent on NAD by an organism that can grow aerobically, may seem like an exception, but several of these stable oxygen-tolerant enzymes have been identified.
The molybdenum-containing/NAD-dependent FDH (Mo/NAD-FDH) from the obligate methanotroph Methylosinus trichosporium was found to be a complex heteromultimer, (αβγδ)2 (≈400 kDa), containing FMN, several Fe/S centres (at least one [2Fe–2S] and four [4Fe–4S]) and a molybdenum centre [156]. Facultative methylotrophic bacteria, Pseudomonas sp. 101 [158], Mycobacterium vaccae [159], Methylobacterium sp. RXM [160, 161], as well as Cupriavidus oxalaticus (Pseudomonas oxalaticus) [162–164] were also suggested to hold complex multimeric Mo/NAD-FDH, containing several Fe/S centres and flavin.
The Mo/NAD-FDH from Rhodobacter capsulatus, in particular, was recently characterised [165]. This enzyme (product of the fdsGBACD operon) is a cytoplasmatic dimer of heterotrimers, (αβγ)2 (≈345 kDa), holding a molybdenum centre, one [2Fe–2S] and four [4Fe–4S] centres in the α subunit (≈105 kDa); a second [2Fe–2S] centre is bound by the γ subunit (≈15 kDa), and a fifth [4Fe–4S] centre and one FMN are harboured by the β subunit (≈50 kDa). The products of the FdsC and FdsD genes are not subunits of the mature enzyme. Instead, they are suggested to be involved in molybdenum centre maturation and insertion into FDH (with FdsC protein being probably a sulfurtransferase similar to the E. coli one). The molybdenum centre, responsible for formate oxidation, holds the molybdenum atom coordinated by two PGD cofactor molecules and a cysteine ligand. The β and γ subunits display reversible NADH dehydrogenase activity (both NAD+ reduction and NADH oxidation).
Notably, there are also tungsten-containing/NAD(P)-dependent FDHs (W/NAD-FDH). This is the case not only of the acetogenic Moorella thermoacetica (Clostridium thermoaceticum) [166, 167], C. formicoaceticum [168, 169] or C. carboxidivorans [170], but also of the methylotrophic Methylobacterium extorquens enzymes [164]. One of the FDHs of the aerobic M. extorquens [172] is an heterodimeric (αβ) W/NAD-FDH (product of the fdh1A and fdh1B genes) [164]. The α subunit (≈110 kDa) displays ≈35 % identity with E.coli FDH-H and was suggested to harbour at least one Fe/S centre and one tungsten centre, coordinated by two PGD molecules and a cysteine residue. The β subunit (≈60 kDa) contains the putative binding motifs for an Fe/S centre and FMN. The NADP-dependent W-FDH from M. thermoacetica is a dimer of a heterodimer, (αβ)2 (≈95 and 75 kDa), containing, besides the tungsten centre with a selenocysteine ligand, several Fe/S centres (at least two [2Fe–2Fe] and two [4Fe–4S]) [166, 167]. In fact, M. thermoacetica FDH was the first enzyme shown to have tungsten [173, 174]. The presence of W-FDH in strictly aerobic bacteria may indicate that tungstoenzymes are not restricted to anaerobic organisms and are probably more widespread than previously thought.
F420-dependent formate dehydrogenases (Mo-FDH and W-FDH)
F420-dependent FDHs are by far less well characterised. Methanobacterium formicicum F420-dependent FDH (product of the fdhCAB operon) is a dimeric (αβ; ≈85 and 55 kDa) molybdenum-containing enzyme that harbours FAD and several Fe/S centres; the enzyme was described to contain also zinc [175–180]. The molybdenum centre is coordinated by two PGD molecules, a sulfur atom (as suggested by cyanide inhibition), but no selenocysteine ligand. The anaerobe Methanococcus vannielii holds two F420-dependent FDH, a 105 kDa iron–molybdoenzyme, with no selenocysteine, and a high molecular mass iron–molybdo/tungstoenzyme, with a selenocysteine in which replacement of molybdenum with tungsten appears to occur [181, 182].
Hydrogen-dependent carbon dioxide reductase (Mo-FDH)
The acetogenic bacterium Acetobacterium woodii was recently described to hold a new type of FDH, a hydrogen-dependent carbon dioxide reductase complex, that catalyses the reduction of carbon dioxide to formate with the simultaneous and direct oxidation of dihydrogen [76]. The reductase complex (product of the gene cluster Awo_c08190-08260) is a tetramer (≈169 kDa) comprising one selenium-containing Mo-FDH (FdhF2), an iron–iron hydrogenase (HydA2) and two small electron-transfer subunits (HycB2 and HycB3) (Fig. 6). The reductase complex was described to contain also zinc and a total of eleven [4Fe–4S] centres [76]. Besides the hydrogen-dependent activity, the reductase complex also catalyses the reduction of carbon dioxide using reduced ferredoxin instead of dihydrogen as electron donor (Fig. 6).
A model for the hydrogen-dependent carbon dioxide reductase complex from A. woodii [76]. The reductase complex catalyses the carbon dioxide reduction at the FdhF2 subunit. The necessary electrons are provided by the hydrogenase subunit (HydA2), where dihydrogen oxidation takes place, and are delivered via the electron-transferring subunits HycB2 and HycB3. Alternatively, the electrons may be provided by reduced ferredoxin (Fd); ferredoxin can be reduced using, e.g. carbon monoxide and carbon monoxide dehydrogenase, also present in A. woodii. The eleven [4Fe–4S] centres are represented. Adapted from Ref. [76]. Reprinted with permission from AAAS
Molybdenum and tungsten-containing formate dehydrogenases: spectroscopic properties
Several spectroscopic methods, namely X-ray absorption fine structure (EXAFS), resonance Raman, electron nuclear double resonance (ENDOR) and electron paramagnetic resonance (EPR) spectroscopies, have been widely used to probe the metal redox centres of metalloproteins, providing valuable information regarding the structures and reactivity of those centres. FDHs have been particularly explored using XAS and EPR spectroscopy.
Initial molybdenum K-edge X-ray absorption spectroscopic studies on oxidised E. coli FDH-H pointed towards a molybdenum centre with a hydroxyl group and a selenocysteine coordinated with the molybdenum atom, as found by X-ray crystallography (Fig. 2c) [183], and similar results were obtained with the D. desulfuricans enzyme [184]. But selenium K-edge data obtained with dithionite-reduced FDH-H suggested the presence of an unexpected Se–S bond (2.2 Å), which is very interesting for the FDH reaction mechanism (as will be discussed below; Fig. 8b) [183]. The molybdenum centre structure of the formate-reduced E. coli FDH-H was also probed by EPR spectroscopy. EPR studies revealed an anisotropic, nearly axial Mo5+ signal, with a high g z value of 2.094, suggesting a nearly square–pyramidal geometry for that reduced molybdenum species (g y and g x of 2.001 and 1.990, respectively) [130, 185, 186]. Simultaneously, the high hyperfine coupling constant observed with 77Se-labelled enzyme (A z, A y and A x of 4.4, 25 and 80 × 10−4cm−1, respectively) suggested the direct selenocysteine coordination to the molybdenum atom [186]. Together, those EPR data suggested a global coordination, once more, similar to that initially determined by X-ray crystallography [131] (Fig. 2e), with an apical selenium atom (from the selenocysteine) and the four sulfur atoms of the two PGD molecules at the pyramid bottom.
EPR spectroscopy was also employed to probe the FDH reaction mechanism. Assays with 2H-labelled formate revealed that the hyperfine structure of the E. coli FDH-H molybdenum signal is due to the coupling with a solvent exchangeable proton derived from formate (A z, A y and A x of 2.5, 6.3 and 7.0 × 10−4cm−1, respectively) [186]. To convert formate into carbon dioxide, the formate Cα hydrogen has to be transferred to a proton acceptor in the enzyme active site (as will be discussed below). The hyperfine interaction observed demonstrated that the proton acceptor is located within magnetic contact to the molybdenum centre, but additional photolysis assays suggested that the selenium atom is not the proton acceptor.
Recent EPR studies with the D. desulfuricans FDH, however, demonstrated that the nearly axial signal, with the g z of 2.094, only arises in the presence of azide, a strong FDH inhibitor (which was employed as a protective additive during the E. coli FDH-H purifications) (Fig. 7) [146]. In the absence of inhibitors, formate-reduced D. desulfuricans FDH develops a rhombic, with small anisotropy, Mo5+ signal (g z, g y and g x of 2.012, 1.996 and 1.985, respectively). The D. desulfuricans Mo5+ signal showed the molybdenum hyperfine interaction with two protons, one not solvent exchangeable (with an A max of 11.7 × 10−4 cm−1 and A mid and A min non-detectable) and another solvent exchangeable (A max, A mid and A min of 7.7, 10 and 9.3 × 10−4 cm−1, respectively) (Fig. 7) [146]. These protons were assigned to the selenocysteine Cβ hydrogen atoms and to the molybdenum sulfhydryl (or hydroxyl) ligand, respectively.
Mo5+ EPR signals of D. desulfuricans FDH. Left panel experimental, acquired at 100 K (black lines), and simulated (grey lines) spectra. a Spectrum of as-prepared enzyme after 30 min reduction with formate. b The same as a, but with the enzyme exchanged into 2H-labelled buffer. c Azide-inhibited enzyme reduced with formate for ≈5 s. d The same as c, but reduced with 2H-labelled formate. e Simulation of the axial EPR signal obtained with E. coli FDH-H (g z = 2.094). Spectra were simulated with the following parameters (A values in ×10−4 cm−1; A H1 and A H2 represent the non-solvent and solvent exchangeable protons, respectively; g values position indicated by arrows): a g max = 2.012, g mid = 1.996, g min = 1.985, A H1max = 11.7, A H2max = 7.7, A H2mid = 10.0, A H2min = 9.3; b g max = 2.012, g mid = 1.996, g min = 1.984, A H1max = 11.7; c g max = 2.092, g mid = 2.000, g min = 1.989, A max = n.d, A mid = A min = 7.0; d g max = 2.092, g mid = 2.000, g min = 1.988; e g max = 2.094, g mid = 2.000, g min = 1.989, A max = 2.5, A mid = 6.3, A H2min = 7.0. Right panel experimental spectra, acquired at 100 K, of f FDH exchanged into 2H-labelled buffer reduced with formate and g as-prepared FDH reduced with 2H-labelled formate. Spectra given in black and grey lines correspond to ≈5 s and 15 min of incubation time, respectively. Adapted (with permission from Elsevier) from [146]
The EPR species detected in FDH, as well as in many other enzymes, probably do not arise from a true intermediate state of the catalytic cycle, but, instead, are “side-products” that accumulate under non-turnover conditions (as in the absence of external electron acceptors). Nevertheless, the information provided is extremely relevant for our knowledge about the structure of the redox centres of the enzyme, their reactivity and interaction. EPR spectroscopy has been often and successfully employed to determine which redox centre is closer to another and was also used to assign the proximal Fe/S centre to the PGD molecule in FDH from sulfate-reducing bacteria [148]. Moreover, and most importantly, EPR spectroscopy was essential to demonstrate the incorporation of either molybdenum or tungsten in D. alaskensis FDH [148], one of the few examples of an enzyme of the large Mo/W-bis PGD family that can incorporate both metal atoms and retain the activity.
Molybdenum and tungsten-containing formate dehydrogenases: mechanistic strategies for formate handling
FDH-catalysed formate oxidation occurs at the enzyme’s molybdenum or tungsten centre (Eq. 10). However this reaction is not an oxygen atom transfer reaction, as is characteristic of many Mo/W-bis PGD family enzymes: the reaction product is carbon dioxide and not hydrogen carbonate (Eq. 10 versus 11) [187]. In fact, oxidation of 13C-labelled formate in 18O-enriched water demonstrated clearly the formation of 13CO2 gas with no 18O atoms [186].


To form carbon dioxide (Eq. 10), FDH has to abstract one proton and two electrons from formate. To accomplish this, different reaction mechanisms have been proposed. Overall, the formate oxidation mechanism is believed to be similar in Mo-FDH and W-FDH. Molybdenum and tungsten have similar chemical properties and the Mo/W-FDH of Desulfovibrio alaskensis [147, 148] and of D. vulgaris [151], were shown to be active with either molybdenum or tungsten.
The first mechanism proposed was based on the structure initially described for E. coli FDH-H [131], where the molybdenum atom was assumed to be coordinated by a hydroxyl group, instead of a terminal sulfur atom (se above; Fig. 2c). The subsequent identification of a terminal sulfo group in the molybdenum coordination sphere and the observation that the loop containing the SeCys140 was shifted away (9 Å) from the molybdenum centre in the formate-reduced FDH (as described above; Fig. 2f) [132], led to a reformulation of that mechanism. Assuming that the sulfo ligand would have to remain bound to the molybdenum throughout the catalytic cycle (i.e. that it can not be released, as a hydroxyl group can), it was proposed that the selenocysteine ligand would be unbound to create a vacant position for formate to bind to molybdenum [132]. Accordingly, it was suggested that, when formate enters in the active site, it triggers the SeCys140 movement (Fig. 8a). After formate binding, through an oxygen atom, the now uncoordinated SeCys140, stabilised by interaction with the positively charged Arg333, would abstract the formate Cα hydrogen. The two electrons are transferred to molybdenum (Mo6+ → Mo4+) and the proton is subsequently transferred to His141. After carbon dioxide release, the molybdenum would end up in a penta-coordinated form, with the sulfo group as the fifth axial ligand, in accordance with the X-ray data. The catalytic cycle would be closed with the oxidation of Mo4+ to Mo6+, via intramolecular electron transfer to the Fe/S centre, and re-binding of the selenocysteine to the molybdenum centre. Theoretical calculations of the activation energy for C–H bond cleavage showed that the proton abstraction is much more favoured with an unbound selenocysteine (19 kcal/mol compared to 36 kcal/mol for bounded selenocysteine), thus supporting the suggested selenocysteine dissociation [188]. In this mechanistic proposal, the conserved histidine and arginine roles would be to facilitate the formate binding and the histidine residue would also act as the final proton acceptor; the molybdenum terminal sulfo ligand, on the contrary, would have no active role in formate oxidation.
More recently, a second mechanism was proposed based on theoretical calculations involving also the selenocysteine dissociation from the molybdenum, but, in this case, through a sulfur-shift [189, 190]. In this mechanism, it is proposed that the oxidised molybdenum, hexa-coordinated by the two PGD molecules plus the selenocysteine and the terminal sulfo group, is an inactive form (Fig. 8b). The enzyme would be activated only after formate reaches the active site. When formate enters in the active site, oriented by the positively charged arginine residue, the repulsive environment generated would trigger the insertion of the sulfur atom into the Mo–SeCys bond, to yield a Mo–S–SeCys moiety—that is, it would trigger a sulfur-shift. In this process, the molybdenum is formally reduced to Mo4+ and a new binding position is created that can, now, coordinate the formate molecule. Subsequently, the S–SeCys bond is cleaved and the formed selenol anion is stabilised by a hydrogen bond with the histidine residue. The selenol anion is, then, in position to abstract the formate Cα proton, to yield a “carbon dioxide moiety” coordinated in a bidentate mode to molybdenum, via the formate oxygen (Mo–O–Ccarbon dioxide) and molybdenum sulfur (Mo–S–Ccarbon dioxide) atoms (Fig. 8b). Carbon dioxide is eventually released, leaving a Mo4+=S centre. The catalytic cycle would be closed with the oxidation of Mo4+ to Mo6+, via intramolecular electron transfer to the Fe/S centre, and deprotonation of the selenocysteine residue. The molybdenum centre can, now, bind a new formate molecule and start a new catalytic cycle. Theoretical calculations showed that the catalysis of the second, and following, formate molecules would have a lower activation energy than the first activation cycle [189]. In the absence of formate, the selenocysteine-containing loop is reoriented, the Mo–S–SeCys bond reformed and the enzyme returns to the inactive form.
In this model, the conserved positively charged arginine is suggested to have a key role in driving the formate anion into the active site and subsequent binding to the molybdenum in the correct position; it would also have a role in product releasing. The conserved histidine would lower the activation energy for the selenocysteine dissociation, through the formation of a hydrogen bond with the selenol anion; this would facilitate the formate proton abstraction by the selenocysteine. Accordingly, in this mechanistic proposal, the histidine is not directly involved in the proton abstraction, a role that is suggested to be played by the selenocysteine residue. This proposal is in agreement with the selenocysteine side chain pK a value (≈5.2), which allows the existence of a selenol anion at physiological pH values (7.0). The much higher pK a value of a cysteine side chain (≈8.2) would prevent a cysteine residue having such a function. The pK a values of both amino acids have been evoked to explain why the replacement of the SeCys140 by a cysteine resulted in an activity decrease of ≈300 times in E. coli FDH-H [191]. In addition, the high activation energy calculated for the proton transfer from formate to selenol is in agreement with the isotopic effect studies that showed that the formate C–H bond cleavage is the rate-limiting step of the catalytic cycle [189]. The terminal sulfur role is still a matter of debate.
The sulfur-shift-based mechanism to oxidise formate, described above, has also been evoked to explain the nitrate reduction catalysed by periplasmatic nitrate reductases [190]. The similarity of the overall 3D structure of the catalytic subunits of periplasmic nitrate reductases and FDH is noteworthy [192]. Moreover, both molybdenum active sites are surprisingly superimposable (Fig. 9b) [192]. Periplasmatic nitrate reductase active site (Fig. 9a) harbours a molybdenum atom coordinated by the two characteristic PGD molecules, one terminal sulfo group and a cysteine sulfur atom (replaced by a selenocysteine selenium, in D.gigas and E.coli FDH; Fig. 9c); in addition, the nitrate reductase active site also comprises conserved arginine and methionine residues (arginine and histidine, in FDH). In the oxidised active site of both enzymes, the molybdenum or tungsten atom is hexa-coordinated (Fig. 9b) and a sulfur-shift is needed to displace the selenocysteine or cysteine residue to create a vacant position for substrate (formate or nitrate) binding [190]. This mechanism is similar to the carboxylate-shift observed in zinc-dependent enzymes [193, 194], which was recently detected by experimental means, suggesting that this type of ligand-shift could be more common in nature than initially thought.
Comparison of the FDH and periplasmatic nitrate reductases active sites. Comparison of the active site of a D. desulfuricans (colour coded) and R. sphaeroides (blue) nitrate reductases (D. desulfuricans enzyme numbering), and of c E. coli FDH-H (red), E.coli FDH-N (gold) and D. gigas W-FDH (colour coded; D. gigas enzyme numbering). b Comparison of D. gigas W-FDH (blue) and D. desulfuricans nitrate reductase (light blue). The structures shown are based on the PDB files 2NAP (D. desulfuricans nitrate reductase), 1OGY (R. sphaeroides nitrate reductase), 1FDO (E. coli FDH-H), 1KQF (E. coli FDH-N) and 1H0H (D. gigas FDH). The images were produced with Accelrys DS Visualizer, Accelrys Software Inc. Adapted (with permission) from [192]
The FDH comparison with periplasmatic nitrate reductase can be taken further to highlight the key role of the FDH active site in oxidising formate to carbon dioxide. Nitrate reductase and FDH catalyse different types of reaction, oxygen atom transfer (Eq. 3) and hydrogen atom abstraction (Eq 1), respectively, and their reactivity and substrate specificity must be tuned by the small but significant differences found between their active sites (Fig. 9a, c). Notably, in both enzymes, the molybdenum centre and the key amino acid residues occupy roughly the same position, but the FDH histidine is replaced by the conserved nitrate reductase methionine (Fig. 9b) [192]. This replacement supports the mechanism described above, that suggests that a histidine residue would be essential to stabilise the selenol anion formed (to facilitate the subsequent abstraction of a proton from formate by the selenocysteine). This function could not be carried out by a methionine residue, but the nitrate reduction does not need the generation of such an anionic species and nitrate reductases do not seem to harbour a histidine in the active site [192]. A similar reasoning has been employed to explain the presence of a cysteine in nitrate reductases and of a selenocysteine in D. gigas FDH. As described above, a selenocysteine, in the form of a selenol anion, would be crucial to abstract the formate Cα hydrogen, but the oxygen atom transfer reaction of nitrate reductase does not need it and nitrate reductases harbour, instead a cysteine.
In accord with this suggested mechanism (and to the comparison between FDH and periplasmatic nitrate reductases), the presence of a selenocysteine residue in the active site would be mandatory for a FDH enzyme to be able to catalyse the formate oxidation to carbon dioxide. Nevertheless, there are Mo-FDH and W-FDH that display formate oxidation activity in spite of having a cysteine, and not a selenocysteine, in the active site (Table 1). This is the case, e.g. of the R. capsulatus Mo-FDH [165]. R. capsulatus FDH, with a cysteine at the active site, is ≈10 times faster (≈90 s−1 for fully active enzyme [165]) than the SeCys140Cys-mutated E. coli FDH-H (9 s−1 [191]); yet it is considerably slower than the native E. coli enzyme (2,800 s−1 [191]). In comparison to other FDH, the R. capsulatus FDH k cat value is consistent with the one determined for P. oxalaticus FDH and ≈3 times lower than the one described for R. eutropha FDH, another enzyme with a cysteine ligand in the active site. In this context, it is possible that the high R. capsulatus FDH activity pH optimum of 9 [165] is a consequence of having a cysteine (pK a ≈ 8.2) instead of a selenocysteine in the active site. Overall, it is likely that the formate oxidation proceeds in a similar way in both R. capsulatus and E. coli FDH-H [165], in spite of the fact that the less covalent Mo–S–Cys bond may alter the chemical reaction mechanism [186]. Future mechanistic work will determine if this is the case.
Finally, a distinct third mechanistic model was proposed, in which the formate oxidation would occur through an initial hydride transfer from formate to the molybdenum atom, with the formation of a Mo–H intermediate, followed by proton transfer from molybdenum to the selenocysteine (the final proton acceptor) [195]. Hence, in this proposal, the molybdenum centre would have the unprecedented role of mediating the proton transfer from formate to the selenocysteine (contrary to other proposals, where proton transfer occurs directly between formate and selenocysteine). Nonetheless, the selenocysteine is also suggested to be an essential residue, because it would be involved in shuttling protons from the molybdenum atom.
Future work will certainly shed light on the mechanistic details that are still poorly understood. Particularly relevant is the suggested selenocysteine-containing loop movement. High-resolution structures would be needed to confirm the existence of the two alternating conformations of the molybdenum centre. Can this protein loop really exist in two different conformations? How is the conformational change triggered? Is it part of the catalytic cycle? Or, on the contrary, has only one of the two conformations catalytic activity? Is the conformational change just an experimental artefact, resulting from an aerobic purification procedure? Or is it an in vivo regulatory strategy to control the enzyme activity (mechanism “on/off” to respond to cellular needs)?
Also decisive would be the evaluation of the role of the terminal sulfo group. Why can a hydroxyl group not do the job? Moreover, why should a selenocysteine be incorporated if a “suflo-cysteine” seems to be able to assist formate oxidation? Although several theoretical works can describe the formate oxidation, only once the conformational and other issues have been resolved, will it be possible to evaluate the different mechanistic proposals critically.
Also very interesting is the question of why a few enzymes, as the Mo/W-FDH from D. alaskensis and D. vulgaris (or the R. capsulatus DMSOR [65] and E. coli trimethylamine N-oxide reductase [196] ), can be active with molybdenum and tungsten, while many others, of different functions, including other FDH, are active with only one of the two metals. The chemical similarities between molybdenum and tungsten and the fact that both metals are coordinated by the same pyranopterin cofactor seem to suggest that the enzymes should be catalytically active with either metal. However, as far as is presently known, this is an exception, rather than the rule. In fact, the growth of different organisms in the presence of tungstate leads to the formation of either metal-free inactive (molybdo)enzymes or tungsten-substituted (molybdo)enzymes with insignificant or no activity [110, 111]. The opposite is also true and, e.g. the molybdenum-substituted Pyrococcus furiosus aldehyde:ferredoxin oxidoreductase (a tungstoenzyme) is inactive [113, 197]. In this scenario of “metal selectivity”, why some enzymes display activity with both metals? The easiest answer is to suggest that it would be an advantage to incorporate both metals. This could be particularly relevant in sulfate-reducing bacteria, for which the molybdenum bioavailability can be decreased as a result of sulfide accumulation. But, if this is the case, why are the other molybdo- and tungstoenzymes from these organisms active with only one of the two metals? D. gigas, e.g. holds a W-FDH and a molybdenum-containing aldehyde oxidoreductase, both active only with the respective metal. Clearly, the incorporation of both or of only one of the two metals in active enzymes is far from being elucidated.
Formate dehydrogenase-catalysed carbon dioxide reduction
FDH-catalysed formate oxidation is, theoretically, a reversible reaction and several FDHs are, in fact, able to catalyse the carbon dioxide reduction, either in vivo or in vitro. In vivo, the most obvious example is provided by the acetogens that fix carbon dioxide, using a FDH to reduce it to formate and eventually form acetate (see Sect. 2); in vitro, the list of enzymes that are able to catalyse the carbon dioxide reduction, under appropriate conditions, is more extensive and some examples will be described below.
Carbon dioxide is a thermodynamically and kinetically stable molecule and, consequently, it is difficult to activate and reduce it. As a result, some of the studies aimed to characterise the FDH-catalysed carbon dioxide reduction employed enzymes that are known to be involved in carbon dioxide reduction in vivo. The two W-FDHs of the anaerobic syntrophic S. fumaroxidans, e.g. were shown to interconvert formate and carbon dioxide. FDH1 was demonstrated to be an efficient carbon dioxide reductase, with a higher rate for carbon dioxide reduction (≈2.5 × 103 s−1) than for formate oxidation (≈1.9 × 103 s−1; values reported as 900 and 700 Umg−1, respectively) [88]; FDH2, on the other hand, is a preferential formate dehydrogenase, with a formate oxidation rate (≈5.6 × 103 s−1) 30 times higher than that of carbon dioxide reduction (≈0.2 × 103 s−1; values reported as 2,700 and 90 Umg−1, respectively) [88]. The S. fumaroxidans FDH1 was also used to demonstrate the feasibility of interconverting carbon dioxide and formate electrochemically [198]. In that work, FDH1, adsorbed to an electrode surface, was shown to efficiently (≈0.5 × 103 s−1) catalyse the electrochemical reduction of carbon dioxide to formate (the only product formed), under thoroughly mild conditions and application of small overpotentials. Nonetheless, carbon dioxide reduction was, in this study, found to be more than five times slower than formate oxidation (≈0.5 × 103 versus ≈3 × 103s−1) [198].
In addition, also the acetogenic M. thermoacetica [166] and C. carboxidivorans [170] W-FDHs were shown to have carbon dioxide reductase activity, but, in these cases, dependent on NAD(P)H and with the C. carboxidivorans enzyme displaying a k cat of only 0.08 s−1 [170]. Notably, the reduction of carbon dioxide with NAD(P)H (either by W-FDH or Mo-FDH) is thermodynamically quite unfavourable, with reduction potentials of −0.43 and −0.32 V, respectively. This thermodynamic constraint highlights the key role played by these enzymes in overcoming the reaction energy barrier, allowing those organisms to effectively reduce carbon dioxide to formate. Other acetogens, as the A. woodii, developed a specific and remarkable hydrogen-dependent carbon dioxide reductase complex that couples the carbon dioxide reduction directly with the dihydrogen oxidation (see Sects. 2, 3.3.) [76]. This notable reductase complex allows the carbon dioxide reduction by dihydrogen with a k cat of 28 s−1 (reported as a turnover frequency of 101,600 h−1 [76]). Even though, the reverse reaction (formate oxidation and dihydrogen formation) is 1.4 times faster (reported as 10 versus 14 U/mg [76]).
Although the W-FDHs are suggested to be more efficient at reducing carbon dioxide than the Mo-FDH counterparts, because of the lower reduction potential of W4+ compared to Mo4+ [110, 111, 199], different molybdenum-dependent carbon dioxide reductases were already described. The Mo/NAD-FDH of Cupriavidus oxalaticus, e.g. is long known to be able to reduce carbon dioxide, although in a reaction ≈30 times slower than the formate oxidation [162, 200]. Also the R. capsulatus Mo-FDH is able to catalyse both formate oxidation and carbon dioxide reduction in solution, but, once more, carbon dioxide reduction reaction is ≈20 times slower than the formate oxidation (1.5 versus 36.5 s−1) [165].
Undoubtedly, carbon dioxide activation, for its fixation and utilisation by living organism, is a “difficult task”. The same problem is also experienced by d-ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBbisCO), with its promiscuous (CO2 versus O2, energy wasting) and slow (<10 s−1 [201]) catalytic performance. This common catalytic “inefficiency” places the FDH carbon dioxide reductase activity in another perspective, suggesting that its alleged “poor” efficiency might be quite good instead. This is particularly relevant in S. fumaroxidans FDH1 (with its odd carbon dioxide reduction rate of ≈2.5 × 103 s−1), which seems to suggest a remarkable selective pressure in this anaerobic syntrophic organism.
Outlook
Mo-FDH and W-FDH are a group of heterogeneous enzymes that catalyse the reversible 2-electron formate oxidation to carbon dioxide. They are involved in prokaryotic energy metabolism in numerous pathways, where they catalyse the electron transfer to/from different electron acceptors/donors. Besides the clear biological and biochemical interest in FDH, the enzyme’s ability to reduce carbon dioxide is of great interest for the sequestration of carbon dioxide and for the production of formic acid as a “stabilised” and “safe” form of hydrogen fuel [33, 69, 70, 76, 77, 202–209]. As a result, there is an increasing biotechnological interest in FDH. These enzymes have the advantage over chemical catalysts of being specific, yielding only one product (formate), and of working as homogeneous catalysts. The FDH dioxygen sensitivity is certainly the major bottleneck for the biotechnological use of FDH. In addition, the issue of long-term stability of the enzyme and the slow reaction rates would also have to be addressed in future work aimed to apply FDH in practical uses [210–212].
Notes
The Oligotropha carboxidovorans CO dehydrogenase with its unique binuclear copper–molybdenum cofactor is, however, presently classified under the xanthine oxidase family of mononuclear molybdoenzymes.
Respiratory nitrate reductases are membrane-bound cytoplasm-faced molybdoenzymes and, as the name indicates, are used by the organisms to generate a proton motive force across the cytoplasmatic membrane [135–137]. They are also called NarGHI, because they are the product of the narG, H, and I genes. These enzymes, belonging to the DMSOR family, are heterotrimers, comprising: (i) a cytoplasmatic nitrate-reducing NarG subunit (≈125 kDa) that holds one molybdenum centre and one [4Fe–4S] centre; the molybdenum atom is coordinated by four sulfur atoms (from the two pyranopterin cofactor molecules) and two oxygen atoms (both from an aspartate residue or one from a terminal oxo group plus another one from an aspartate residue [136]); (ii) an electron-transfer NarH subunit (≈60 kDa) that holds one [3Fe–4S] and three [4Fe–4S] centres; and (iii) a membrane-bound quinol-oxidising NarI subunit (≈22 kDa) that holds two b-type haems.
Abbreviations
- DMSOR:
-
Dimethylsulfoxide reductase
- EPR:
-
Electron paramagnetic resonance spectroscopy
- FDH:
-
Formate dehydrogenase
- FDH-H:
-
E. coli formate dehydrogenase H, from the formate-hydrogen lyase system
- FDH-N:
-
E. coli formate dehydrogenase N, from the anaerobic nitrate–formate respiratory pathway
- FDH-O:
-
E. coli formate dehydrogenase O, from the aerobic respiratory pathways
- Fe/S:
-
Iron–sulfur centre
- Mo-FDH:
-
Molybdenum-dependent formate dehydrogenase
- Mo/W-FDH:
-
Formate dehydrogenase that incorporates either molybdenum or tungsten
- Mo/NAD-FDH:
-
Molybdenum-dependent/NAD-dependent formate dehydrogenase
- Mo/W-bis PGD:
-
Molybdenum/tungsten-bis pyranopterin guanosine dinucleotide-containing enzymes
- NAD-FDH:
-
NAD-dependent formate dehydrogenase
- NarGHI:
-
Respiratory nitrate reductase, after the name of the encoding genes, narG, H, and I
- PGD:
-
Pyranopterin guanosine dinucleotide cofactor
- W/NAD-FDH:
-
Tungsten-dependent/NAD-dependent formate dehydrogenase
- W-FDH:
-
Tungsten-dependent formate dehydrogenase
References
Thauer RK, Jungermann K, Decker K (1977) Bacteriol Rev 41:100–180
Sakami W (1948) J Biol Chem 176:995–1003
Hartman SC, Buchanan JM (1959) Ann Rev Biochem 28:365–410
Tibbetts AS, Appling DR (2010) Ann Rev Nutr 30:57–81
Cook RJ, Champion KM, Giometti CS (2001) Arch Biochem Biophys 393:192–198
Krupenko NI, Dubard ME, Strickland KC, Moxley KM, Oleinik NV, Krupenko SA (2010) J Biol Chem 285:23056–23063
des Francs-Small CC, Ambard-Bretteville F, Darpas A, Sallantin M, Huet J-C, Pernollet J-C, Remy R (1992) Plant Physiol 98:273–278
Igamberdiev AU, Bykova NV, Kleczkowski LA (1999) Plant Physiol Biochem 37:503–513
David P, des Francs-Small CC, Sevignac M, Thareau V, Macadre C, Langin T, Geffroy V (2010) Theor Appl Genet 121:87–103
Hourton-Cabassa C, Ambard-Bretteville F, Moreau F, Davy de Virville J, Remy R, des Francs-Small CC (1998) Plant Physiol 116:627–635
Suzuki K, Itai R, Suzuki K, Nakanishi H, Nishizawa N-K, Yoshimura E, Mori S (1998) Plant Physiol 116:725–732
Thompson P, Bowsher CG, Tobin AK (1998) Plant Physiol 118:1089–1099
Andreadeli A, Flemetakis E, Axarli I, Dimou M, Udvardi MK, Katinakis P, Labrou NE (2009) Biochim Biophys Acta 1794:976–984
Thauer RK, Fuchs G, Jungermann K (1977) In: Lovenber W (ed) Iron–sulfur proteins. Academic, New York, pp 121–156
Stubbe JA, van der Donk WA (1998) Chem Rev 98:705–762
Maden BEH (2000) Biochem J 350:609–629
Adams MWW, Mortenson LE (1985) In: Spiro TG (ed) Molybdenum enzymes. Wiley, New York, pp 519–593
Ferry JG (1990) FEMS Microbiol Rev 7:377–382
Unden G, Bongaerts J (1997) Biochim Biophys Acta 1320:217–234
Richardson DJ (2000) Microbiology 146:551–571
Richardson D, Sawers G (2002) Science 295:1842–1843
Vorholt JA, Thauer RK (2002) Metals ions in biological system. In: Sigel A, Sigel H (eds) Molybdenum and tungsten: their roles in biological processes, vol 39. CRC Press, USA, pp 571–619
Sawers RB (2005) Biochem Soc Trans 33:42–46
Trchounian K, Poladyan A, Vassilian A, Trchounian A (2012) Crit Rev Biochem Mol Biol 47:236–249
Bagramyan K, Trchounian A (2003) Biochem Moscow 68:1159–1163
Grimaldi S, Schoepp-Cothenet B, Ceccaldi P, Guigliarelli B, Magalon A (2013) Biochem Biophys Acta 1827:1048–1085
Sawers G (1994) A v Leeuwenhoek 66:57–88
Andrews SC, Berks BC, McClay J, Ambler A, Quail MA, Golby P, Guest JR (1997) Microbiology 143:3633–3647
Trchounian AA, Bagramyan KA, Vassilian AV, Poladian AA (1999) Biol Membr 16:416–428
Berg BL, Li J, Heider J, Stewart V (1991) J Biol Chem 266:22380–22385
Blasco F, Guigliarelli B, Magalon A, Asso M, Giordano G, Rothery RA (2001) Cell Mol Life Sci 58:179–189
Jormakka M, Tornroth S, Byrne B, Iwata S (2002) Science 295:1863–1868
Jormakka M, Byrne B, Iwata S (2003) Curr Opin Struct Biol 13:418–423
Jones RW, Lamont A, Garland PB (1980) Biochem J 190:79–89
Jormakka M, Byrne B, Iwata S (2003) FEBS Lett 545:25–30
Jormakka M, Yokoyama K, Yano T, Tamakoshi M, Akimoto S, Shimamura T, Curmi P, Iwata S (2008) Nat Struct Mol Biol 15:730–745
Sawers G, Heider J, Zehelein E, Bock A (1991) J Bacteriol 173:4983–4993
Pommier J, Mandrand MA, Holt SE, Boxer DH, Giodano G (1992) Biochim Biophys Acta 1107:305–313
Abaibou H, Pommier J, Benoit S, Giordano G, Mandrandberthelot MA (1995) J Bacteriol 177:7141–7149
Benoit S, Abaibou H, Mandrand-Berthelot MA (1998) J Bacteriol 180:6625–6637
Kroger A, Dorrer E, Winkler E (1980) Biochim Biophys Acta 589:118–138
Bokranz M, Gutmann M, Kortner C, Kojro E, Fahrenholz F, Lauterbach F, Kroger A (1991) Arch Microbiol 156:119–128
Sebban C, Blanchard L, Bruschi M, Guerlesquin F (1995) FEMS Microbiol Lett 133:143–149
Costa C, Teixeira M, LeGall J, Moura JJG, Moura I (1997) J Biol Inorg Chem 2:198–208
Lenger R, Herrmann U, Gross R, Simon J, Kroger A (1997) Eur J Biochem 246:646–651
Simon J (2002) FEMS Microbiol Rev 26:285–309
Simon J, Klotz MG (2013) Biochim Biophys Acta 1827:114–135
Yagi T (1979) Biochim Biophys Acta 548:96–105
Sebban-Kreuzer C, Blackledge M, Dolla A, Marion D, Guerlesquin F (1998) Biochemistry 37:8331–8340
Sebban-Kreuzer C, Dolla A, Guerlesquin F (1998) Eur J Biochem 253:645–656
Morelli X, Guerlesquin F (1999) FEBS Lett 460:77–80
Matias PM, Pereira IA, Soares CM, Carrondo MA (2005) Prog Biophys Mol Biol 89:292–312
Silva SM, Voordouw J, Leitão C, Martins M, Voordouw G, Pereira IA (2013) Microbiology 159:1760–1769
Pereira IA, Ramos AR, Grein F, Marques MC, Silva SM, Venceslau SS (2011) Front Microbiol 2:69–91
Silva SM, Pacheco I, Pereira IA (2012) J Biol Inorg Chem 17:831–838
Steenkamp DJ, Peck HD Jr (1981) J Biol Chem 256:5450–5458
Barton LL, LeGall J, Odom JM, Peck HD Jr (1983) J Bacteriol 153:867–871
Marietou A, Richardson D, Cole J, Mohan S (2005) FEMS Microbiol Lett 248:217–225
Vorholt JA (2002) Arch Microbiol 178:239–249
Pomper BK, Saurel O, Milon A, Vorholt JA (2002) FEBS Lett 523:133–137
Friedrich CG, Bowien B, Friedrich B (1979) J Gen Microbiol 115:185–192
Bowien B, Schlegel HG (1981) Annu Rev Microbiol 35:405–452
Friedebold J, Bowien B (1993) J Bacteriol 175:4719–4728
Thauer RK (1998) Microbiology 144:2377–2406
Stewart LJ, Bailey S, Bennett B, Charnock JM, Garner CD, McAlpine AS (2000) J Mol Biol 299:593–600
Costa KC, Wong PM, Wang T, Lie TJ, Dodsworth JA, Swanson I, Burn JA, Hackett M, Leigh JA (2010) Proc Natl Acad Sci USA 107:11050–11055
Costa KC, Yoon SH, Pan M, Burn JA, Baliga NS, Leigh JA (2013) J Bacteriol 195:1456–1462
Ljungdahl LG, Wood HG (1969) Annu Rev Microbiol 23:515–538
Thauer RK (1972) FEBS Lett 27:111–115
Scherer PA, Thauer RK (1978) Eur J Biochem 85:125–135
Ragsdale SW (1997) BioFactors 6:3–11
Liou JS-C, Balkwill DL, Drake GR, Tanner RS (2005) Int J Syst Evol Microbiol 55:2085–2091
Ragsdale SW, Pierce E (2008) Biochim Biophys Acta 1784:1873–1898
Bruant G, Levesque M-J, Peter C, Guiot SR, Masson L (2010) PLoS One 5:e13033. doi:10.1371/journal.pone.0013033
Paul D, Austin FW, Arick T, Bridges SM, Burgess SC, Dandass YS, Lawrence ML (2010) J Bacteriol 192:5554–5555
Schuchmann K, Müller V (2013) Science 342:1382–1385
Pereira I (2013) Science 342:1329–1330
Stams AJM (1994) A v Leeuwenhoek 66:271–294
Schink B (1997) Microbiol Mol Biol Rev 61:262–280
Thiele JH, Zeikus JG (1988) Appl Environ Microbiol 54:20–29
Zindel U, Freudenberg W, Rieth M, Andreesen JR, Schnell J, Widdel F (1988) Arch Microbiol 150:254–266
Boone DR, Johnson RL, Liu Y (1989) Appl Environ Microbiol 55:1735–1741
Dong XZ, Plugge CM, Stams AJM (1994) Appl Environ Microbiol 60:2834–2838
Stams AJM, Dong XZ (1995) A v Leeuwenhoek 68:281–284
McInerney MJ, Sieber JR, Gunsalus RP (2009) Curr Opin Biotechnol 20:623–632
Stams AJ, Plugge CM (2009) Nat Rev Microbiol 7:568–577
Bok FAM, Luijten MLGC, Stams AJM (2002) Appl Environ Microbiol 68:4247–4252
Bok FA, Hagedoorn PL, Silva PJ, Hagen WR, Schiltz E, Fritsche K, Stams AJ (2003) Eur J Biochem 270:2476–2485
Lovley DR, Holmes DE, Nevin KP (2004) Adv Microb Physiol 49:219–286
Shi L, Squier TC, Zachara JM, Fredrickson JK (2007) Mol Microbiol 65:12–20
Niggemyer A, Spring S, Stackebrandt E, Rosenzweig RF (2001) Appl Environ Microbiol 67:5568–5580
Kato N (1990) Meth Enzymol 188:459–462
Vinals C, Depiereux E, Feytmans E (1993) Biochem Biophys Res Commun 192:182–188
Popov VO, Lamzin VS (1994) Biochem J 301:625–643
Lamzin VS, Dauter Z, Popov VO, Harutyunyan EH, Wilson KS (1974) J Mol Biol 236:759–785
Blanchard JS, Cleland WW (1980) Biochemistry 19:3543–3547
Rotberg NS, Cleland WW (1991) Biochemistry 30:4068–4076
Mesentsev AV, Ustinnikova TB, Tikhonova TV, Popov VO (1996) Appl Biochem Microbiol 32:529–537
Tishkov VI, Matorin AD, Rojkova AM, Fedorchuk VV, Savitsky PA, Dementieva LA, Lamzin VS, Mezentzev AV, Popov VO (1996) FEBS Lett 390:104–108
Filippova EV, Polyakov KM, Tikhonova TV, Stekhanova TN, Boiko KM, Popov VO (2005) Crystallogr Rep 50:796–801
Castillo R, Oliva M, Marti S, Moliner V (2008) J Phys Chem B 112:10012–10022
Shabalin IG, Polyakov KM, Tishkov VI, Popov VO (2009) Acta Nat 1:89–93
Hille R (1996) Chem Rev 96:2757–2816
Hille R (2002) Trends Biochem Sci 27:360–367
Schwarz G, Mendel R, Ribbe M (2009) Nature 460:839–847
Hille R, Mendel R (2011) Coord Chem Rev 255:991–992
Mendel R, Kruse T (2012) Biochim Biophys Acta 1823:1568–1579
Hille R (2013) Dalton Trans 42:3029–3042
Anbar AD (2008) Science 322:1481–1483
Johnson MK, Rees DC, Adams MW (1996) Chem Rev 96:2817–2840
Kletzin A, Adams MW (1996) FEMS Microbiol Rev 18:5–63
Andreesen JR, Makdessi M (2008) Ann NY Acad Sci 1125:215–229
Bevers LE, Hagedoorn PL, Hagen WR (2009) Coord Chem Rev 253:269–290
George GN, Pickering IJ, Yu EY, Prince RC, Bursakov SA, Gavel OY, Moura I, Moura JJG (2000) J Am Chem Soc 122:8321–8323
Bursakov SA, Gavel OY, Di Rocco G, Lampreia J, Calvete J, Pereira AS, Moura JJ, Moura I (2004) J Inorg Biochem 98:833–840
Rivas MG, Carepo MS, Mota CS, Korbas M, Durand MC, Lopes AT, Brondino CD, Pereira AS, George GN, Dolla A, Moura JJG, Moura I (2009) Biochemistry 48:873–882
Carepo MS, Pauleta SR, Wedd AG, Moura JJG, Moura I (2014) J Biol Inorg Chem 19:605–614
Rothery RA, Workun GJ, Weiner JH (2008) Biochim Biophys Acta 1778:1897–1929
Roy R, Adams MW (2002) Met Ions Biol Syst 39:673–697
Hille R, Nishino T (1995) FASEB J 9:995–1003
Hille R (2005) Arch Biochem Biophys 433:107–116
Hille R (2006) Eur J Inorg Chem 1913–1926
Nishino T, Okamoto K, Eger BT, Pai EF, Nishino T (2008) FEBS J 275:3278–3289
Hille R, Nishino T, Bittner F (2011) Coord Chem Rev 255:1179–1205
Okamoto K, Kusano T, Nishino T (2013) Curr Pharm Des 19:2606–2614
Schink B (1985) Arch Microbiol 142:295–301
Messerschmidt A, Niessen H, Abt D, Einsle O, Schink B, Kroneck PM (2004) Proc Natl Acad Sci USA 101:11571–11576
Zinoni F, Birkmann A, Stadtman TC, Böck A (1986) Proc Natl Acad Sci USA 83:4650–4654
Axley MJ, Grahame DA, Stadtman TC (1990) J Biol Chem 265:18213–18218
Gladyshev VN, Boyington JC, Khangulov SV, Grahame DA, Stadtman TC, Sun PD (1996) J Biol Chem 271:8095–8100
Boyington JC, Gladyshev VN, Khangulov SV, Stadtman TC, Sun PD (1997) Science 275:1305–1308
Raaijmakers HCA, Romao MJ (2006) J Biol Inorg Chem 11:849–854
Thome R, Gust A, Toci R, Mendel R, Bittner F, Magalon A, Walburger A (2012) J Biol Chem 287:4671–4678
Jormakka M, Tornroth S, Abramson J, Byrne B, Iwata S (2002) Acta Crystallogr D Biol Crystallogr 58:160–162
Bertero MG, Rothery RA, Palak M, Hou C, Lim D, Blasco F, Weiner J, Strynadka NC (2003) Nat Struct Biol 10:681–687
Jormakka M, Richardson D, Byrne B, Iwata S (2004) Structure 12:95–104
Gonzaalez PG, Correia C, Moura I, Brondino CD, Moura JJG (2006) J Inorg Biochem 100:1015–1023
Berks BC (1996) Mol Microbiol 22:393–404
Sargent F, Bogsch EG, Stanley NR, Wexler M, Robinson C, Berks BC, Palmer T (1998) EMBO J 17:3640–3650
Sargent F, Stanley NR, Berks BC, Palmer T (1999) J Biol Chem 274:36073–36083
Abaibou H, Pommier J, Benoit JP, Giordano G, Mandrand M (1995) J Bacteriol 177:141–7149
Plunkett G, Burland V, Daniels DL, Blattner FR (1993) Nucleic Acids Res 21:3391–3398
Almendra MJ, Brondino CD, Gavel O, Pereira AS, Tavares P, Bursakov S, Duarte R, Caldeira J, Moura JJ, Moura I (1999) Biochemistry 38:16366–16372
Raaijmakers H, Teixeira S, Dias JM, Almendra MJ, Brondino CD, Moura I, Moura JJ, Romao MJ (2001) J Biol Inorg Chem 6:398–404
Raaijmakers H, Macieira S, Dias JM, Teixeira S, Bursakov S, Huber R, Moura JJG, Moura I, Romao MJ (2002) Structure 10:1261–1272
Rivas M, Gonzalez P, Brondino CD, Moura JJG, Moura I (2007) J Inorg Biol 101:1617–1622
Mota CS, Valette O, Gonzalez PJ, Brondino CD, Moura JJG, Moura I, Dolla A, Rivas MG (2011) J Bacteriol 193:2917–2923
Brondino CD, Passeggi MCG, Caldeira J, Almendra MJ, Feio MJ, Moura JJG, Moura I (2004) J Biol Inorg Chem 9:145–151
Heidelberg JF, Seshadri R, Haveman SA, Hemme CL, Paulsen IT, Kolonay JF, Eisen JA, Ward N, Methe B, Brinkac LM, Daugherty SC, Deboy RT, Dodson RJ, Durkin AS, Madupu R, Nelson WC, Sullivan SA, Fouts D, Haft DH, Selengut J, Peterson JD, Davidsen TM, Zafar N, Zhou L, Radune D, Dimitrov G, Hance M, Tran K, Khouri H, Gill J, Utterback TR, Feldblyum TV, Wall JD, Voordouw G, Fraser CM (2004) Nat Biotechnol 22:554–559
Bursakov S, Liu M-Y, Payne WJ, LeGall J, Moura I, Moura JJG (1995) Anaerobe 1:55–60
Silva SM, Pimentel C, Valente FMA, Rodrigues-Pousada C, Pereira IAC (2011) J Bacteriol 193:2909–2917
ElAntak L, Dolla A, Durand MC, Bianco P, Guerlesquin F (2005) Biochemistry 44:14828–14834
Friedebold J, Mayer F, Bill E, Trautwein AX, Bowien B (1995) Biol Chem Hoppe Seyler 376:561–568
Oh JI, Bowien B (1998) J Biol Chem 273:26349–26360
Oh JI, Bowien B (1999) Mol Microbiol 34:365–376
Jollie DR, Lipscomb JD (1991) J Biol Chem 266:21853–21863
Sazanov LA, Hinchliffe P (2006) Science 311:1430–1434
Karzanov VV, Bogatsky YuA, Tishkov VI, Egorov AM (1989) FEMS Mivrobiol Lett 60:197–200
Karzanov VV, Correa CM, Bogatsky YG, Netrisuoc AI (1991) FEMS Microbiol Lett 81:95–100
Girio FM, Amaral-Collaco MT, Attwood M (1994) Appl Microbiol Biotechnol 40:898–903
Duarte RO, Reis AR, Girio F, Moura I, Moura JJG, Collaco TA (1997) Biochem Biophys Res Com 230:30–34
Muller U, Willnow P, Ruschig U, Hopner T (1978) Eur J Biochem 83:485–498
Chistoserdova L, Laukel M, Portais J-C, Vorholt JA, Lidstrom ME (2004) J Bacteriol 186:22–28
Laukel M, Chistoserdova L, Lidstrom ME, Vorholt JA (2003) Eur J Biochem 270:325–333
Hartmann T, Leimkuhler S (2013) FEBS J 280:6083–6096
Yamamoto I, Saikit T, Liu S-M, Ljungdahlg L (1983) J Biol Chem 258:1826–1832
Deaton JC, Solomon EI, Watt GD, Wetherbee PJ, Durfor CN (1987) Biochem Biophys Res Commun 149:424–430
Ljungdahl LG, Andreesen JR (1975) FEBS Lett 54:279–282
Leonhardt U, Andreesen JR (1977) Arch Microbiol 115:277–284
Alissandratos A, Kim HK, Matthews H, Hennessy JE, Philbrook A, Easton CJ (2013) Appl Environ Microbiol 79:741–744
Hille R, Hall J, Basu P (2014) Chem Rev 114:3963–4038
Chistoserdova L, Crowther GJ, Vorholt JA, Skovran E, Portais JC, Lidstrom ME (2007) J Bacteriol 189:9076–9081
Andreesen JR, Ljungdahl LG (1973) J Bacteriol 116:867–873
Andreesen JR, Ljungdahl LG (1975) J Bacteriol 120:6–14
Schauer NI, Ferry JF (1982) J Bacteriol 150:1–7
Barber MJ, Siege LM, Schauer NL, May HD, Ferry JG (1983) J Biol Chem 258:10839–10845
Schauert NI, Ferry JG (1986) J Bacteriol 165:405–411
Shuber AP, Orr EC, Recny MA, Schendel P, May HD, Schauer NL, Ferry JG (1986) J Biol Chem 261:12942–12947
Johnson JL, Bastian NR, Schauer NL, Ferry JG, Rajagopalan KV (1991) FEMS Microbiol Lett 77:213–216
Noolling K, Reeve JN (1997) J Bacteriol 179:899–908
Jones JB, Stadtmang TC (1980) J Biol Chem 255:1049–1053
Jones JB, Stadtmant TC (1981) J Biol Chem 256:656–663
George GN, Colangelo CM, Dong J, Scott RA, Khangulov SV, Gladyshev VN, Stadtman TC (1998) J Am Chem Soc 120:1267–1273
George GN, Costa C, Moura JJG, Moura I (1999) J Am Chem Soc 121:2625–2631
Gladyshev VN, Khangulov SK, Axley MJ, Stadtman TC (1994) Proc Natl Acad Sci USA 91:7708–7711
Khangulov SV, Gladyshev VN, Dismukes GC, Stadtman TC (1998) Biochemistry 37:3518–3528
Thauer RK, Kaufer B, Fuchs G (1975) Eur J Biochem 55:111–117
Leopoldini M, Chiodo SG, Toscano M, Russo N (2008) Chemistry 14:8674–8681
Mota CS, Rivas MG, Brondino CD, Moura I, Moura JJ, González PJ, Cerqueira NM (2011) J Biol Inorg Chem 16:1255–1268
Cerqueira NMFSA, Fernandes PA, Gonzalez PJ, Moura JJG, Ramos MJ (2013) Inorg Chem 52:10766–10772
Axley MJ, Bock A, Stadtman TC (1991) Proc Natl Acad Sci USA 88:8450–8454
Moura JJG, Brondino CD, Trincão J, Romão MJ (2004) J Biol Inorg Chem 9:791–799
Demsar A, Kosmrlj J, Petricek S (2002) J Am Chem Soc 124:3951–3958
Sousa SF, Fernandes PA, Ramos MJ (2007) J Am Chem Soc 129:1378–1383
Tiberti M, Papaleo E, Russo N, Gioia L, Zampella G (2012) Inorg Chem 51:8331–8339
Buc J, Santini CL, Giordani R, Czjzek M, Wu LF, Giordano G (1999) Mol Microbiol 32:159–168
Sevcenco AM, Bevers LE, Pinkse MWH, Krijger GC, Wolterbeek HT, Verhaert PDEM, Hagen WR, Hagedoorn PL (2010) J Bacteriol 192:4143–4152
Reda T, Plugge CM, Abram NJ, Hirst J (2008) Proc Natl Acad Sci USA 105:10654–10658
Callis GE, Wentworth RA (1977) Bioinorg Chem 7:57–70
Ruschig U, Muller U, Willnow P, Hopner T (1976) Eur J Biochem 70:325–330
Tcherkez GGB, Farquhar GD, JohnAndrews TJ (2006) Proc Natl Acad Sci USA 103:7246–7521
Parkinson BA, Weaver PF (1984) Nature 309:148–149
Miyatani R, Amao Y (2002) Biotechnol Lett 24:1931–1934
Lu Y, Jiang ZY, Xu SW, Wu H (2006) Catal Today 115:263–268
Minteer SD, Liaw BY, Cooney MJ (2007) Curr Opin Biotecnol 18:1–7
Joó F (2008) ChemSusChem 1:805–808
Crable BR, Plugge CM, McInerney MJ, Stams AJM (2011) Enzyme Res 532536. doi:10.4061/2011/532536
Yadav RK, Baeg J-O, Oh GH, Park N-J, Kong K-J, Kim J, Hwang DW, Biswas SK (2012) J Am Chem Soc 134:11455–11461
Yoshimoto M, Kunihiro N, Tsubomura N, Nakayama M (2013) Colloids Surf B Biointerfaces 109:40–44
Slusarczyk H, Felber S, Kula MR, Pohl M (2000) Eur J Biochem 267:1280–1289
Yamamoto H, Mitsuhashi K, Kimoto N, Kobayashi Y, Esaki N (2005) Appl Microbiol Biotechnol 67:33–39
Tishkov VI, Popov VO (2006) Biomol Eng 23:89–110
Acknowledgments
This work was supported by Grants and Project PEst-C/EQB/LA0006/2013 from Fundação para a Ciência e Tecnologia (FCT)/MEC, Portugal.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editors: José Moura and Paul Bernhardt.
Rights and permissions
About this article
Cite this article
Maia, L.B., Moura, J.J.G. & Moura, I. Molybdenum and tungsten-dependent formate dehydrogenases. J Biol Inorg Chem 20, 287–309 (2015). https://doi.org/10.1007/s00775-014-1218-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00775-014-1218-2